Abstract
Postsegregational killing (PSK) systems consist of a tightly linked toxin–antitoxin pair. Antitoxin must be continually produced to prevent the longer lived toxin from killing the cell. PSK systems on plasmids are widely believed to benefit the plasmid by ensuring its stable vertical inheritance. However, experimental tests of this “stability” hypothesis were not consistent with its predictions. We suggest an alternative hypothesis to explain the evolution of PSK: that PSK systems have been selected through benefiting host plasmids in environments where plasmids must compete during horizontal reproduction. In this “competition” hypothesis, success of PSK systems is a consequence of plasmid–plasmid competition, rather than from an adaptive plasmid–host relationship. In support of this hypothesis, a plasmid-encoded parDE PSK system mediated the exclusion of an isogenic ΔparDE plasmid. An understanding of how PSK systems influence plasmid success may provide insight into the evolution of other determinants (e.g., antibiotic resistance and virulence) also rendering a cell potentially dependent on an otherwise dispensable plasmid.
Postsegregational killing (PSK) systems are probably ubiquitous amongst conjugative plasmids (1–3). If a plasmid encoding a PSK system fails to segregate to both daughter cells during cell division (“vertical” reproduction), then the fate of the siblings is dramatically different. The plasmid-containing daughter remains viable, through continued expression of the antitoxin gene. In contrast, the cell not inheriting the plasmid becomes vulnerable to the effects of the more stable toxin (4–7). PSK thereby ensures that the majority of cells in a population remain plasmid-containing (8, 9). The effect of killing plasmid-free daughter cells appears, at the population level, to “addict” the host to the plasmid. It is widely held that this apparent contribution to plasmid vertical stability has caused the success of plasmid-borne PSK systems (7, 10, 11).
Plasmid stability is a measure of the likelihood with which a plasmid is inherited by daughter cells at cell division (10). In the absence of any additional fitness cost, a stability system increases not only the frequency but also the number of plasmid-containing cells in a population. Stability is selected when cell division is the primary means of plasmid reproduction. However, PSK is not predicted to increase either the likelihood of plasmid inheritance or the number of plasmid-containing cells (12). It is therefore not obvious how the stability hypothesis can explain the apparent success of PSK-encoding plasmids.
Recently, Naito et al. (11, 13), compared a series of psk+ and psk− plasmids during competition for host cells. They found that plasmids with a PSK system, in this case a restriction–modification (rm) toxin–antidote pair, inhibited establishment of a competing plasmid within host cells initially containing an r+m+ plasmid (11, 13). The mechanism proposed to explain this “competitive exclusion” was the PSK-mediated death of those cells from which the r+m+ (psk+) plasmid was displaced. This proposal was dubbed the “selfish gene” hypothesis (11). PSK systems were considered selfish genetic entities, sacrificing some hosts to ensure remaining hosts (and therefore themselves) a competitor-free environment (11, 13–15). In a manner thought reminiscent of the altruistic suicide of bacterial and eukaryotic cells after viral infection (14, 16), the success of psk+ plasmids derived from the death of the few, so that the many may live.
Systems similar to PSK are carried by both plasmids and bacteriophage (17, 18), suggesting that PSK function is relevant to the success of elements that reproduce horizontally, i.e., by a process of infectious transfer. An understanding of the nature of the selection of PSK systems may therefore be instrumental to an understanding of the role of plasmids and viruses in the evolution of bacteria (19), virulence (20–22), and antibiotic resistance (23, 24).
Here we test the mechanisms proposed by the stability and selfish-gene hypotheses to explain the evolution and continued success of PSK systems. We also present additional evidence showing that a psk+ plasmid can displace a resident psk− plasmid from host cells. In accord with theoretical analysis (12), we conclude that the stability model is unable to explain the success of PSK-encoding plasmids. Our results are consistent with expectations of the selfish-gene hypothesis; however, we consider them to better support the competition hypothesis of PSK evolution (25). In this view, psk+ plasmids are successful not because they better protect the cell–plasmid relationship, but because they kill competing plasmids per se. Other examples of traits which may have evolved by contributing to success in horizontal competition are discussed.
Materials and Methods
Bacteria and Plasmids.
Bacteria and plasmids used in this study are listed in Table 1. Escherichia coli strains TC102 and TC103 differ from their RR1 progenitor by the insertion of a miniTn10 element encoding only resistance to gentamicin (Gmr), or both Gmr and the parDE PSK system. MiniTn10 elements were mobilized on pBSL vectors from S17.1-λpir as described (26). pTP100 was constructed by introducing a miniTn10 element encoding resistance to chloramphenicol (27) into the psk+ plasmid pRK2526. pTP100 and pRK21526 (psk−) are self-transferable on solid media only (9). Hereafter, TC102/TC103 and pRK21526/pTP100 are denoted psk−C/psk+C and psk−P/psk+P to distinguish otherwise essentially isogenic psk− and psk+ chromosome (C) and plasmid (P) derivatives, respectively. pBSL182-parDE was constructed by the insertion of a BamHI/EcoRI fragment encoding the parDE PSK system of pOU82-parDE into the multiple-cloning site of pBSL182.
