Abstract
To address the dual needs for improved methods to assess potential health risks associated with chemical exposure in aquatic environments and for new models for in vivo mutagenesis studies, we developed transgenic fish that carry multiple copies of a bacteriophage λ vector that harbors the cII gene as a mutational target. We adapted a forward mutation assay, originally developed for λ transgenic rodents, to recover cII mutants efficiently from fish genomic DNA by λ in vitro packaging. After infecting and plating phage on a hfl− bacterial host, cII mutants were detected under selective conditions. We demonstrated that many fundamental features of mutation analyses based on λ transgenic rodents are shared by transgenic fish. Spontaneous mutant frequencies, ranging from 4.3 × 10−5 in liver, 2.9 × 10−5 in whole fish, to 1.8 × 10−5 in testes, were comparable to ranges in λ transgenic rodents. Treatment with ethylnitrosourea resulted in concentration-dependent, tissue-specific, and time-dependent mutation inductions consistent with known mechanisms of action. Frequencies of mutants in liver increased insignificantly 5 days after ethylnitrosourea exposure, but increased 3.5-, 5.7- and 6.7-fold above background at 15, 20, and 30 days, respectively. Mutants were induced 5-fold in testes at 5 days, attaining a peak 10-fold induction 15 days after treatment. Spontaneous and induced mutational spectra in the fish were also consistent with those of λ transgenic rodent models. Our results demonstrate the feasibility of in vivo mutation analyses using transgenic fish and illustrate the potential value of fish as important comparative animal models.
Keywords: medaka, ethylnitrosourea
A major challenge to the detection of spontaneous and induced mutations is the difficulty with which mutant genes can be efficiently recovered and accurately identified in vivo. Considering that mutations must be detected at low frequencies (e.g., ≈1 spontaneous mutation/105–107 loci), and that sufficient DNA sequence information must be available to distinguish mutant from nonmutant genes, the problem of efficiently detecting and quantifying mutations in whole animals can be formidable. Transgenic animals that carry specific genes for quantitation of spontaneous and induced mutations have been developed to assist in improving in vivo mutation analyses (1). In this approach, a transgenic animal carries a prokaryotic vector that harbors a gene that serves as a mutational target. After mutagen exposure, the vector is separated from the animal's genomic DNA and shuttled into indicator bacteria where mutant and nonmutant genes are readily quantified (2, 3). Transgenic mutation assays offer numerous benefits for in vivo mutation detection not available by using other approaches. Benefits include the ability to screen rapidly statistically meaningful numbers of genetically neutral mutational targets in a variety of tissues and the ability to characterize mutations to aid in disclosing possible mechanisms of mutagen action.† A significant additional attribute is the potential adaptability of the mutation assays to different strains or species, thereby facilitating comparisons of an identical mutation target among cells, tissues, organs, and species to an extent that was not possible otherwise.
Recognizing the value of this approach, we reasoned that fish had excellent potential to meet the dual needs for improved methods to assess health hazards associated with exposure to chemicals in aquatic environments and for alternative nonmammalian animal models in mutagenesis and carcinogenesis studies. Increasingly, fish have been embraced as valuable animal models in genetics, developmental biology, and toxicology (5–7). In some applications, such as in the assessment of health hazards associated with exposure to complex chemical mixtures or in low-dose chronic exposure regimens, fish are recognized not merely as alternatives to traditional rodent models, but as having distinct and superior benefits providing insights to fundamental mechanisms of disease processes (8). Recent advances in fish transgenesis have made it possible to enhance the utility of fish by generating new animal models (9).
In the present study, we introduce transgenic medaka (Oryzias latipes), which carry multiple copies of a λ bacteriophage vector as a new animal model for in vivo mutation detection. We recovered mutations from fish by using procedures adapted from a recently developed positive-selection assay for transgenic rodents that uses the cII gene as a logistically simpler and cost-effective alternative to the lacI mutational target (10). This assay is based on the role the cII protein plays in the commitment of bacteriophage λ to the lysogenic cycle in Escherichia coli host cells. A specialized E. coli strain (hfl−) extends the longevity of the cII gene product, facilitating selection of mutant cII λ. The λ phage with wild-type cII produce lysogens such that they are indistinguishable in the E. coli lawn, whereas λ phage that carry a mutation in cII are selected by forming plaques on the bacterial lawn when incubated at 24°C. We demonstrate many of the fundamental features of mutation analyses based on λ transgenic rodents are shared by the λ-based transgenic fish mutation assay. We rescued λ bacteriophage from fish genomic DNA with high efficiency, facilitating analyses of spontaneous and induced mutations in individual fish and various tissues. We show frequencies of spontaneous cII mutants are tissue-specific, comparable to the ranges seen in λ transgenic mice, and are of sufficiently low range to permit sensitive detection of induced mutations. We show cII mutants are induced in concentration-dependent, tissue-specific, and time-dependent relationships consistent with known mechanisms of mutagen action after treatment of fish with ethylnitrosourea (ENU). We also demonstrate that spontaneous and ENU-induced cII mutational spectra in fish are similar to λ transgenic rodents, providing further support to the use of the fish as comparative animal models for in vivo mutagenesis.
Materials and Methods
Animals.
