Abstract
The accumulation of unfolded proteins in the endoplasmic reticulum (ER) triggers the unfolded protein response (UPR)-signaling pathway. The UPR coordinates the induction of ER chaperones with decreased protein synthesis and growth arrest in the G1 phase of the cell cycle. Three ER transmembrane protein kinases (Ire1α, Ire1β, and PERK) have been implicated as proximal effectors of the mammalian UPR. We now demonstrate that activation of PERK signals the loss of cyclin D1 during the UPR, culminating in cell-cycle arrest. Overexpression of wild-type PERK inhibited cyclin D1 synthesis in the absence of ER stress, thereby inducing a G1 phase arrest. PERK expression was associated with increased phosphorylation of the translation elongation initiation factor 2α (eIF2α), an event previously shown to block cyclin D1 translation. Conversely, a truncated form of PERK lacking its kinase domain acted as a dominant negative when overexpressed in cells, attenuating both cyclin D1 loss and cell-cycle arrest during the UPR without compromising induction of ER chaperones. These data demonstrate that PERK serves as a critical effector of UPR-induced growth arrest, linking stress in the ER to control of cell-cycle progression.
The accumulation of unfolded proteins in the endoplasmic reticulum (ER) triggers the unfolded protein response (UPR)-signaling pathway (1, 2). In mammalian cells, the UPR signals the increased expression of genes encoding ER chaperones like BiP/GRP78 and GRP94, as well as CHOP (C/EBP homologous protein), a transcription factor also known as growth arrest and DNA damage gene-153 (GADD153) (2, 3). The induction of ER chaperones is coordinated with a decreased rate of protein synthesis and G1 phase arrest (4–6). Although this coordinated response likely serves as a stress-induced checkpoint that allows cells time to reestablish homeostasis, chronic ER stress culminates in apoptosis (7). The respective roles of the various components of the UPR in maintaining tissue homeostasis both during and after ER stress remain to be established.
Mammalian cells contain at least three ER transmembrane protein kinases (Ire1α, Ire1β, and PERK) that function as effectors of the UPR. Ire1α/β are comprised of an ER luminal domain and a cytosolic tail containing both a serine/threonine kinase module and an RNase domain (7, 8). Ire1 activation may lie upstream of stress-induced expression of ER chaperones and CHOP (8, 9). Ire1 proteins have also been linked to the activation of cJUN NH2-terminal kinases, consistent with the notion that the UPR may regulate gene expression through modulation of transcription factors such as cJUN and ATF2 (10). PERK consists of an ER luminal domain and a cytosolic serine/threonine kinase domain that shares homology with the cytosolic RNA-dependent protein kinase (PKR) (11, 12). The down-regulation of protein synthesis in cells challenged with ER stress is accompanied by increased phosphorylation of eIF-2α, a modification that hinders assembly of 40S translation-initiation complexes and inhibits translation (7). PERK is activated by ER stress (11) and can phosphorylate eIF-2α (11, 13), implicating PERK in UPR-mediated repression of protein synthesis. Although it seems clear that Ire1 molecules and PERK modulate distinct components of the UPR, the events downstream of these ER kinases and their precise roles in determining the fate of stressed cells are unclear.
Cell-cycle arrest after UPR activation occurs primarily in the G1 phase (14). Progression through G1 phase requires the activities of one or more of the D-type cyclins (D1, D2, or D3) in association with either CDK4 or CDK6, followed by activation of the cyclin E- and A-dependent kinase CDK2, as cells near the G1/S transition (15). Cell-cycle arrest can be achieved through degradation of cyclin subunits, specific posttranslational modifications of the CDK subunits, or association of active cyclin-bound CDKs with polypeptide CDK inhibitors (CKIs) (16, 17). Members of the Cip/Kip family of CKIs (including p21Cip1, p27Kip1, and p57Kip2) positively regulate cyclin D–CDK assembly (18, 19) and remain stably bound to catalytically active cyclin D–CDK complexes (18, 20). Mobilization of Cip/Kip proteins through loss of cyclin D1 results in the subsequent Cip/Kip-mediated inhibition of cyclin E– and A–CDK2 complexes, thereby ensuring G1 phase arrest (21).
Recent evidence indicates that ER stress-induced G1 arrest results from the specific loss of cyclin D1 protein via inhibition of cyclin D1 translation (14). We now demonstrate that activation of PERK is sufficient to mediate loss of cyclin D1 and promote cell-cycle arrest. PERK, therefore, serves as a proximal effector of the UPR-signaling pathway, linking stress in the ER to the regulation of cell-cycle progression.
Materials and Methods
Tissue Culture Conditions.
Cell lines were maintained in DMEM supplemented with 10% FCS, antibiotics, and glutamine (GIBCO Life Sciences). D1-T286A-3T3 cells were previously described (22). PERKΔC-3T3 cells were established by infection of NIH 3T3 cells with retrovirus encoding PERKΔC and the puromycin resistance gene followed by selection with 7.5 μg/ml puromycin. PURO-3T3 cells were established by transfection (23) of NIH 3T3 cells with a plasmid encoding only the puromycin resistance gene followed by selection with 7.5 μg/ml puromycin.