Table 1.
Bacteria and plasmids used in this study
| Strain or plasmid | Abbreviation | Relevant description | Reference or source |
|---|---|---|---|
| Bacteria | |||
| RR1 | rpsL hsdR | (64) | |
| TC102 | psk−c | RR1∷aac2 (Gmr) | This work |
| TC103 | psk+c | RR1∷aac2-parDE (Gmr, psk+) | This work |
| S17.1-λpir | ∷RP4 | (26) | |
| Plasmids | |||
| pRK21526 | psk−p | IncP, ΔparDE, ant3" (Spr) | (9) |
| pRK2526 | IncP, parDE (psk+) | (9) | |
| pTP100 | psk+p | IncP, parDE (psk+), ∷cat (Cmr) | This work |
| pBSL182 | Tn10∷aac2 (Gmr) | (26) | |
| pBSL182-parDE | Tn10∷aac2-parDE (Gmr, psk+) | This work | |
| pOU82-parDE | parDE (psk+) | (30) |
Spr, spectinomycin resistance; Cmr, chloramphenicol resistance.
Media.
Liquid and solid media were supplemented with antibiotics at the following concentrations: 20 μg/ml chloramphenicol, 5 μg/ml gentamicin, 150 μg/ml spectinomycin, and 100 μg/ml streptomycin. Then 5-bromo-4-chloro-3-indolyl β-D-galactoside was used at 40 μg/ml to distinguish plasmid-containing from plasmid-free cells. Cells were grown to saturation in Luria–Bertani–Herskowitz (LBH) medium (28) supplemented with antibiotics as appropriate to maintain plasmids, diluted 100-fold in fresh media, and grown to mid-log before all assays.
Mating Conditions.
Matings were performed for 2 h on solid media as described previously (29). “Resident” plasmids were those contained in psk−C or psk+C hosts. These cells were introduced in 100-fold excess to donor RR1 cells containing “incoming” plasmids. After mating, cells were resuspended in LBH media. To measure plasmid transfer frequency, an aliquot was plated immediately on media supplemented with antibiotics to select psk−C- or psk+C-recipient cells having received the incoming plasmid. To form colonies on this media, the incoming plasmid need only be inherited at a frequency >0.5/cell division. Therefore, this enumeration gives a measure of plasmid transfer to recipient cells relatively insensitive to imperfect transmission to daughter cells. Transfer frequencies are reported as transconjugants per limiting parent.
To measure incoming plasmid transmission frequency, remaining cells were grown with vigorous shaking for ≈10 generations in LBH media without antibiotics, and then plated onto selective media to enumerate incoming plasmid-containing recipient cells as above. Under these conditions, de novo plasmid transfer is not observed (9). The number of recipient cells that contain the incoming plasmid after incubation in this environment is dependent on both the number to which the incoming plasmid had initially transferred and on the subsequent transmission of transferred plasmids to daughter cells during cell division. Incoming plasmid transmission efficiency is defined as the ratio of the observed final number of incoming plasmid-containing recipient cells to that expected if the resident plasmid had no effect on posttransfer transmission of the incoming plasmid (i.e., if incoming plasmid and host-cell generations were coincident).
Plasmid Stability Assays.
Monocultures of psk+ and psk− plasmid-containing cells were diluted 106-fold into 10 ml of LBH media, incubated with shaking until cell titer was ≈108 colony-forming unit/ml, and then diluted 104- to 106-fold into fresh media. This cycle was repeated to allow cells to complete 200 generations of unsaturated growth. To ensure that bacteria never exited log growth phase, cell density was monitored by OD600 measurements and confirmed retrospectively by plate counts at each dilution point. This regime provided an environment in which resources did not become limiting. At the time of each dilution, the fraction of cells carrying a plasmid in the psk+ and psk− populations was measured by comparing the number of blue Lac+ (plasmid-containing) and white Lac− (plasmid-free) colonies on LBH agar plates supplemented with 5-bromo-4-chloro-3-indolyl β-D-galactoside (9). This comparison gives a measure of apparent plasmid stability. An alternative estimate of plasmid stability, the number of generations undergone by plasmid-containing cells between each dilution time point, also was measured.
The vertical competition assay was performed similarly except equal numbers of RR1 containing either psk−P or psk+P were coincubated and allowed to reach saturation densities before dilution. In this environment, psk+ and psk− plasmid-containing cells were forced to compete for resources. Plasmid-containing cells were measured as above. The psk−P- and psk+P-containing cells were enumerated on media supplemented with either spectinomycin or chloramphenicol, respectively.
Results
Evaluation of the Effect of PSK on Plasmid Stability.