Medaka were obtained from in-house populations originally derived from Gulf Coast Research Laboratory stocks (Ocean Springs, MS). Fish were maintained within recirculating dechlorinated freshwater culture systems maintained on artificial light (16 h light:8 h dark) at 20–22°C, or 26–27°C during breeding, using protocols approved by the Institutional Animal Care and Use Committee in accordance with the National Academy of Sciences Guide for the Care and Use of Laboratory Animals. The transgene consisted of a ≈45.5-kb λLIZ bacteriophage vector (Stratagene), which contains both cII and lacI mutation target genes flanked by cos sites to allow for excision and packaging (11). To enhance genomic integration of multiple copies of the vector and possibly increase the numbers of phage recovered per genome (12), λLIZ DNA was linearized, cos-ligated to form linear concatamers (T4 ligase, GIBCO/BRL, 4 min, 10°C), and filter-dialyzed over buffer before use (5 mM Tris/0.1 mM EDTA, 40 min, 0.025-μm filter pore size, Millipore). Zygotes collected within 15 min after fertilization were microinjected via the cytoplasm with the concatenated λLIZ DNA (50–100 ng/μl) and incubated (26°C) until hatching (≈10 days) by using procedures described with minor modifications (13).
λ Transgene Integration.
Genomic DNA was isolated from excised caudal fins (4–6 weeks old) by digestion (0.3 ml 1 × SSC/0.5% SDS/10 mg/ml proteinase K, GIBCO/BRL) for 1–3 h at 55°C and extraction twice with methylene chloride/isoamyl alcohol (24:1) containing NaCl (0.15 M). DNA was precipitated with 2 vol ethanol and resuspended in TE (10 mM Tris/1 mM EDTA, pH 7.5). The presence of λ sequences in fish was tested with PCR using primers specific for the λ transgene, 5′-GATGAGTTCGTGTCCGTACAACTGG-3′, and 5′-GGTTATCGAAATCAGCCACAGCGCC-3′, to generate a 500-bp product. Fish showing positive for λ sequences in fin tissue were mated with nontransgenic fish, and their offspring (>20 offspring/founder) were analyzed by PCR as described above. Numbers of integrated λ copies in germ-line transmitting lineages were estimated by quantitative PCR using the ABI 7700 Sequence Detection System and methods recommended by the manufacturer with minor modifications (Applied Biosystems). A standard curve was prepared by using 6–8 replicate serial dilutions of a ≈1 λ copy DNA standard. A minimum of four replicate 100-ng DNA samples from fish of each lineage (F1 and F2 generation) were amplified by using primers specific to the lacI transgene, 5′-ATGCGCCCATCTACACCAA-3′ and 5′-GGATTCTCCGTGGGAACAAA-3′, which generated a 70-bp product. An oligonucleotide, 5′-AACCTATCCCATTACGGTCAATCCGCC-3′, that annealed within the amplified product served as a probe. FAM (6-carboxy-fluorescein) was used as the reporter dye linked 5′ on the oligonucleotide, and TAMRA (6-carboxy-tetramethylrhodamine) was the quencher dye attached on the 3′ terminus (Applied Biosystems). The number of λ copies in each lineage was estimated by using supplied software.
λcII Mutagenesis Assay.
Genomic DNA was obtained by using procedures to optimize isolation of high molecular weight DNA required for in vitro packaging. Whole fish were disaggregated with a dounce homogenizer before proteinase digestion (1 × SSC/1% SDS/0.6 mg/ml proteinase K) at 37°C for 1½-4 h. Liver and testes were prepared similarly without disaggregation. Samples were extracted twice with equal volume phenol/chloroform by using wide-bore pipette tips to minimize DNA shearing. Potassium acetate was added to a final concentration of 1 M followed by a final extraction with equal volume chloroform. DNA was ethanol-precipitated, removed with a flame-sealed glass pipette, and resuspended in Tris-EDTA. The cII mutagenesis assay was conducted by using the λ Select-cII Mutation System kit according to the manufacturer's protocols with minor modifications (Stratagene). The assay is a positive mutant-screening method based on the ability of λ bacteriophage to multiply through either the lytic or lysogenic cycle in E. coli G1250 host strain, which carries mutant hfl− genes that increase the stability of the cII protein facilitating a lysogenic response (14). The λ vector was recovered from fish genomic DNA (≈10–20 μg) by incubation with Transpack packaging extracts (30°C, 3 h), which simultaneously excised and packaged the vector as viable phage particles. To select cII mutants, the individually packaged phage were mixed with G1250 cells and TB1 top agar and plated on 10 TB1 plates at 24°C (±0.5°C) for 40 h. λ Phage with wild-type cII underwent lysogenization and were indistinguishable in the bacterial lawn, whereas λ phage with detectable mutations in the cII gene multiplied through the lytic cycle, forming plaques. To determine the total number of packaged phage, a subsample of the packaged phage solution was infected in G1250 cells, mixed with top agar, and incubated on three TB1 titer plates at 37°C overnight. Mutant frequencies were calculated by dividing the total number of cII mutant plaque-forming units (pfu) on selective mutant screening plates by the estimated total λ+ and cII phage on the titer plates. A minimum of two samples/treatment were packaged and plated simultaneously in a blocked design (15). To verify mutant λ cII phenotypes, plaques (30–100% total plaques) were cored, incubated in SM buffer (0.1 M NaCl/0.01 M MgSO4/0.05 M Tris⋅HCl, pH 7.5/0.01% gelatin) at room temperature for 1 h, and replated at low density under selective conditions. The minimum number of animals required to detect a significant induction of mutations was estimated by using a spontaneous mutant frequency of 3 × 10−5, recovery of >300,000 pfu per animal, a power of 0.80, and α = 0.05 (16, 17). Comparisons of mutation frequencies were tested for significance by using the generalized Cochran–Armitage test.
Mutagen Exposure.