For virus production, the mouse PERK cDNA, engineered to express a C-terminal c-myc epitope tag, was inserted into the pBabe-puro retroviral vector as an EcoRI/XhoI fragment. The PERK deletion mutant was engineered as previously described (11). Human kidney 293T cells were transfected with 15 μg of ecotropic helper retrovirus plasmid plus 15 μg of pBabe-puro vector encoding myc epitope-tagged PERK, PERKΔC, or Ire1β. Supernatants collected every 6 h for 48 h were pooled and filtered. Virus infections were carried out on exponentially growing cells in an 8% CO2 atmosphere in the presence of 10 μg/ml polybrene (Sigma).
Immunoprecipitation, Immunoblotting, and Kinase Assays.
For direct Western analysis, cells were lysed in EBC buffer (50 mM Tris⋅HCl, pH 7.5/120 mM NaCl/0.5% Nonidet P-40/1 mM PMSF/20 units/ml of aprotinin/0.4 mM NaF). Proteins were resolved on polyacrylamide gels, transferred to nitrocellulose membranes (Millipore), and blotted with the indicated primary antibodies. For detection of eIF2α, lysates prepared as above were initially blotted with a serine 51 phospho-specific antibody (KAP-CP130; StressGen Biotechnologies, Victoria, Canada) or an antibody that recognizes total eIF2α (sc-7629; Santa Cruz Biotechnology). Ectopic expression of myc-tagged PERK, PERKΔC, and Ire1β was detected by immunoblot analysis with the 9E10 monoclonal antibody (24). Immunoblot analysis of cyclin D1 and BiP was performed as previously described (14). Sites of antibody binding were visualized by chemiluminescence detection (NEN). Immune complex kinase assay of cyclin D1- or CDK2-associated catalytic activity was performed as previously described (14).
Immunofluorescence.
For detection of BrdUrd positive cells, cells seeded on coverglass and treated as indicated were fixed and permeabilized with ice-cold methanol–acetone (1:1) for 10 min at −20°C. After treatment of cells with 1.5 M HCL, cells were stained with a BrdUrd-specific antibody (Amersham Pharmacia Biotech) and FITC conjugated anti-mouse antibody (1:100; Amersham Pharmacia Biotech). All incubations were performed in PBS containing 10% FCS. After a final wash in PBS, DNA was stained with Hoechst dye 33258 (Sigma). Cells were visualized by using a Nikon microscope fitted with the appropriate filters.
Northern Analysis.
Total RNA was isolated from cultured cells by using the RNeasy kit (Qiagen, Chatsworth, CA) and analyzed by Northern blotting with random prime labeled probes corresponding to mouse cyclin D1 and human γ-actin. Hybridization conditions were as previously described (14).
Biosynthetic Labeling.
Subconfluent cells treated as indicated in the figure legend (Fig. 2) were incubated for 30 min in methionine-free DMEM (BioWhittaker) and shifted to medium containing 150 μCi/ml Trans-35S-label (ICN) for the indicated periods of time. Proteins were precipitated from cell lysates as previously described (25), and radiolabeled proteins were resolved on denaturing polyacrylamide gels and visualized by autoradiography.
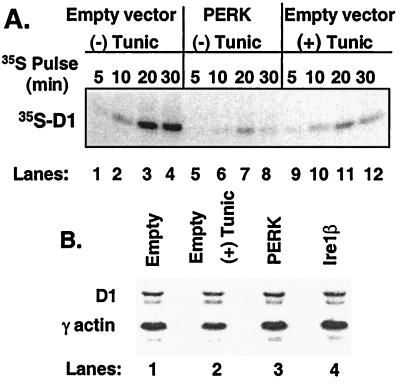
Figure 2.
Inhibition of cyclin D1 translation by PERK. (A) After infection of NIH 3T3 cells with virus encoding PERK (lanes 5–8) or empty virus (lanes 1–4 and 9–12), cells were left untreated or were treated with 0.5 μg/ml tunicamycin for 5 h and were subsequently pulse labeled with 35S-methionine for the indicated intervals. Cyclin D1 was immunoprecipitated from lysates, resolved on a denaturing gel, and visualized by autoradiography. (B) Total RNA was isolated from NIH 3T3 cells infected with PERK (lane 3), Ire1β (lane 4), empty virus (lane 1), or empty virus with tunicamycin treatment for 6 h and subjected to Northern blot analysis with probes specific for cyclin D1 and γ-actin.
Results
PERK Antagonizes Cyclin D1 Accumulation.