Plasmid stability is a measure of the fidelity with which a plasmid is vertically inherited during cell division. If PSK contributes to plasmid stability, a psk+ plasmid is expected to be inherited more frequently and consequently increase in number faster than an otherwise isogenic psk− plasmid during cell division. To test this prediction, we monitored plasmid dynamics in two parallel monocultures of RR1, harboring either psk−P or psk+P. Monocultures were incubated under conditions previously shown to constrain these plasmids to vertical replication (9). Cultures were periodically diluted to ensure cells never reached saturation density.
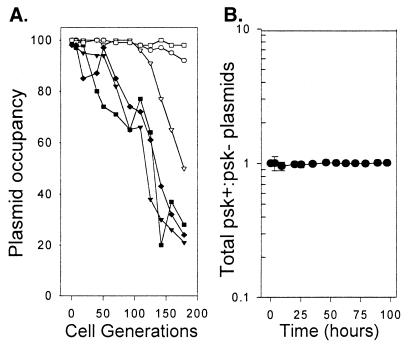
The monoculture grown from cells containing the psk− plasmid quickly accumulated plasmid-free cells. In the second monoculture, started from psk+ plasmid-containing cells, a higher percentage of cells retained the plasmid throughout growth (Fig. 1A). These results were consistent with previous observations of these and other psk+/psk− plasmid pairs (9, 30). This increased frequency of occupancy has been used to infer a contribution by PSK to plasmid reproductive success. However, the death of those daughter cells not inheriting the psk+ plasmid is likely to obscure the relationship between plasmid stability and frequency of plasmid occupancy. For this reason, a second corollary of plasmid stability, accumulation of plasmid-containing cells, also was measured. If PSK is selected as a plasmid stability system, more plasmid-containing cells were expected to be present in the psk+ plasmid-containing population. A comparison of plasmid-containing cells in the populations described above revealed that the total number of psk+ and psk− plasmids remained similar throughout ≈200 generations of growth (Fig. 1B). Therefore, different occupancy rates did not indicate different rates of plasmid reproduction.
Figure 1.
(A) Apparent stability of psk+ and psk− plasmids measured in RR1 in the absence of selection. The fraction of cells carrying a plasmid was measured at ≈20 generation intervals as described in Materials and Methods. RR1 containing psk−p-empty symbols, ○, □, and ▵; psk+p-filled symbols, ●, ■, and ▴. Each line represents a single experiment. (B) Realized stability of psk+ and psk− plasmids in the absence of selection. The total number of psk+ and psk− plasmids inherited in A was estimated by extrapolating the number of plasmid generations between each dilution time point. Shown is the average ratio (and SD) of psk+:psk− plasmid generations.
Freed Resource Model for Stabilization.
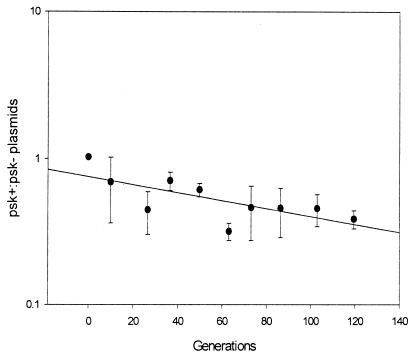
PSK may provide an advantage to psk+ plasmids by freeing nutrients, otherwise shared with plasmid-free segregants, for use by plasmid-containing cells (31–33). The “freed resource” hypothesis differs from the stability model in predicting PSK to increase only relative, not absolute, plasmid vertical reproduction. This mechanism requires the biased allocation of freed resources to psk+ plasmid-containing cells if they are to be advantaged over a competing psk− plasmid (12). If psk+ plasmids out-compete psk− plasmids because of an increase in relative reproduction rate, then in mixed populations, those encoding PSK systems should ultimately dominate. To test this prediction, equal numbers of cells containing either psk+P or psk−P were placed in direct competition during 120 generations of clonal growth (Fig. 2). The ratio of psk+:psk− plasmids should have increased if bacteria with psk+ plasmids captured more resources. However, contrary to freed resource expectations, psk+ plasmids did not outcompete psk− plasmids in this environment (Fig. 2). In fact, the ratio of psk+:psk− plasmids decreased during the course of competition indicating that the psk+ plasmid was marginally less fit (slope = −0.00386, R2 = 0.547, P = 0.016).
Figure 2.
Vertical competition between psk+ and psk− plasmids contained in RR1. Cells containing either psk−p or psk+p were coincubated at 37°C in LBH broth with shaking as described in Materials and Methods. Relative success of the two plasmid types is expressed as the ratio of psk+p-containing cells to psk−p-containing cells. Values reported are the average (and SD) of three independent experiments.
Demonstration of PSK-Mediated Plasmid Competitive Exclusion.
Kusano et al. (34) and Naito et al. (11, 13) have described the “competitive exclusion” of competing plasmids from cells occupied by a r+m+ encoding (psk+) plasmid. In their experiments, two incompatible plasmids competed for vertical reproduction (11, 13, 34). We sought to extend the generality of this phenomenon to elements that reproduce naturally by horizontal transfer by comparing the conjugation frequency of the self-transferable, incompatible plasmids, psk−P, and psk+P, to host cells initially occupied by the other.