Fish (6–10 hemizygous λ310 fish/treatment, 2–6 months old) were immersed 1 h in water (pH 7.0) containing the direct-acting mutagen ethylnitrosourea (Sigma) at 0, 60, or 120 mg/liter (ppm). The fish were rinsed, transferred to clean water, and held for 5, 15, 20, or 30 days until euthanized with an overdose of tricane methylsulfonate (MS222, Argent, Redmond, VA). Whole fish, or dissected tissues (liver, testes), were frozen in liquid nitrogen and stored at −80°C until processed for DNA isolation. Fish treated similarly without mutagen exposure served as controls.
cII Sequence Analysis.
cII mutant plaques were cored at random and purified individually on G1250 E. coli cells. A 446-bp λ DNA fragment, including the entire 294-bp cII gene, was sequenced after PCR amplification using the phage lysate with the primers: 5′-AAAAAGGGCATCAAATTAAACC-3′ and 5′-CCGAAGTTGAGTATTTTTGCTGT-3′. The product was labeled with a BigDye Terminator Cycling DNA Sequencing Kit, and sequences were analyzed with an ABI PRISM 310 Genetic Analyzer (Applied Biosystems).
Results
Transgenic Fish.
To identify fish containing λ sequences, DNA isolated from excised fin tissue of 401 presumptive founders was analyzed by PCR. Sixty-two fish that were positive for the λ sequence (16%) were mated with nontransgenic fish, and germ-line transmission was confirmed in progeny from nine founders (15%). Frequencies of transgene transmission ranged from 5% to 47%, indicating mosaic integration of transgenes commonly observed in founders (9). Mendelian inheritance of the transgene in offspring from subsequent sibling crosses (>3 generations) supports the conclusion that the λ vector was integrated in a single chromosomal site in each transgenic lineage.
Recovery of λ from Fish.
To test whether the λ bacteriophage vector could be efficiently recovered from fish, genomic DNA from six lineages was mixed with in vitro packaging extracts and plated on hfl− bacteria. Approximately 5,000 pfu per packaging reaction were obtained from lineage λ203, whereas 300,000 to 2,500,000 pfu were recovered from lineage λ310, and no λ phage were recovered from the remaining lineages. In subsequent recovery trials, an average of 2,281,945 pfu per packaging reaction (≈60,000–70,000 pfu/μg) was obtained from lineage λ310 whole fish, 831,667 pfu from liver, and 1,701,667 pfu from testes (Table 1). To determine whether vector recovery was related to the number of genomically integrated λ copies, the copy number in each lineage was estimated by quantitative PCR. Copy numbers ranged from ≈74 λ copies in lineage λ310, five copies in λ203, to less than four λ copies in the remaining lineages. Similar results showing enhanced integration of multiple vector copies and improved phage recovery associated with the use of ligated DNA constructs have been reported in rodents (12) and fish (13). The exceptionally high numbers of pfu recovered from λ310 fish indicated this lineage would satisfy the minimum 100,000 pfu/packaging reaction recommended in transgenic rodent assays to obtain adequate statistical power (17).
Table 1.
Spontaneous mutant frequencies in whole fish, liver, and testes for lineage λ310
| Tissue | No. fish | Total mutants | Total pfu | Mean pfu | Mean mutants × 10−5pfu ± SEM |
|---|---|---|---|---|---|
| Whole fish | 6 | 395 | 13,691,667 | 2,281,945 | 2.9 (0.3) |
| Testes | 5 | 139 | 8,508,333 | 1,701,667 | 1.8 (0.3) |
| Liver | 11 | 405 | 9,148,333 | 831,667 | 4.3 (0.6) |
Spontaneous and ENU-Induced cII Mutant Frequencies.
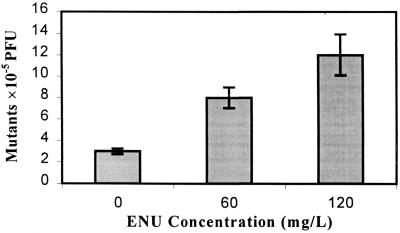
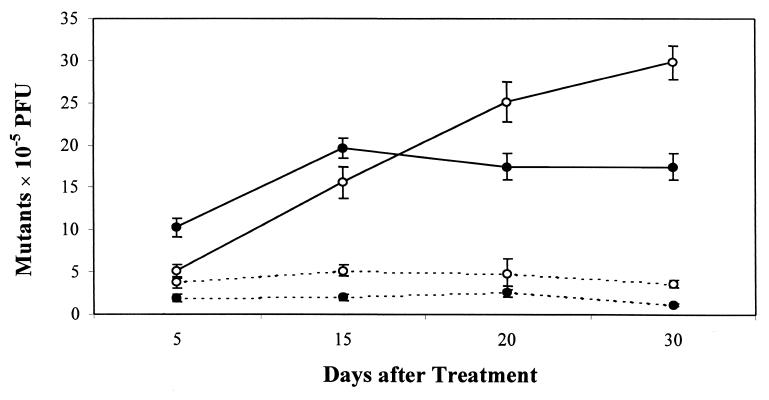
To test the feasibility of cII for detecting fish-derived in vivo mutations, we determined the frequencies of spontaneous cII mutants in λ310 whole fish, liver, and testes. The lowest frequencies of spontaneous mutants were 1.8 ± 0.3 × 10−5 in testes, followed by 2.9 ± 0.3 × 10−5 in whole fish, and 4.3 ± 0.6 × 10−5 in liver (Table 1). Mutant frequencies among the tissues all were significantly different (P < 0.03). The observed mutant frequencies were sufficiently low, comparable to ranges in λ transgenic rodents (18, 19), indicating sensitive detection of induced mutations in the fish was feasible. Statistical analyses revealed that as few as 6–7 animals were required to detect a 50% induction above background (3 × 10−5), and 2–3 animals were required to detect a 100% induction (power = 0.80, α = 0.05). A total of 5–10 animals/treatment are recommended in transgenic rodent assays to give similar power of detection (16, 17). To determine whether the cII gene in the fish was responsive to chemical mutagen exposure, we