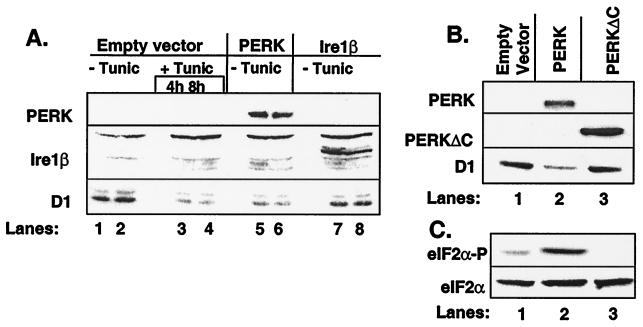
UPR-mediated cyclin D1 loss in NIH 3T3 cells results from the inhibition of cyclin D1 protein synthesis (14). Because PERK mediates translational repression after treatment of cells with ER stressing agents (11, 26), we reasoned that PERK might inhibit cyclin D1 accumulation after activation of the UPR. Overexpression of PERK promotes its autoactivation in the absence of ER stress through enforced oligomerization (11, 27). NIH 3T3 cells were infected with retrovirus encoding PERK or, as a control, retrovirus encoding Ire1β or empty vector. Both PERK and Ire1β were engineered to encode a C-terminal c-myc epitope tag to facilitate their detection (8, 11). Expression of PERK (Fig. 1, lanes 5–6) and Ire1β (lanes 7–8) as measured by immunoblot analysis revealed nearly equivalent levels of expression. Immunoblot analyses further revealed that overexpression of PERK, but not Ire1β, resulted in a significant decrease in the level of cyclin D1 protein (Fig. 1A, third panel, compare lanes 5–6 with 7–8). The loss of cyclin D1 in PERK-infected cells was similar to that achieved by treatment of cells with tunicamycin, an agent that triggers the UPR because of its capacity to inhibit N-linked glycosylation (Fig. 1A, third panel, compare lanes 5–6 with 3–4). As previously noted with tunicamycin, PERK expression did not result in the loss of cyclins A or E (14) and only modestly reduced levels of cyclin D2 (data not shown).
Figure 1.
PERK regulates accumulation of cyclin D1. (A) NIH 3T3 cells infected with empty virus (lanes 1–4), virus encoding PERK (lanes 5–6), or virus encoding Ire1β (lanes 7–8) were left untreated or were treated with 0.5 μg/ml tunicamycin (lanes 3–4). Equivalent amounts of cellular protein were resolved on denaturing polyacrylamide gels and subjected to immunoblot analysis with the 9E10 antibody, which recognizes a c-myc epitope tag expressed at the C terminus of both PERK and Ire1β or a cyclin D1-specific monoclonal antibody. Sites of antibody binding were visualized by enhanced chemiluminescence. (B and C) After infection of NIH 3T3 cells with virus encoding PERK (lane 2), PERKΔC (lane 3) or empty virus (lane 1), equivalent amounts of total protein were resolved on a denaturing polyacrylamide gel and transferred to nitrocellulose membrane. Membranes were subsequently probed with the 9E10 antibody (PERK and PERKΔC) or antibodies specific for cyclin D1, serine 51-phosphorylated eIF2α, or total eIF2α. Sites of antibody binding were visualized by enhanced chemiluminescence.
To determine whether PERK kinase activity is required for down-regulation of cyclin D1, NIH 3T3 cells were infected with virus encoding either wild-type PERK or a kinase-defective PERK lacking the C-terminal kinase domain (PERKΔC) (11). Both PERK and PERKΔC contain the c-myc epitope tag to facilitate their detection. Immunoblotting revealed expression of PERKΔC (Fig. 1B Middle, lanes 2–3) to be greater than wild-type PERK. Yet the kinase-defective PERKΔC failed to reduce cyclin D1 levels (Fig. 1B Lower, lanes 2–3).
PERK phosphorylates eIF2α on serine 51 (11, 13), an event associated with inhibition of translation (7). To determine whether PERK overexpression is associated with increased phosphorylation of eIF2α on serine 51 in our system, lysates prepared above were subjected to immunoblot analysis with an antibody that recognizes serine 51-phosphorylated eIF2α. Lysates from PERK-infected cells contained elevated levels of phosphorylated eIF2α relative to cells infected with empty control virus (Fig. 1C, lanes 1–2). Strikingly, expression of PERKΔC reduced eIF2α phosphorylation below the basal levels observed in control lysates (lanes 1–3). Immunoblotting with an antibody that recognizes both phosphorylated and nonphosphorylated eIF2α revealed equal amounts of total eIF2α (Fig. 1C Lower). These results suggest that PERK acts as an eIF2α kinase in vivo and implicate PERK-dependent phosphorylation of eIF2α in UPR-dependent cyclin D1 loss. Furthermore, the ability of PERKΔC to reduce eIF2α phosphorylation suggests that this kinase-defective protein may function as a dominant negative (see below).
PERK Inhibits Cyclin D1 Translation and Promotes Cell-Cycle Arrest.
PERK overexpression can trigger the inhibition of reporter gene translation in the absence of an ER stress-inducing agent (11). This presumably reflects the propensity of PERK to phosphorylate eIF2α and thereby to inhibit protein synthesis. To determine whether PERK can inhibit cyclin D1 translation, NIH 3T3 cells were infected with virus encoding PERK or virus encoding only a CD8 cell-surface marker (empty virus). Forty-eight hours after infection, cells were pulse labeled with [35S]methionine, and cyclin D1 was precipitated from cellular lysates and resolved on a polyacrylamide gel. A significant reduction in radiolabeled cyclin D1 was detected in PERK-infected cells relative to control-infected cells (Fig. 2A). Synthesis of cyclin D1 in PERK-infected cells was similar to that in cells treated with tunicamycin for 5 h (lanes 5–8 with 9–12). Northern analysis was performed on total RNA isolated from cells infected with empty virus, empty virus and treated tunicamycin, PERK, or Ire1β. Cyclin D1 mRNA was not affected in cells overexpressing PERK (Fig. 2B). Thus, enforced expression of PERK signals the inhibition of cyclin D1 protein synthesis without affecting accumulation of cyclin D1 message.