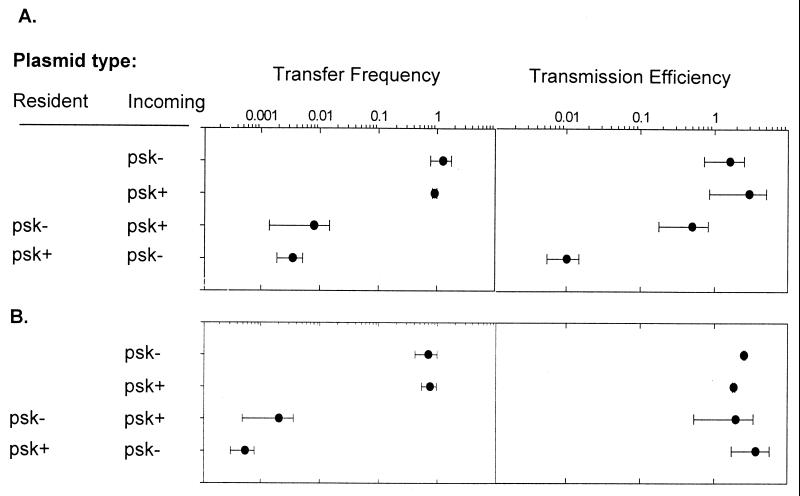
Consistent with previous results (11, 13, 34), we found that the presence of a PSK system on a resident plasmid reduced maintenance (plasmid transmission) of a psk− plasmid within recipient cell populations containing a psk+ plasmid (Fig. 3, row 4). In contrast, the transmission of a psk+ plasmid was not affected by the presence of a resident psk− plasmid (Fig. 3A, row 3). psk+P and psk−P were equally well transmitted within a plasmid-free host population (Fig. 3A, rows 1 and 2). Thus psk+ plasmids were able to both resist invasion by psk− plasmids and to colonize hosts previously occupied by psk− plasmids.
Figure 3.
(A) Effect of PSK on the transfer and transmission of incoming plasmids. (B) Effect of recipient cell immunity to PSK action on competitive exclusion. Transconjugant number was measured immediately after conjugation and after 10 generations of nonselective growth in LBH broth with vigorous shaking to give initial transfer frequency and final transmission frequency, respectively (as described in Materials and Methods). Transmission efficiency is the ratio of observed transmission to that expected if the resident plasmid had no effect on incoming plasmid stability. Values reported are the average (and SD) of at least four independent experiments.
Effect of Host-Cell Immunity on PSK-Mediated Plasmid Exclusion.
Although the proposed competitive exclusion model seems the most likely explanation for the inability of psk−P to establish, it also is possible that exclusion could be caused by the death of psk+ plasmid-containing cells immediately after psk− plasmid transfer. Perturbation of PSK expression, e.g., as occurs after phage infection, exposure to the antibiotic rifampicin, or when cells are being starved, has previously been observed to induce PSK (33, 35, 36). This suggestion makes two unique predictions. Firstly, that transfer of a psk− plasmid to cells occupied by a psk+ plasmid will be lower than vice versa. Secondly, that the presence of a chromosomal PSK system, allowing continued ParD (antitoxin) production after loss of a psk+ plasmid, will not alleviate PSK-mediated exclusion of a psk− plasmid. Contrary to the first of these predictions, we found that the transfer frequency of psk−P or psk+P to cells already occupied by the other plasmid was not significantly different (Fig. 3A).
To test the second prediction, the parDE PSK system was inserted into the RR1 chromosome by using the miniTn10 encoded by pBSL182-parDE (to give psk+C). The presence of this chromosomally borne PSK system rendered the cell immune to death after the loss of the psk+ plasmid (data not shown). When a psk+ plasmid was resident in this strain before psk− plasmid transfer, the incoming plasmid was able to establish (Fig. 3B). Therefore, it seems likely that PSK-mediated exclusion depends on death of host cells after psk+ plasmid loss. The ability of psk− plasmids to establish correlated with a reduced frequency of psk+ plasmid co-inhabitance. The psk+ resident plasmid was present in all (400/400) psk− plasmid-containing cells in PSK toxin-sensitive hosts, but in <0.1% when the host cell carried a chromosomal copy of the PSK system. This decrease in co-inhabitance is expected if the incoming psk− plasmid is no longer dependent on the continued presence of the resident psk+ plasmid for host-cell viability.