measured mutant frequencies in fish 15 days after exposure to 0, 60, or 120 mg/liter ENU. cII mutant frequencies in whole fish were induced significantly, 2.7-fold (8.0 ± 1.0 × 10−5, P = 0.01) and 4-fold (12.0 ± 1.9 × 10−5, P < 0.01) over untreated fish (3.0 ± 0.3 × 10−5) at 60 and 120 mg/liter ENU, respectively (Fig. 1). To test whether the time between mutagen exposure and analysis influenced mutant frequencies, the frequencies of cII mutants were determined in liver and testes from fish held 5, 15, 20, or 30 days after exposure (0 or 120 mg/liter ENU). Mutant frequencies in liver from treated fish did not increase significantly above background (4.4 ± 0.7 × 10−5) at 5 days (5.1 ± 0.8 × 10−5, P = 0.23), but increased significantly 3.5-fold at 15 days (15.6 ± 1.8 × 10−5, P < 0.01), 5.7-fold at 20 days (25.1 ± 2.3 × 10−5), and 6.7-fold at 30 days (29.8 ± 2.0 × 10−5) after mutagen treatment (Fig. 2). There was no significant difference between mutant frequencies at days 20 and 30 (P = 0.16). In contrast, mutant frequencies in testes were elevated 5.2-fold over the mean background mutant frequency (2.0 ± 0.6 × 10−5) at 5 days (10.3 ± 1.1 × 10−5), and reached a peak 10-fold induction at 15 days (19.6 ± 1.2 × 10−5), followed by an insignificant decline to 8.8-fold above background at 20 and 30 days after ENU treatment (17.5 ± 1.6 × 10−5, P = 0.30).
Figure 1.
Mutant frequencies (± standard error) in whole fish exposed 1 h to 0, 60, or 120 mg/liter ENU and held 15 days before analysis. cII mutants were induced 3- and 4-fold above the mutant frequency in untreated fish at each concentration.
Figure 2.
Mutant frequencies (± standard error) in liver and testes from fish analyzed 5, 15, 20, or 30 days after 1-h exposure to 0 or 120 mg/liter ENU. Mutant frequencies in liver increased insignificantly over mean background (4.4 ± 0.7 × 10−5) at 5 days (5.1 ± 0.8 × 10−5), 4-fold at 15 days (15.6 ± 1.8 × 10−5), 6-fold at 20 days (25.1 ± 2.3 × 10−5), and 7-fold at 30 days (29.8 ± 2.0 × 10−5). Mutant frequencies in testes increased 5-fold over mean background (2.0 ± 0.6 × 10−5) at 5 days (10.3 ± 1.1 × 10−5), 10-fold at 15 days (19.6 ± 1.2 × 10−5), and 9-fold at 20 and 30 days (17.5 ± 1.6 × 10−5). –○–, ENU-exposed liver; –●–, ENU-exposed testes; - -○- - control liver; - -●- -, control testes.
cII Mutational Spectra.
To determine whether spontaneous and induced cII mutants in the fish could be characterized, and to determine to what extent mutational spectra differed from that of rodent models, we performed sequence analyses on mutant phage recovered from untreated and ENU-treated fish. Spontaneous cII plaques (89 plaques) from whole fish, liver, and testes were combined for analyses after relatively minor differences were observed among the tissues (Table 2). Five mutations found outside the cII protein-coding region, and 10 duplicate mutations suggestive of clonal origin within a single animal, were excluded from further analysis. Of the remaining 74 independent mutations, one sequence contained a double substitution: G→C (nucleotide 125) and G→T (nucleotide 126). Single base substitutions were the most numerous mutations (74%), with transversions (42%) more frequent than transitions (32%). Frequencies of G:C→T:A transversions and G:C→A:T transitions were equal (20%), with 47% occurring at CpG sites. The predominance of mutations at CpG sites in transgenes is observed also in rodent models (10, 20) and has been attributed to cytosine methylation at CpG sites, eventually leading to G:C→A:T transitions during replication (17, 21–23). Frameshifts accounted for 24% of the mutations with the majority (89%) being either insertions or deletions within the homonucleotide run of guanosines (sense strand nucleotides 179–184), a known mutation hotspot (20, 24). The majority of plaques (36 of 39 mutations) from liver of ENU-treated fish were independent mutations within the cII coding region, including one sequence with a 3-nt deletion (AAC, nucleotides 55–57). Single base substitutions were the most frequent mutations (94%), with transitions (53%) somewhat more numerous than transversions (42%). However, the proportion of mutations at A:T base pairs was elevated from 31% in untreated fish to 59% in treated liver, which is highly characteristic of ENU exposure (20, 25). Also indicative of ENU exposure, the percentage of frameshift mutations was lower in the induced spectra (3%) compared with that of the spontaneous spectrum (24%), reflecting the low frequency of insertions or deletions at the homonucleotide string of guanosines at nucleotides 179–184 (sense strand).
Table 2.