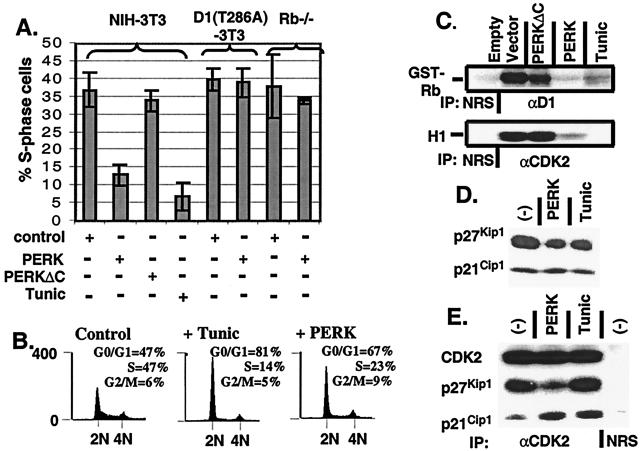
Cyclin D1-dependent kinase is rate limiting for cell-cycle progression in cells containing wild-type Rb (28–30). Its inhibition by overexpression of dominant-negative cyclins (31) or by microinjection of inhibitory antibodies (32, 33) results in cell-cycle arrest in G1 phase. The ability of PERK to inhibit cyclin D1 synthesis, thereby reducing steady-state levels of cyclin D1, suggested that PERK activation might modulate cell-cycle progression. To test this hypothesis, NIH 3T3 cells were infected with retrovirus encoding wild-type PERK, PERKΔC, or empty virus. Forty-eight hours after infection, the fraction of S-phase cells was determined. As a control, one set of empty virus-infected cells was treated with tunicamycin for 20 h. Treatment with tunicamycin resulted in a 4- to 5-fold reduction in the fraction of S-phase cells (Fig. 3A), with a concomitant increase in the percentage of G1 phase cells (Fig. 3B). Similarly, PERK expression reduced the percentage of cells in S-phase 3- to 4-fold (Fig. 3A), also accompanied by an increase in G1 phase cells (Fig. 3B). In contrast, PERKΔC did not effect the fraction of cells replicating their DNA (Fig. 3A). Cells overexpressing the proteasome resistant cyclin D1 mutant, D1-T286A, fail to arrest in G1 phase upon activation of the UPR with tunicamycin (14). Similar to these findings, PERK expression failed to arrest cells that express D1-T286A (Fig. 3A). Rb−/− murine embryonic fibroblasts (MEFs), which no longer require cyclin D1 for cell-cycle progression (21), were also refractory to PERK overexpression (Fig. 3A). It should be noted that overexpression of cyclin D1 or loss of Rb impacts G1 phase progression at multiple levels. Thus, although these results suggest that PERK mediates cell-cycle arrest by virtue of its ability to eliminate cyclin D1, complete G1 arrest likely requires inhibition of the CDK2 kinase as well (see below).
Figure 3.
Enforced overexpression of cyclin D1 prevents PERK-dependent cell-cycle arrest. (A) NIH 3T3 cells, cyclin D1-T286A derivatives, and Rb−/− MEFs were infected with the viruses indicated at the bottom of the graph. Empty virus-infected cells were left untreated or treated with tunicamycin for 20 h. During the last 2 h of tunicamycin treatment, all cell populations were pulsed with BrdUrd and processed for immunofluorescence with a BrdUrd-specific monoclonal antibody except for Rb−/− MEFs in which the S-phase fraction was determined by flow cytometry, as in B. The number of BrdUrd-positive cells from a minimum of three independent experiments were quantitated and are expressed relative to the total population of cells. (B) NIH 3T3 cells were infected with virus encoding PERK or empty virus (control) and left uninfected or treated with 0.5 μg/ml tunicamycin for 20 h. Forty-eight hours after infection (or 20 h after addition of tunicamycin), cells were stained with propidium iodide and assayed for DNA content by flow cytometry (14). (C) NIH 3T3 cells were infected the indicated retroviruses, lysed, and immune complexes were recovered with either a cyclin D1 antibody or a CDK2 antibody and analyzed for protein kinase activity by using retinoblastoma protein or histone H1 as the substrate respectively. (D) Whole-cell lysates prepared from NIH 3T3 cells left untreated, treated with 0.5 μg/ml tunicamycin for 20 h, or infected with virus encoding PERK were subjected to a direct immunoblot analysis by using either p21Cip1- or p27Kip1-specific antibodies. (E) Whole-cell lysates prepared from cells treated as in D were precipitated with antibodies to CDK2. Denatured immune complexes separated on gels were blotted with antibodies to CDK2, p21Cip1, or p27Kip1.
Concomitant with PERK-dependent loss of cyclin D1 protein, treatment of cells with tunicamycin or overexpression of PERK, but not PERKΔC, reduced cyclin D1-dependent kinase activity to background levels (Fig. 3C). PERK expression also dramatically inhibited CDK2 catalytic activity (Fig. 3C). Because CDK2 activation is associated with late G1 phase and S-phase progression, these data are consistent with cell-cycle arrest in G1 phase.