Discussion
The canonical “stability” model explains the evolution and maintenance of plasmid-borne PSK systems by asserting that the action of killing plasmid-free daughter cells increases the vertical stability of linked plasmids (10, 37, 38). Indeed, we also observed a generally higher percentage of plasmid-containing cells in populations started with bacteria containing psk+ rather than psk− plasmids (Fig. 1A). In the stability model, plasmid vertical stability is seen as a corollary of high plasmid occupancy in viable daughter cells. However, the presence of a PSK system did not increase the effective frequency of plasmid vertical inheritance measured in the monoculture environment (Fig. 1B). The decrease in the ratio of psk+:psk− plasmids shown in Fig. 2 probably reflects a cost of carriage of the PSK system (Fig. 2). It would appear that different plasmid occupancy rates do not indicate different reproductive potential. These experimental observations agree with a previous theoretical analysis (12). In one psk+ replicate plasmid-free cells did accumulate (Fig. 1A). We consider the most likely explanation for this observation to be the rare “escape” of some psk+ segregants from the action of PSK, as has been noted previously (30).
In all experiments, plasmid-free segregants eventually became significantly represented. However, because the experimental environment was either effectively limitless (such that plasmid-free and plasmid-containing cells did not have to compete for resources) (Fig. 1) or both plasmid-containing competitors were coexisting (Fig. 2), plasmid-free cells did not affect estimation of relative success of the two plasmid types. We also note that the outcomes observed may not be transitive to competition in structured environments, e.g., biofilms, in which differential use of freed resources may be more likely to occur (39).
It has been suggested that PSK systems might affect plasmid competition during horizontal reproduction (13, 15, 25). The results presented here demonstrate the inability of psk− plasmids to establish in populations of cells occupied by a psk+ plasmid. The presence of a resident psk+ plasmid did not significantly affect psk− plasmid transfer, but did affect subsequent transmission to transconjugant daughter cells (Fig. 3A, row 4). Both psk+ and psk− plasmids were equally well able to establish in plasmid-free cells (Fig. 3A, rows 1 and 2). Therefore, PSK increases relative, but not absolute, plasmid horizontal reproduction. Difference in the transfer frequency of either plasmid to cells already containing a plasmid, relative to transfer to plasmid-free cells, was likely the result of PSK-independent plasmid-encoded surface exclusion (40).
To account for these results, we proposed the competition model. After transfer of psk−P to a cell containing psk+P, replication incompatibility results in a high frequency of plasmid missegregation (41). In those cells losing psk+P, dilution/degradation of the ParD antitoxin rendered descendants increasingly vulnerable to the effect of the ParE toxin (42). psk−p plasmids present in these cells are also “killed.” Exclusion is therefore not due to the inability of psk− plasmids to transfer to, or stabilize in, cells containing a psk+ plasmid, but to the death of those cells in which displacement occurs. Consistent with this model, the exclusion phenomena disappeared when host cells were made immune to the effect of psk+ plasmid loss (Fig. 3B, rows 3 and 4).
The competition model also predicts the death of cells remaining infected by a resident psk−p but which have failed to inherit a copy of the psk+ plasmid. Observation of this outcome was masked by a high background of psk−p containing cells never infected by psk+p. Nevertheless, the inability of psk−p to be maintained alone in cells whose ancestors have contained psk+p supports the idea that the psk−p host was killed. This reciprocal competition provides a mechanism for the initial success, as well as the maintenance, of psk+ plasmids (43).
The competition model is similar to that proposed as evidence for the selfish gene hypothesis of r+m+ (restriction–modification) system evolution (11, 13). Those researchers hypothesized that the inability of a plasmid to establish in cells containing an incompatible r+m+ plasmid was due to the restriction endonuclease-mediated death of cells in which the r+m+ plasmid had been displaced (11, 13). In that work, transformation was used to mediate plasmid horizontal transfer. Our work extends the phenomena of PSK-mediated plasmid exclusion to include the exclusion of incompatible plasmids after conjugal transfer. This type of transfer has the advantage of allowing direct comparison of plasmid transfer and transmission frequencies, and of allowing the effect of PSK to be assessed in situ on the plasmids upon which they have evolved. In addition, conjugation is likely to be the most frequent and widespread form of plasmid horizontal reproduction (44). Thus, demonstration of PSK-mediated competitive exclusion after conjugal plasmid transfer is ecologically relevant.
Although agreeing on the mechanism by which PSK systems are successful, the selfish gene and competition hypotheses disagree on how to interpret this mechanism to give insight into their ecology and evolution. Under the selfish gene hypothesis, PSK systems are thought of as selfish entities because of their detrimental effect on host-cell populations. In this respect, PSK systems have been viewed as analogs of maternal-effect selfish genes (11, 16), which also are subject to the apparent paradox of success despite a detrimental effect on their host (45–47). In both cases, an increase in vertical reproduction due to biased access to resources freed by the death of competitors is predicted to allow the success of selfish genes (13, 48). PSK is thought to protect the cellular niche of a psk+ plasmid and increase its frequency in the next generation through vertical reproduction (15, 34). Therefore, like the stability hypothesis, the selfish gene hypothesis relies upon a cell-level competition to determine the most fit cell–plasmid entities. The selfish gene hypothesis is, therefore, unable to explain the rapid initial success of psk+ plasmids when they are rare because in vertical competition between cells carrying psk− and psk+ plasmids, PSK confers no advantage (Fig. 2). In contrast, the competition model posits that plasmid–plasmid horizontal competition has driven the selection of PSK. Under this hypothesis, psk+ plasmid-mediated death of host cells is secondary to the death of competing psk− plasmids (25). Subsequent establishment in cells initially occupied by competing plasmids confers an advantage to psk+ plasmids independent of cell-level vertical competition and is only dependent on horizontal reproduction. Thus, optimization of the plasmid–host relationship is also a secondary consideration to the origin, and possibly maintenance, of PSK systems.