Spontaneous and ENU-induced mutational spectra
| Spontaneous | ENU-exposed liver | |
|---|---|---|
| Total mutations | 89 | 39 |
| Mutations outside cII | 5 | 3 |
| Independent mutations | 74 | 36 |
| % (n) | % (n) | |
| Transitions | ||
| G:C→A:T | 20 (15) | 28 (10) |
| % CpG | 47 (7) | 30 (3) |
| A:T→G:C | 12 (9) | 25 (9) |
| Transversions | ||
| G:C→T:A | 20 (15) | 6 (2) |
| G:C→C:G | 11 (8) | 6 (2) |
| A:T→T:A | 4 (3) | 28 (10) |
| A:T→C:G | 7 (5) | 3 (1) |
| Frameshift | ||
| (+) | 14 (10) | 3 (1) |
| (−) | 11 (8) | 0 |
| Other | 1 (1) | 3 (1) |
Discussion
We generated transgenic fish that carry multiple copies of a λ bacteriophage vector and used a positive-selection mutation assay based on the cII gene as the mutable target to detect spontaneous and ENU-induced in vivo mutations. Our results show many fundamental features of mutation analyses based on λ transgenic rodents are shared by the λ transgenic fish, providing support to the use of fish for assessing health risks associated with chemical exposure in aquatic environments and for comparative animal models for in vivo mutagenesis studies.
λ-based mutation assays rely on in vitro packaging extracts to excise simultaneously the intact vector DNA from transgenic animal genomic DNA and package the vector into viable phage particles to facilitate subsequent infection of bacterial host cells. The efficiency with which the vector can be recovered is therefore a fundamental practical requirement of a transgenic mutation assay. Low vector recovery decreases the utility of the assay by increasing the numbers of animals needed to meet statistical requirements, as well as increasing costs of reagents and labor. The unprecedented recovery of high numbers of λ plaques from lineage λ310 fish provided distinct practical benefits for conducting in vivo mutation analyses, particularly considering the small size of medaka (≈2–4 cm adult length). Comparably high pfu numbers obtained from whole adult and juvenile fish and from liver and testes precluded multiple platings or pooling of samples. To enhance genomic integration of multiple copies in the fish, we ligated linear λ vector at the cohesive termini to form linear concatamers before injection. Apparently cos ligation improves the number of rescued phage per genome by protecting the integrity of intervening cohesive termini, which are important in phage assembly (12). Our analyses confirmed an association between efficiency of vector recovery and the number of λ copies carried by the lineages, consistent with λ transgenic rodents (12). In comparison to lineage λ310, which carries ≈74 λ copies, and yielded greater than 300,000 pfu/packaging reaction (≈60,000-70,000 pfu/μg DNA), typical recoveries from Big Blue mice carrying 30–40 λ copies are ≈10,000–20,000 pfu/ug DNA (11, 12, 18, 26, 27). The smaller genome size of medaka (2.2 pg/diploid genome) (28) as compared with the mouse (≈6 pg/diploid genome) (29) also contributed to the relatively high efficiency in vector recovery from the fish.
Determination of the spontaneous mutant frequencies was essential to establishing the basis for comparing mutations induced after mutagen exposure, as well as for comparing mutagenesis in the identical transgene target in rodent models. Spontaneous mutant frequencies varied significantly among tissues with the lowest frequency observed in testes, followed by whole fish and liver. The observed lower mutant frequency in testes compared with somatic tissues is consistent with rodent studies suggesting that fish may provide an excellent model for comparative studies of the mechanisms in germ-line mutagenesis (30–32). Spontaneous mutant frequencies in the fish were lower, but comparable, to ranges of cII mutant frequencies in spleen (5.7 × 10−5), and liver (13.8 × 10−5) from Big Blue mice (18, 19). The low spontaneous mutant frequencies indicated de novo disruption of the target gene, which would possibly obscure detection of induced responses, did not occur during genomic integration of the transgene. An example of this phenomenon was observed in a Big Blue transgenic rat lineage that carried ≈200 λ copies and had a heritable mutation at a frequency of approximately 10−2, which reduced the practical application of the lineage (12). Sensitivity of a mutation assay is defined by the magnitude of the induced mutations compared with the background mutation frequencies (33), indicating that fish have at least equivalent, if not somewhat greater, sensitivity compared with rodents in detecting induced mutations.
The cII target in the fish was shown to be highly responsive after mutagen treatment, reflecting mutation induction consistent with tissue specificity, mutation manifestation time, mutational spectra, and known modes of ENU action. cII mutants were induced 3- and 4-fold above background after exposure to 60 and 120 mg/liter ENU. In comparison, 3-fold induction of cII mutants (from 13.8 to 36.8 × 10−5) was reported in mice after i.p. injection of 100 mg/kg ENU (18). Different frequencies of mutations observed in testes, liver, and whole fish at equivalent mutagen treatments underscore the utility of the assay for examining tissue-specific mutagenesis. Our results reveal the interval between mutagen treatment and analyses, termed mutation manifestation time (34–36), influenced the frequency of mutations and were specific for individual tissues. cII mutant frequencies in liver were not significantly elevated above background at 5 days but increased significantly at 15 and 30 days after ENU exposure. In contrast, mutant frequencies in testes increased 5-fold above background at 5 days, reached a peak 10-fold induction by day 15, followed by a slight, but not significant, decline 20 and 30 days after exposure. The relatively higher magnitude of mutation induction and shorter mutation manifestation time observed in testes compared with liver may reflect differences in cell proliferation rates and ENU action in these tissues. ENU is well-characterized as a direct acting alkylating agent and potent germ-cell mutagen, which produces O6-ethylguanine, O4-ethylthymidine, and O2-ethylthymidine in DNA, promoting G:C→AT and A:T→G:C transitions, as well as A:T→T:A transversions (37, 38). It is understood that DNA replication is necessary for DNA damage to become fixed as a mutation, and that this time is affected by several variables, including tissue/cell type, mutagen, and mutagen treatment regimen.‡ In rodent tissues with rapid cell turnover, such as bone marrow (40, 41) and intestinal epithelium (42), sampling times as short as 7 days, roughly coinciding with stem cell turnover in these tissues, may be sufficient to detect a significant induction of mutations. Tissues with slower cell turnover, such as liver, may require a manifestation time greater than 35 days (43). As a consequence, a sampling time of 35 days has been recommended as suitable for several rodent tissues (33). Our results indicate that 15 days may be sufficient time to detect a significant 2-fold induction in most fish tissues, although >30 days may be required to detect induction by weak mutagens, and 5 days is probably suitable only for the most potent mutagens. These results also demonstrate that detection of induced mutations may be obscured in applications that rely solely on mutation analyses using DNA isolated from whole fish rather than specific tissues.