Tunicamycin inhibits CDK2 via mobilization of p21Cip1 and p27Kip1 into cyclin E and A/CDK2 complexes (14). Because PERK-dependent inhibition of CDK2 activity was not accompanied by loss of cyclins A or E, or with a loss of CDK-activating kinase-phosphorylated CDK2 (data not shown), we considered the possibility that a similar mechanism underlies PERK-dependent loss of CDK2 activity. CDK2 complexes were isolated from mock-infected cells either treated or untreated with tunicamycin or PERK-infected cells. Immunoblot analysis revealed that PERK infection resulted in the recruitment of p21Cip1 but not p27Kip1 into CDK2 complexes (Fig. 3E). As PERK expression did not result in a net increase of p21Cip1 (Fig. 3D), this likely reflects the redistribution of p21Cip1 from D1/CDK4 complexes and into CDK2 complexes. These data are consistent with the notion that PERK inhibits cell-cycle progression via down-regulation of cyclin D1, resulting in the inhibition of cyclin D1/CDK4 activity and an indirect inhibition of CDK2 activity because of the redistribution of p21Cip1.
PERKΔC Inhibits UPR-Dependent Cell-Cycle Arrest.
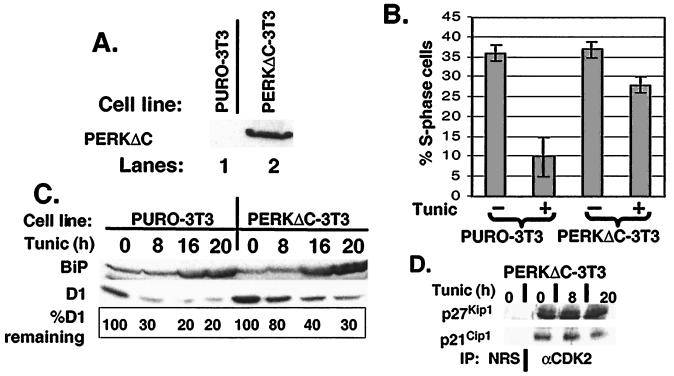
The ability of PERKΔC to attenuate eIF2α phosphorylation (Fig. 1C) suggested that PERKΔC could function as a dominant negative (11). If so, cells expressing PERKΔC should maintain levels of cyclin D1 and should be refractory to UPR-triggered cell-cycle arrest. We therefore established NIH 3T3 cell lines that constitutively express PERKΔC (PERKΔC-3T3; Fig. 4A). Consistent with the notion that PERKΔC functions as a dominant negative, PERKΔC-3T3 cells maintained levels of protein synthesis after tunicamycin treatment, whereas parental control cells (PURO-3T3) exhibited at least a 2-fold decrease in protein synthesis (data not shown). Similar results were reported for PERK nullizygous embryonic stem cells (26).
Figure 4.
PERK is required for UPR-dependent cell-cycle arrest. (A) NIH 3T3 cells were infected with virus encoding PERKΔC and selected with 7.5 μg/ml puromycin for 4 days. Expression of PERKΔC was confirmed by immunoblot analysis. (B) Parental NIH 3T3 or cells expressing PERKΔC were treated with tunicamycin for 20 h and pulsed with BrdUrd during the last 2.5 h of tunicamycin treatment. The percentage of S-phase cells is expressed as the number of BrdUrd-positive cells vs. the total number of cells. (C) Equivalent amounts of total protein prepared from cells treated as above were resolved on a denaturing polyacrylamide gel and blotted with antibodies specific for BiP and cyclin D1. Sites of antibody binding were visualized by enhanced chemiluminescence. (D) Whole-cell lysates prepared from PERKΔC-3T3 cells treated with 0.5 μg/ml tunicamycin for the indicated intervals were precipitated with antibodies to CDK2. Denatured immune complexes were separated on gels and blotted with antibodies to p21Cip1 or p27Kip1.
To assess the effect of PERKΔC on UPR-dependent cell-cycle arrest, asynchronously proliferating PERKΔC-3T3 or PURO-3T3 cells were left untreated or were challenged with tunicamycin for 20 h. Cell-cycle arrest was monitored by determining the fraction of cells able to incorporate BrdUrd during the last 2.5 h of treatment. Tunicamycin treatment resulted in a significant loss in S-phase PURO-3T3 cells (Fig. 4B Left). In contrast, the percentage of BrdUrd-positive PERKΔC-3T3 cells did not decrease substantially over this time course (Fig. 4B Right). Western analysis revealed that PERKΔC attenuated cyclin D1 loss after tunicamycin treatment (Fig. 4C Center) but did not prevent BiP induction (Fig. 4C Upper). On the basis of the failure of tunicamycin to efficiently down-regulate cyclin D1 in the PERKΔC-3T3 cells, we considered the possibility that p21Cip1 and p27Kip1 might not be mobilized into CDK2 complexes (15). Immunoblotting of anti-CDK2 precipitates with antibodies specific for p21Cip1 and p27Kip1 revealed that tunicamycin did not drive a redistribution of p21Cip1 or p27Kip1 into CDK2 complexes (Fig. 4 D and E). These data implicate PERK as the mediator of UPR-dependent inhibition of cyclin D1 translation and the associated G1 phase arrest.