Both the competition hypothesis and the selfish gene hypothesis are consistent with the widespread occurrence of multiple, unrelated PSK systems on genetic elements (2, 38, 49). However, only the competition hypothesis is consistent with the observation that psk+ plasmids are only advantaged when cell death is accompanied by the “death” of a competing psk− plasmid (compare Fig. 3A to Fig. 2 and 3B). Thus, although PSK systems may be “genetically” selfish, in that they are maintained despite no contribution to cell reproduction (50, 51), it seems unlikely that selfishness has been the driving force for their evolution. In contrast, because the success of maternal-effect killing systems depends directly on their detrimental effect on the host (52) they represent truly “evolutionarily” selfish entities (48, 53). Moreover, both that PSK systems are commonly found on horizontally mobile elements (HMEs) (37, 38), and that these elements can be directly involved in PSK-mediated competition (36, 54, 55) are consistent with the proposal that the autonomy from host replication offered by a horizontal lifestyle (56–59) is influential in determining PSK system success.
Additional support for the competition hypothesis comes from an experimental study of evolution in populations of the bacteriophage φ6 (60). In this study, prediction of bacteriophage success was shown to depend critically not on absolute rates of reproduction (i.e., HME/host–HME/host competition), but on the outcome of phage–phage intrahost competition (i.e., HME–HME competition) (60). That the evolution of HMEs is not predictable from knowledge of the host–HME relationship alone is a unique prediction of the competition hypothesis.
Systems rendering a cell dependent on carriage of an otherwise dispensable plasmid are probably not limited to PSK. In the presence of the cognate intra- or extra-cellular toxin, any antitoxin-encoding gene likely can be thought of as a PSK system (16). Examples of such systems include bacteriocins (39, 61), antibiotic resistance determinants (62), and virulence determinants (63). By not constraining predictions of their success to environments allowing cell-level replication and thus cell-level competition, the HME–HME competition model provides an alternative insight into the selection and evolution of such systems. The observed bias in accumulation of these potential addiction systems to replicons able to reproduce horizontally is consistent with this interpretation (22, 23).
Acknowledgments
We thank K. Gerdes for the gift of several plasmids. I. Kobayashi and D. Thaler provided helpful comments on the manuscript. This work was supported by the University of Canterbury Grants U6275 and U6107 (to J.A.H.) and a Ph.D. scholarship (to T.F.C.) and the New Zealand Lotteries Grant Board Grants AP64928 and AP52713 (to J.A.H.).
Abbreviations
- PSK
postsegregational killing
- rm
restriction–modification
- Gmr
resistance to gentamicin
- HME
horizontally mobile element
- LBH
Luria–Bertani–Herskowitz medium
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.220077897.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.220077897
References
- 1.Poulsen L K, Larsen N W, Molin S, Andersson P. Mol Microbiol. 1989;3:1463–1472. doi: 10.1111/j.1365-2958.1989.tb00131.x. [DOI] [PubMed] [Google Scholar]
- 2.Gronlund H, Gerdes K. J Mol Biol. 1998;285:1401–1415. doi: 10.1006/jmbi.1998.2416. [DOI] [PubMed] [Google Scholar]
- 3.Loh S, Cram D, Skurray R. Mol Gen Genet. 1989;219:177–186. doi: 10.1007/BF00261174. [DOI] [PubMed] [Google Scholar]