Sequence analyses revealed spontaneous and induced cII mutational spectra in fish and rodent models were quite similar. As observed in rodent models, single base substitutions comprised the majority of spontaneous and ENU-induced mutations in fish, with a large percentage of the G:C→A:T mutations at CpG sites. The proportion of mutations at A:T base pairs increased from 31% in untreated fish to 59% in treated livers, with the bulk of the increase being A:T→T:A transversions, consistent with the greater mutagenic effect of ENU at A:T base pairs (20, 25). Frameshifts in the spontaneous mutation spectra accounted for 24% of the mutations, with the majority being either insertions or deletions within a known cII hotspot (20). The proportion of frameshifts coincides most closely with 27% observed in Big Blue Rat2 cell lines (20), compared with 11% in mice (24). The percentage of frameshift mutations decreased in the ENU-exposed fish, which is also consistent with studies showing ENU does not induce high numbers of frameshift mutations (31, 44).
Our results show that λ transgenic fish have many benefits and limitations in common with the transgenic rodent mutation models. Results from fish and rodent studies, however, collectively support the use of λ-based transgenic animals in mutation analyses. A distinct benefit of transgenic animal mutation assays is the ability to compare mutational responses of identical mutation target loci in different cells, tissues, organs, and species. We selected medaka for this application because it is widely used in environmental toxicology and is the fish species of choice in carcinogenesis bioassays (45) and germ-cell mutagenesis studies (46). Small size, sensitivity, well-characterized histopathology, short generation time, and cost-effective husbandry contribute to the utility of this species in routine testing of a wide variety of compounds (8). The development of transgenic fish models for mutation analyses should enhance the value of medaka as an important comparative animal model. The relative ease of performing the cII assay facilitates efficient screening of large numbers of spontaneous and induced mutations in virtually any tissue from which DNA may be isolated. Because of high efficiency of vector recovery and low variability in mutant frequencies among animals, as few as six fish per treatment are required to detect significant induction of mutations. The ease of sequencing the cII locus may be particularly useful in examining whether small increases in mutant frequencies after exposure to chemicals at low environmental concentrations may be accompanied by shifts in mutational spectra. In addition to detection of mutations at the cII locus, mutations can be detected in the widely used lacI gene, which also is contained within the λLIZ vector. Using identical in vitro packaging procedures, mutant plaques are scored by blue-white screening on a Δlac E. coli lawn on agar plates containing 5-bromo-4-chloro-3-indolyl β-d-galactoside (11). Preliminary analyses indicate that lacI and cII respond similarly in the λ transgenic medaka with regards to fold increases in mutant frequencies above background and in mutational spectra (unpublished data). Two additional transgenic fish mutation assays have been introduced: the mummichog (Fundulus heteroclitus) based on the bacteriophage φX174 vector (13) and zebrafish carrying the plasmid (pML4) containing the rspL gene as the mutational target gene (47). Although limited use of these systems does not permit extensive evaluation of their relative utility, it is apparent these approaches also have merit.
A fundamental assumption of transgenic mutation assays is that the mutation target sequence in the animal accurately represents the mutagenic response of endogenous DNA. It is understood, however, that transgenes and endogenous loci differ in several features that can affect mutational response. Compared with most mammalian genes, and probably fish genes as well, bacterial transgenes have higher GC content, higher density of dinucleotide CpG and associated 5-methylcytosine, are highly methylated, and lack transcription-coupled repair (33). An advantage of using transgenes that are genetically neutral is that, unlike assays involving endogenous genes that are limited to very specific tissues or developmental stages, mutations in transgene targets persist and accumulate without being subjected to selection in the animal, thereby facilitating mutation analyses in virtually any tissue or developmental stage (40, 48, 49). As a consequence, accumulation of mutations over time in neutral transgenes indicates that repeated or chronic mutagen treatments should increase the sensitivity of the assay (39). The ease with which fish can be applied to a wide range of treatment regimens makes them ideally suited for such evaluations. Transgene mutation targets have mutational spectra that may or may not reflect those of endogenous genes. Mutations detected in the λ-based systems consist primarily of base pair substitutions with a few frameshifts and small insertions/deletions, but are limited in their ability to detect large rearrangements. This may not be a significant limitation as the relative proportion of these mutations in transgenic rodents has been shown to be similar to the endogenous hprt gene (19). Compared with some endogenous loci, spontaneous mutant frequencies in transgenes are higher, suggesting bacterial mutational target genes may not measure significant induction by weak mutagens (4). In light of the extreme paucity of endogenous genes, particularly in fish, that are readily amenable to mutation quantitation, it can be argued that if the differences between transgene loci and endogenous genes are considered when interpreting mutation analyses, transgenic mutation assays have significant utility (33). Although the challenge remains to identify to what extent such factors as metabolism, DNA repair, and cell proliferation play a role in mutagenesis in fish, and how these factors compare in rodent models, the availability of transgenic models makes such pursuits feasible.
Acknowledgments
This work was supported by Grant R24RR11733 from the National Institutes of Health National Center for Research Resources and Grant RR251139 from the Georgia Advanced Technology Development Center. Equipment support was provided by Grant RR380030 from the Georgia Research Alliance.