Discussion
PERK Mediates UPR-Dependent Loss of Cyclin D1 and Cell-Cycle Arrest.
The capacity of the stressed ER to elicit a cellular response is mediated by the activity of at least three resident ER protein kinases: Ire1α, Ire1β, and PERK (7). Our results now implicate PERK as a critical mediator of UPR-dependent cell-cycle arrest. We have found that PERK overexpression inhibits the accumulation of cyclin D1 protein, thereby effectively eliminating the cyclin D-dependent kinase and inducing cell-cycle arrest. Analogous to previous work demonstrating that cells overexpressing cyclin D1 were resistant to UPR-dependent cell-cycle arrest (14), PERK could not arrest cells that either overexpressed cyclin D1 or no longer required cyclin D1 for proliferation (Rb−/− MEFs). These results are consistent with cyclin D1 being the critical downstream target of UPR- and PERK-dependent cell-cycle arrest.
Our results also demonstrate that PERK-dependent loss of cyclin D1 results from the inhibition of cyclin D1 synthesis, mechanistically reminiscent of UPR-dependent down-regulation of cyclin D1 (14). Importantly, the capacity of PERK, but not of Ire1β, to down-regulate cyclin D1 synthesis demonstrates this is not a nonspecific response that ensues upon overexpression of proteins targeted to the ER membrane. Finally, overexpression of the dominant-negative PERKΔC attenuated UPR-dependent inhibition of cyclin D1 and the associated cell-cycle arrest. Taken together, these data suggest that PERK is the resident ER protein kinase that mediates UPR-dependent inhibition of cyclin D1 translation and cell-cycle arrest.
Overexpression of PERK not only eliminated cyclin D1/CDK4 activity but also significantly reduced CDK2 activity. Inhibition was associated with loss of neither cyclin A nor E (negative data not shown). We therefore reasoned that inhibition might be caused by CDK2 sequestration in complexes containing either p21Cip1 or p27Kip1. Consistent with this hypothesis, overexpression of PERK resulted in significant increase in the level of p21Cip1 coprecipitating with CDK2. As PERK expression did not result in a net increase in the level of p21Cip1, its recruitment into CDK2 complexes likely reflects the mobilization of p21Cip1 from cyclin D1 complexes. In contrast to tunicamycin treatment where the level of p27Kip1 bound CDK2 also increased, PERK overexpression resulted in decreased levels of CDK2-associated p27Kip1. The precise mechanism for this difference is not clear, because both PERK expression and tunicamycin treatment resulted in decreased p27Kip1 levels. One possibility is that the differential modulation of p27Kip1 may reflect the potentially pleiotropic affects of impaired protein transport resulting from tunicamycin treatment.
The capacity of PERKΔC to attenuate UPR-dependent D1 loss and cell-cycle arrest argues that PERK activation is required for the manifestation of this UPR-mediated checkpoint. Yet it is formally possible that overexpression of PERKΔC results in the sequestration of nonphysiologic substrates that also impact on cell-cycle progression. It is clear that multiple protein kinases (Ire1α/β and PERK) are activated by ER stress to synergistically regulate cellular responses. Thus, overexpression of a dominant-negative isoform of any one could impinge on signals emanating from other ER kinases. It should be noted that PERKΔC failed to block UPR induction of BiP, which is thought to lie downstream of Ire1α/β (8, 9), arguing for a high degree of specificity. Evaluation of mice nullizygous for one or more components of the ER-signaling pathway will be necessary to fully appreciate its role in regulating UPR-dependent growth arrest.
PERK Regulates Translation of Cyclin D1.
Although cyclin D1 accumulation is regulated at the level of transcription, protein degradation, and subcellular localization, we have found that UPR- (14) and likewise PERK-dependent loss of cyclin D1 results from inhibition of protein synthesis. Rates of translation are most frequently regulated at the level of initiation, and translation initiation is subject to regulation via phosphorylation of either eIF4E or eIF2α (7). eIF4E, or the cap-binding protein, functions in the context of a ternary complex composed of eIF4B and eIF4G (34). The activity of the mature cap-binding complex (eIF4F) is, in turn, regulated by eIF4E-binding proteins that bind directly to eIF4E in a phosphorylation-sensitive fashion (34). The function of the cap-binding complex is to promote the ATP-dependent unwinding of mRNA (7). The inability of ectopic eIF4E to overcome either UPR-dependent inhibition of cyclin D1 translation or cell-cycle arrest (negative data not shown) argues that eIF4E is not targeted by the UPR.
eIF2α is also a component of a ternary complex whose function is to recruit the initiator tRNAMet (7). Because initiation of polypeptide translation requires GTP hydrolysis, conditions that promote stabilization of eIF2:GDP decrease mRNA translation. Phosphorylation of eIF2α at serine 51 has been shown to inhibit GDP:GTP exchange, resulting in high levels of eIF2:GDP (7). The following observations suggest that phosphorylation of eIF2α determines the rate of cyclin D1 translation after activation of the UPR or overexpression of PERK. First, activation of the UPR is associated with increased eIF2α phosphorylation (13, 26). Second, PERK, which is activated by ER stress (11), can phosphorylate eIF2α in vitro and in vivo (13, 26), and PERK-dependent inhibition of cyclin D1 translation correlates with increased eIF2α phosphorylation. Finally, cyclin D1 loss and eIF2α phosphorylation is attenuated in cells expressing PERKΔC.