- 4.Tsuchimoto S, Nishimura Y, Ohtsubo E. J Bacteriol. 1992;174:4205–4211. doi: 10.1128/jb.174.13.4205-4211.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Thisted T, Nielsen A K, Gerdes K. EMBO J. 1994;13:1950–1959. doi: 10.1002/j.1460-2075.1994.tb06464.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jaffe A, Ogura T, Hiraga S. J Bacteriol. 1985;163:841–849. doi: 10.1128/jb.163.3.841-849.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gerdes K, Rasmussen P B, Molin S. Proc Natl Acad Sci USA. 1986;83:3116–3120. doi: 10.1073/pnas.83.10.3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ogura T, Hiraga S. Proc Natl Acad Sci USA. 1983;80:4784–4788. doi: 10.1073/pnas.80.15.4784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sia E A, Roberts R C, Easter C, Helinski D R, Figurski D H. J Bacteriol. 1995;177:2789–2797. doi: 10.1128/jb.177.10.2789-2797.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nordstrom K, Austin S J. Annu Rev Genet. 1989;23:37–69. doi: 10.1146/annurev.ge.23.120189.000345. [DOI] [PubMed] [Google Scholar]
- 11.Naito T, Kusano K, Kobayashi I. Science. 1995;267:897–899. doi: 10.1126/science.7846533. [DOI] [PubMed] [Google Scholar]
- 12.Mongold J A. Am Nat. 1992;139:677–689. [Google Scholar]
- 13.Naito Y, Naito T, Kobayashi I. Biol Chem. 1998;379:429–436. doi: 10.1515/bchm.1998.379.4-5.429. [DOI] [PubMed] [Google Scholar]
- 14.Nakayama Y, Kobayashi I. Proc Natl Acad Sci USA. 1998;95:6442–6447. doi: 10.1073/pnas.95.11.6442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kobayashi I. Trends Genet. 1998;14:368–374. doi: 10.1016/s0168-9525(98)01532-7. [DOI] [PubMed] [Google Scholar]
- 16.Yarmolinsky M. Science. 1995;267:836–837. doi: 10.1126/science.7846528. [DOI] [PubMed] [Google Scholar]
- 17.Yu Y N, Snyder L. Proc Natl Acad Sci USA. 1994;91:802–806. doi: 10.1073/pnas.91.2.802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Snyder L. Mol Microbiol. 1995;15:415–420. doi: 10.1111/j.1365-2958.1995.tb02255.x. [DOI] [PubMed] [Google Scholar]
- 19.Lawrence J G, Roth R R. Genetics. 1996;143:1843–1860. doi: 10.1093/genetics/143.4.1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hayes F. J Bacteriol. 1998;180:6415. doi: 10.1128/jb.180.23.6415-6418.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bull J J, Molineaux I J, Rice W R. Evolution. 1991;45:875–882. doi: 10.1111/j.1558-5646.1991.tb04356.x. [DOI] [PubMed] [Google Scholar]
- 22.Levin B R, Svanborg-Eden C. Parasitology. 1990;100:103–115. doi: 10.1017/s0031182000073054. [DOI] [PubMed] [Google Scholar]
- 23.Eberhard W G. Quart Rev Biol. 1990;65:3–22. doi: 10.1086/416582. [DOI] [PubMed] [Google Scholar]
- 24.Heinemann J A. In: Encyclopedia of Microbiology. 2nd Ed. Lederberg J, editor. Vol. 2. New York: Academic; 2000. pp. 698–707. [Google Scholar]
- 25.Heinemann J A. In: Horizontal Gene Transfer. Kado C, Syvanen M, editors. London: Thomson; 1998. pp. 11–24. [Google Scholar]
- 26.Alexeyev M F, Shokolenko I N. Gene. 1995;160:59–62. doi: 10.1016/0378-1119(95)00141-r. [DOI] [PubMed] [Google Scholar]
- 27.Kleckner N, Bender J, Gottesman S. Methods Enzymol. 1991;204:139–180. doi: 10.1016/0076-6879(91)04009-d. [DOI] [PubMed] [Google Scholar]
- 28.Herskowitz I, Signer E R. J Mol Biol. 1970;47:545–556. doi: 10.1016/0022-2836(70)90321-9. [DOI] [PubMed] [Google Scholar]
- 29.Heinemann J A, Scott H E, Williams M. Genetics. 1996;143:1425–1435. doi: 10.1093/genetics/143.3.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jensen R B, Grohmann E, Schwab H, Diaz-Orejas R, Gerdes K. Mol Microbiol. 1995;17:211–220. doi: 10.1111/j.1365-2958.1995.mmi_17020211.x. [DOI] [PubMed] [Google Scholar]
- 31.Wu K, Wood T K. Biotech Eng. 1994;44:912–921. doi: 10.1002/bit.260440807. [DOI] [PubMed] [Google Scholar]
- 32.Wu K, Jahng D, Wood T K. Biotechnol Progr. 1994;10:621–629. doi: 10.1021/bp00030a600. [DOI] [PubMed] [Google Scholar]
- 33.Aizenman E, Engelberg-Kulka H, Glaser G. Proc Natl Acad Sci USA. 1996;93:6059–6063. doi: 10.1073/pnas.93.12.6059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kusano K, Naito T, Handa N, Kobayashi I. Proc Natl Acad Sci USA. 1995;92:11095–11099. doi: 10.1073/pnas.92.24.11095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nielsen A K, Thorsted P, Wagner E G H, Gerdes K. Mol Microbiol. 1991;5:1961–1973. doi: 10.1111/j.1365-2958.1991.tb00818.x. [DOI] [PubMed] [Google Scholar]