Abbreviations
- pfu
plaque-forming units
- ENU
ethylnitrosourea
Footnotes
Monroe, J. J., Kort, K. & Skopek, T. (1998). Environ. Mol. Mutagen. 31, Suppl. 29, 15 (abstr.).
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.220428097.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.220428097
Paashius-Lew, Y., Zhang, X. B. & Heddle, J. A. (1996) Environ. Mol. Mutagen. Suppl. 27, 53 (abstr.).
References
- 1.Mirsalis J C, Monforte J A, Winger R A. Annu Rev Pharmacol Toxicol. 1995;35:135–164. doi: 10.1146/annurev.pa.35.040195.001045. [DOI] [PubMed] [Google Scholar]
- 2.Summer W C, Glazer P M, Malkevich D. Mutat Res. 1989;220:263–268. doi: 10.1016/0165-1110(89)90030-4. [DOI] [PubMed] [Google Scholar]
- 3.Gossen J A, De Leeuw W J, Molijin A C, Vigj J. BioTechniques. 1993;14:624–629. [PubMed] [Google Scholar]
- 4.Walker V E, Andrews J L, Upton P B, Skopek T R, De Boer J G, Walker D M, Shi X, Sussman H E, Gorelick N J. Environ Mol Mutagen. 2000;34:167–181. [PubMed] [Google Scholar]
- 5.Bailey G S, Hendricks J D, Dashwood R. Mutat Res. 1992;267:243–250. doi: 10.1016/0027-5107(92)90068-d. [DOI] [PubMed] [Google Scholar]
- 6.Powers D A. Science. 1989;246:352–358. doi: 10.1126/science.2678474. [DOI] [PubMed] [Google Scholar]
- 7.Postlelthwait J, Amores A, Force A, Yan Y L. Methods Cell Biol. 1999;60:149–163. [PubMed] [Google Scholar]
- 8.Hawkins W E, Walker W W, Overstreet R M. In: Fundamentals of Aquatic Toxicology: Effects, Environmental Fate, and Risk Assessment. Rand G M, editor. Bristol, VA: Taylor and Francis; 1995. pp. 421–446. [Google Scholar]
- 9.Maclean N. Mutat Res. 1998;399:255–266. doi: 10.1016/s0027-5107(97)00260-1. [DOI] [PubMed] [Google Scholar]
- 10.Jakubczak J L, Merlina G, French J E, Muller W J, Paul B, Adhya S, Garges S. Proc Natl Acad Sci USA. 1996;93:9073–9078. doi: 10.1073/pnas.93.17.9073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kohler S W, Provost G S, Kretz P L, Fieck A, Bullock W O, Sorge J A, Putman D L, Short J M. Environ Mol Mutagen. 1991;18:316–321. doi: 10.1002/em.2850180421. [DOI] [PubMed] [Google Scholar]
- 12.Dycaico M J, Provost G S, Kretz P L, Ransom S L, Moores J C, Short J M. Mutat Res. 1994;307:461–478. doi: 10.1016/0027-5107(94)90257-7. [DOI] [PubMed] [Google Scholar]
- 13.Winn R N, Van Beneden R J, Burkhart J G. Marine Environ Res. 1995;40:247–265. [Google Scholar]
- 14.Kihara A, Akiyama Y, Ito K. Proc Natl Acad Sci USA. 1997;94:5544–5549. doi: 10.1073/pnas.94.11.5544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gorelick N J, Thompson E D. Environ Mol Mutagen. 1994;23:12–16. doi: 10.1002/em.2850230104. [DOI] [PubMed] [Google Scholar]
- 16.Carr G J, Gorelick N J. Environ Mol Mutagen. 1995;25:246–255. doi: 10.1002/em.2850250311. [DOI] [PubMed] [Google Scholar]
- 17.Piergorsch W W, Margolin B H, Shelby N M, Johnson A, French J E, Tennant R W, Tindall K R. Environ Mol Mutagen. 1995;25:231–245. doi: 10.1002/em.2850250310. [DOI] [PubMed] [Google Scholar]
- 18.Zimmer D M, Harbach P R, Mattes W B, Aaron C S. Environ Mol Mutagen. 1999;33:249–256. [PubMed] [Google Scholar]
- 19.Monroe J J, Kort K L, Miller J E, Morino D R, Skopek T R. Mutat Res. 1998;421:121–136. doi: 10.1016/s0027-5107(98)00171-7. [DOI] [PubMed] [Google Scholar]
- 20.Watson D E, Cunningham M L, Tindall K R. Mutagenesis. 1998;13:487–497. doi: 10.1093/mutage/13.5.487. [DOI] [PubMed] [Google Scholar]
- 21.Provost G S, Kretz P L, Hammer R T, Matthews C D, Rogers B J, Lundberg K S, Dycaico M J, Short J M. Mutat Res. 1993;288:133–149. doi: 10.1016/0027-5107(93)90215-2. [DOI] [PubMed] [Google Scholar]
- 22.Hayward J J, Shane B S, Tindall K R, Cunningham M L. Carcinogenesis. 1995;16:2429–2433. doi: 10.1093/carcin/16.10.2429. [DOI] [PubMed] [Google Scholar]
- 23.Shane B S, Lockhart A-M C, Winston G W, Tindall K R. Mutat Res. 1997;377:1–11. doi: 10.1016/s0027-5107(97)00004-3. [DOI] [PubMed] [Google Scholar]
- 24.Harbach P R, Zimmer D M, Filipunas A L, Mattes W B, Aaron C S. Environ Mol Mutagen. 1999;33:132–143. doi: 10.1002/(sici)1098-2280(1999)33:2<132::aid-em5>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- 25.Walker V E, Gorelick N J, Andrews J L, Craft T R, De Boer J G, Glickman B W, Skopek T R. Cancer Res. 1996;56:4654–4661. [PubMed] [Google Scholar]