A growing number of protein kinases that specifically phosphorylate eIF2α are being identified. Each member of this family appears to sense specific cellular stresses and respond by phosphorylating eIF2α, with an ensuing inhibition of protein translation. Although two members of this family are activated by the UPR, PERK and PKR (7) and both are capable of inhibiting cyclin D1 translation (ref. 35 and data herein), PERK is likely to be the primary cellular mediator of UPR-dependent signals. Previous work indicated that PKR is dispensable for UPR-dependent inhibition of protein synthesis (11), whereas our data demonstrate that PERK is critically required. However, as cyclin D1 loss is not completely attenuated in PERKΔC-3T3 cells, we cannot rule out the possibility that other eIF2α protein kinases such as PKR also participate in the regulation of protein translation after activation of the UPR. The capacity of the UPR to activate PKR (7) and the ability of PKR to regulate cyclin D1 translation in response to certain stimuli (35) provide support for this supposition. However, our data are entirely consistent with a model wherein ER stress promotes the activation of PERK via stress-dependent oligomerization (27), resulting in a net increase in eIF2α phosphorylation. Phosphorylation of eIF2α results in the inhibition of cyclin D1 translation, culminating in cell-cycle arrest in G1 phase.
ER Stress, the UPR, and Cell Fate.
What is the function of cell-cycle arrest after ER stress? Inhibition of protein translation after activation of the UPR is thought to effectively limit the accumulation of proteins that transit the stressed ER. This mechanism not only reduces ER traffic but also leads to cell-cycle arrest by decreasing steady-state levels of cyclin D1. Although our studies clearly implicate PERK as a critical mediator of UPR-induced growth arrest, it is not yet clear how cell-cycle arrest influences cell survival and thus tissue homeostasis during conditions of ER stress. Although previous work implies that UPR-triggered cell-cycle arrest is not required for apoptosis (14), cell-cycle exit may, through as yet undetermined mechanisms, coordinate the initiation of apoptosis after ER stress. Alternatively, the induction of cell-cycle arrest after activation of the UPR may provide time necessary to reestablish cellular homeostasis. The increased sensitivity of embryonic stem cells lacking PERK to ER stress-induced apoptosis (26) provides support for the latter hypothesis. Elucidating the relationship between UPR-dependent cell-cycle arrest and apoptosis requires further investigation and will provide important insights into how this UPR-induced checkpoint influences cell fate.
Acknowledgments
We thank David Ron (New York University Medical Center, New York) for providing PERK and Ire1β cDNAs, Charles Sherr (St. Jude Children's Research Hospital, Memphis, TN) for providing NIH 3T3 cells and Rb−/− MEFs, Nahum Sonenberg (McGill University, Montréal, Canada) for providing NIH 3T3 cells overexpressing eIF4E, and Charles Kuszynski for his assistance with flow cytometry. We gratefully acknowledge the excellent technical assistance of Ronald Rimerman and Jamie Fornek. This work was supported in part by American Heart Association Grant 006036Z and The V Foundation (J.A.D.).
Abbreviations
- ER
endoplasmic reticulum
- UPR
unfolded protein response
- CDK cyclin-dependent kinase
eIF2α, elongation initiation factor 2α
- ERK
extracellular regulated protein kinase
- CKI
CDK inhibitors
- PKR
RNA-dependent protein kinase
- MEF
murine embryonic fibroblasts
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
See commentary on page 12396.
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.220247197.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.220247197
References
- 1.Sidrauski C, Chapman R, Walter P. Trends Cell Biol. 1998;8:245–249. doi: 10.1016/s0962-8924(98)01267-7. [DOI] [PubMed] [Google Scholar]
- 2.Lee A S. Curr Opin Cell Biol. 1992;4:267–273. doi: 10.1016/0955-0674(92)90042-b. [DOI] [PubMed] [Google Scholar]
- 3.Wang X Z, Lawson B, Brewer J W, Zinszner H, Sanjay A, Mi L J, Boorstein R, Kreibich G, Hendershot L M, et al. Mol Cell Biol. 1996;16:4273–4280. doi: 10.1128/mcb.16.8.4273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brostrom C O, Brostrom M A. Prog Nucleic Acid Res Mol Biol. 1998;58:79–125. doi: 10.1016/s0079-6603(08)60034-3. [DOI] [PubMed] [Google Scholar]