- 36.Pecota D C, Wood T K. J Bacteriol. 1996;178:2044–2050. doi: 10.1128/jb.178.7.2044-2050.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jensen R B, Gerdes K. Mol Microbiol. 1995;17:205–210. doi: 10.1111/j.1365-2958.1995.mmi_17020205.x. [DOI] [PubMed] [Google Scholar]
- 38.Gerdes K, Gultyaev A P, Franch T, Pederson K, Mikkelsen N D. Annu Rev Genet. 1997;31:1–31. doi: 10.1146/annurev.genet.31.1.1. [DOI] [PubMed] [Google Scholar]
- 39.Chao L, Levin B R. Proc Natl Acad Sci USA. 1981;78:6324–6328. doi: 10.1073/pnas.78.10.6324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Haase J, Kalkum M, Lanka E. J Bacteriol. 1996;178:6720–6729. doi: 10.1128/jb.178.23.6720-6729.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Condit R, Levin B R. Am Nat. 1990;135:573–596. [Google Scholar]
- 42.Jovanovic O S, Ayres E K, Figurski D H. J Mol Biol. 1994;237:52–64. doi: 10.1006/jmbi.1994.1208. [DOI] [PubMed] [Google Scholar]
- 43.Axelrod R, Hamilton W D. Science. 1981;211:1390–1396. doi: 10.1126/science.7466396. [DOI] [PubMed] [Google Scholar]
- 44.Davison J. Plasmid. 1999;42:73–91. doi: 10.1006/plas.1999.1421. [DOI] [PubMed] [Google Scholar]
- 45.Beeman R W, Friesen K S, Denell R E. Science. 1992;256:89–92. doi: 10.1126/science.1566060. [DOI] [PubMed] [Google Scholar]
- 46.Bull J J, Molineux I J, Warren J H. Science. 1992;256:65. doi: 10.1126/science.1566058. [DOI] [PubMed] [Google Scholar]
- 47.Hurst D L. Cell. 1993;75:407–408. doi: 10.1016/0092-8674(93)90375-z. [DOI] [PubMed] [Google Scholar]
- 48.Hurst L D, Atlan A, Bengtsson B O. Quart Rev Biol. 1996;71:317–364. doi: 10.1086/419442. [DOI] [PubMed] [Google Scholar]
- 49.Holcick M, Iyer V N. Microbiology. 1997;143:3403–3416. doi: 10.1099/00221287-143-11-3403. [DOI] [PubMed] [Google Scholar]
- 50.Orgel L E, Crick F H C. Nature (London) 1980;284:604–607. doi: 10.1038/284604a0. [DOI] [PubMed] [Google Scholar]
- 51.Doolittle W F, Sapienza C. Nature (London) 1980;284:601–603. doi: 10.1038/284601a0. [DOI] [PubMed] [Google Scholar]
- 52.Smith N G C. J Theor Biol. 1998;191:173–180. doi: 10.1006/jtbi.1997.0579. [DOI] [PubMed] [Google Scholar]
- 53.Hamilton W D. Nature (London) 1970;228:1218–1220. doi: 10.1038/2281218a0. [DOI] [PubMed] [Google Scholar]
- 54.Engelberg-Kulka H, Reches M, Nrasimhan S, Schoulaker-Schwarz R, Klemes Y, Aizenman E, Glaser G. Proc Natl Acad Sci USA. 1998;95:15481–15486. doi: 10.1073/pnas.95.26.15481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Parma D H, Snyder M, Sobolevski S, Nawroz M, Brody E, Gold L. Genes Dev. 1992;6:497–510. doi: 10.1101/gad.6.3.497. [DOI] [PubMed] [Google Scholar]
- 56.Heinemann J A, Ankenbauer R G. Mol Microbiol. 1993;10:57–62. doi: 10.1111/j.1365-2958.1993.tb00903.x. [DOI] [PubMed] [Google Scholar]
- 57.Heinemann J A. Drug Disc Today. 1999;4:72–79. doi: 10.1016/s1359-6446(98)01294-x. [DOI] [PubMed] [Google Scholar]
- 58.Peters J E, Benson S A. J Bacteriol. 1995;177:847–850. doi: 10.1128/jb.177.3.847-850.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Peters J E, Bartoszyk I M, Dheer S, Benson S A. J Bacteriol. 1996;178:3037–3043. doi: 10.1128/jb.178.11.3037-3043.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Turner P E, Chao L. Genetics. 1998;150:523–532. doi: 10.1093/genetics/150.2.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tan Y, Riley M A. Microbiology. 1996;142:2175–2180. doi: 10.1099/13500872-142-8-2175. [DOI] [PubMed] [Google Scholar]
- 62.Shaw K J, Rather P N, Hare R S, Miller G H. Microbiol Rev. 1993;57:138–163. doi: 10.1128/mr.57.1.138-163.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Finlay B B, Falkow S. Micro Mol Rev. 1997;61:136–169. doi: 10.1128/mmbr.61.2.136-169.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Boyer H W, Roulland-Dussoix D. J Mol Biol. 1969;41:459–472. doi: 10.1016/0022-2836(69)90288-5. [DOI] [PubMed] [Google Scholar]