- 26.Gollapudi B B, Jackson K M, Stott W T. Mutat Res. 1998;419:131–135. doi: 10.1016/s1383-5718(98)00132-6. [DOI] [PubMed] [Google Scholar]
- 27.Zimmer D M, Harbach P R, Mattes W B, Aaron C S. Environ Mol Mutagen. 1998;32:325–330. [PubMed] [Google Scholar]
- 28.Lamatsch D K, Steinlein C, Schmid M, Schartl M. Cytometry. 2000;39:91–95. doi: 10.1002/(sici)1097-0320(20000201)39:2<91::aid-cyto1>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- 29.Hogan B, Beddington R, Costantini F, Lacy E. Manipulating the Mouse Embryo: A Laboratory Manual. Plainview, NY: Cold Harbor Lab. Press; 1994. [Google Scholar]
- 30.Walter C A, Intano G W, McCarrey J R, McMahan C A, Walter R B. Proc Natl Acad Sci USA. 1998;95:10015–10019. doi: 10.1073/pnas.95.17.10015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kohler S W, Provost G S, Fieck A, Kretz P L, Bullock W O, Sorge J A, Putman D L, Short J M. Proc Natl Acad Sci USA. 1991;88:7958–7962. doi: 10.1073/pnas.88.18.7958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Katoh M, Inomata T, Horiya N, Suzuki F, Shida T, Ishioka K, Shibuya T. Mutat Res. 1994;341:17–28. doi: 10.1016/0165-1218(94)90020-5. [DOI] [PubMed] [Google Scholar]
- 33.Heddle J A, Dean S, Nohmi T, Boerrigter M, Casciano D, Douglas G R, Glickman B W, Gorelick N J, Mirsalis J C, Martus H, et al. Environ Mol Mutagen. 2000;35:253–259. doi: 10.1002/(sici)1098-2280(2000)35:3<253::aid-em11>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- 34.Walker V E, Jones I M, Crippen T L, Meng Q, Walker D M, Bauer M J, Reilly A A, Tates A D, Nakamura J, Upton P B, Skopek T R. Mutat Res. 1999;431:371–388. doi: 10.1016/s0027-5107(99)00180-3. [DOI] [PubMed] [Google Scholar]
- 35.Hara T, Sui H, Kawakami K, Shimada Y, Shibuya T. Environ Mol Mutagen. 1999;34:121–123. [PubMed] [Google Scholar]
- 36.Sun B, Shima N, Heddle J A. Mutat Res. 1999;427:11–19. doi: 10.1016/S0027-5107(99)00039-1. [DOI] [PubMed] [Google Scholar]
- 37.Shibuya T, Morimoto K. Mutat Res. 1993;297:3–38. doi: 10.1016/0165-1110(93)90005-8. [DOI] [PubMed] [Google Scholar]
- 38.Shelby M D, Tindall K R. Mutat Res. 1997;388:99–109. doi: 10.1016/s1383-5718(96)00106-4. [DOI] [PubMed] [Google Scholar]
- 39.Heddle J A, Shaver-Walker P, Tao K S, Zhang X B. Mutagenesis. 1995;10:467–470. doi: 10.1093/mutage/10.6.467. [DOI] [PubMed] [Google Scholar]
- 40.Tao K S, Urlando C, Heddle J A. Proc Natl Acad Sci USA. 1993;90:10681–10685. doi: 10.1073/pnas.90.22.10681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang X B, Urlando C, Tao K S, Heddle J A. Mutat Res. 1995;338:189–201. doi: 10.1016/0921-8734(95)00024-z. [DOI] [PubMed] [Google Scholar]
- 42.Hoorn A J W, Custer L L, Myhr B C, Brusick D, Gossen J, Vijg J. Mutagenesis. 1993;8:7–10. doi: 10.1093/mutage/8.1.7. [DOI] [PubMed] [Google Scholar]
- 43.Mirsalis J C, Provost G S, Matthews C, Hammer R, Schindler J E, O'Laughlin K, McGregor J T, Short J M. Mutagenesis. 1993;8:265–271. doi: 10.1093/mutage/8.3.265. [DOI] [PubMed] [Google Scholar]
- 44.Skopek T R, Walker V E, Cochrane J E, Craft T R, Cariello N F. Proc Natl Acad Sci USA. 1992;89:7866–7870. doi: 10.1073/pnas.89.17.7866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bunton T E. In: Data on Genetic Effects in Carcinogenic Hazard Evaluation. McGregor D B, Rice J M, Venitt S, editors. Vol. 146. Lyon, France: IARC Scientific; 1999. pp. 151–184. [Google Scholar]
- 46.Shima A, Shimada A. Environ Health Perspect. 1994;102, Suppl. 12:33–35. doi: 10.1289/ehp.94102s1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Amanuma K, Takeda H, Amanuma H, Aoki Y. Nat Biotechnol. 2000;18:62–65. doi: 10.1038/71938. [DOI] [PubMed] [Google Scholar]
- 48.Cosentino L, Heddle J A. Environ Mol Mutagen. 1996;28:313–316. doi: 10.1002/(SICI)1098-2280(1996)28:4<313::AID-EM3>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 49.Swiger R R, Cosentino L, Shima N, Bielas J H, Cruz-Munoz W, Heddle A. Environ Mol Mutagen. 1999;34:201–207. [PubMed] [Google Scholar]