- 5.Melero J A, Fincham V. J Cell Physiol. 1978;88:355–365. doi: 10.1002/jcp.1040950307. [DOI] [PubMed] [Google Scholar]
- 6.Carlberg M, Larsson O. Anticancer Res. 1993;13:167–171. [PubMed] [Google Scholar]
- 7.Kaufman R J. Genes Dev. 1999;13:1211–1233. doi: 10.1101/gad.13.10.1211. [DOI] [PubMed] [Google Scholar]
- 8.Wang X Z, Harding H P, Zhang Y, Jolicoeur E M, Kuroda M, Ron D. EMBO J. 1998;17:5708–5717. doi: 10.1093/emboj/17.19.5708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tirasophon W, Welihinda A A, Kaufman R J. Genes Dev. 1998;12:2416–2423. doi: 10.1101/gad.12.12.1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Urano F, Wang X, Bertolotti A, Zhang Y, Chung P, Harding H P, Ron D. Science. 2000;287:664–666. doi: 10.1126/science.287.5453.664. [DOI] [PubMed] [Google Scholar]
- 11.Harding H P, Zhang Y, Ron D. Nature (London) 1999;397:271–274. doi: 10.1038/16729. [DOI] [PubMed] [Google Scholar]
- 12.Shi Y, Vattem K M, Sood R, An J, Liang J, Stramm L, Wek R C. Mol Cell Biol. 1998;18:7499–7509. doi: 10.1128/mcb.18.12.7499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sood R, Porter A C, Ma K, Quilliam L A, Wek R C. Biochem J. 2000;346:281–293. [PMC free article] [PubMed] [Google Scholar]
- 14.Brewer J W, Hendershot L M, Sherr C J, Diehl J A. Proc Natl Acad Sci USA. 1999;96:8505–8510. doi: 10.1073/pnas.96.15.8505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sherr C J. Cell. 1994;79:551–555. doi: 10.1016/0092-8674(94)90540-1. [DOI] [PubMed] [Google Scholar]
- 16.Sherr C J, Roberts J M. Genes Dev. 1995;9:1149–1163. doi: 10.1101/gad.9.10.1149. [DOI] [PubMed] [Google Scholar]
- 17.Morgan D O. Nature (London) 1995;374:131–134. doi: 10.1038/374131a0. [DOI] [PubMed] [Google Scholar]
- 18.Cheng C, Olivier P, Diehl J A, Fero M, Roussel M F, Roberts J M, Sherr C J. EMBO J. 1999;18:1571–1583. doi: 10.1093/emboj/18.6.1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.LaBaer J, Garrett M D, Stevenson L F, Slingerland J M, Sandhu C, Chou H S, Fattaey A, Harlow E. Genes Dev. 1997;11:847–862. doi: 10.1101/gad.11.7.847. [DOI] [PubMed] [Google Scholar]
- 20.Blain S W, Montalvo E, Massague J. J Biol Chem. 1997;272:25863–25872. doi: 10.1074/jbc.272.41.25863. [DOI] [PubMed] [Google Scholar]
- 21.Sherr C J. Science. 1996;274:1672–1677. doi: 10.1126/science.274.5293.1672. [DOI] [PubMed] [Google Scholar]
- 22.Diehl J A, Cheng M, Roussel M F, Sherr C J. Genes Dev. 1998;12:3499–3511. doi: 10.1101/gad.12.22.3499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen C, Okayama H. Mol Cell Biol. 1987;7:2745–2752. doi: 10.1128/mcb.7.8.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cheng M, Sexl V, Sherr C J, Roussel M F. Proc Natl Acad Sci USA. 1998;95:1091–1096. doi: 10.1073/pnas.95.3.1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Diehl J A, Zindy F, Sherr C J. Genes Dev. 1997;11:957–972. doi: 10.1101/gad.11.8.957. [DOI] [PubMed] [Google Scholar]
- 26.Harding H P, Zhang Y, Bertolotti A, Zeng H, Ron D. Mol Cell. 2000;5:897–904. doi: 10.1016/s1097-2765(00)80330-5. [DOI] [PubMed] [Google Scholar]
- 27.Bertolotti A, Zhang Y, Hendershot L M, Harding H P, Ron D. Nat Cell Biol. 2000;2:326–332. doi: 10.1038/35014014. [DOI] [PubMed] [Google Scholar]
- 28.Quelle D E, Ashmun R A, Shurtleff S E, Kato J Y, Bar-Sagi D, Roussel M F, Sherr C J. Genes Dev. 1993;7:1559–1571. doi: 10.1101/gad.7.8.1559. [DOI] [PubMed] [Google Scholar]
- 29.Resnitzky D, Gossen M, Bujard H, Reed S I. Mol Cell Biol. 1994;14:1669–1679. doi: 10.1128/mcb.14.3.1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Baldin V, Lukas J, Marcote M J, Pagano M, Draetta G. Genes Dev. 1993;7:812–821. doi: 10.1101/gad.7.5.812. [DOI] [PubMed] [Google Scholar]
- 31.Diehl J A, Sherr C J. Mol Cell Biol. 1997;17:7362–7374. doi: 10.1128/mcb.17.12.7362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mittnacht S, Paterson H, Olson M F, Marshall C J. Curr Biol. 1997;7:219–221. doi: 10.1016/s0960-9822(97)70094-0. [DOI] [PubMed] [Google Scholar]
- 33.Mulcahy L S, Smith M R, Stacey D W. Nature (London) 1995;313:241–243. doi: 10.1038/313241a0. [DOI] [PubMed] [Google Scholar]
- 34.Sonenberg N, Gingras A-C. Curr Opin Cell Biol. 1998;10:268–275. doi: 10.1016/s0955-0674(98)80150-6. [DOI] [PubMed] [Google Scholar]
- 35.Aktas H, Fluckiger R, Acosta J A, Palakurthi S S, Halperin J A. Proc Natl Acad Sci USA. 1998;95:8280–8285. doi: 10.1073/pnas.95.14.8280. [DOI] [PMC free article] [PubMed] [Google Scholar]