Abstract
Drug combinations that include nucleoside reverse transcriptase inhibitors (NRTIs) are remarkably effective in preventing maternal-viral transmission of HIV during pregnancy. However, there may be potential long-term risks for children exposed in utero. Examination of the genotoxic and mutagenic effects of two NRTIs, zidovudine [AZT (3′-azido-3′-deoxythymidine)] and didanosine [ddI (2′,3′-dideoxyinosine)], in cultured human lymphoblastoid cells revealed multiplicative synergistic enhancement of AZT-DNA incorporation and mutant frequency induction in response to the combined drug exposure, as compared with single-drug exposures. Dose-related increases in DNA incorporation of AZT (as measured by a competitive RIA) and mutagenicity at the HPRT and TK loci (as assessed by cell-cloning assays) were observed in cells exposed in culture to AZT, or equimolar combinations of AZT + ddI, at exposure concentrations ranging from 3 to 30 times the maximum plasma levels found in humans. Because mutagenesis is strongly associated with tumor induction in experimental models, children exposed transplacentally to combinations of NRTIs may be at risk for cancer development later in life.
Combinations of antiretroviral drugs have been found to be superior to single agent regimens for effective and durable treatment of HIV-1 infections and for reduction of transmission of HIV from infected pregnant women to their infants (1). Generally, combination antiretroviral therapies consist of one or more nucleoside reverse transcriptase inhibitors (NRTIs), protease inhibitors, and/or non-nucleoside reverse transcriptase inhibitors. Combination treatments should ideally achieve additive or synergistic therapeutic effects without causing serious toxicity. However, researchers in France have hypothesized a link between mitochondrial dysfunction and NRTI exposure of eight HIV-negative children, two of whom died at approximately one year of age (2). Genotoxic events that include incorporation of the NRTI drug into DNA and drug-induced mutagenesis have been shown to accompany transplacental exposure to AZT [zidovudine (3′-azido-2′,3′-deoxythymidine)] in primate models, and are associated with tumor induction in rodents. These events could therefore be biomarkers of cancer risk in children receiving antiretroviral prophylaxis including NRTIs in utero.
AZT and ddI [didanosine (2′,3′-dideoxyinosine)], the first two NRTIs approved by the Food and Drug Administration for AIDS treatment, have different profiles of side effects, and were thus considered favorably as combination drugs to inhibit viral replication and to prevent perinatal transmission of HIV (3–5). In mutagenicity tests, AZT has produced primarily clastogenic effects in human cells in culture and in mice exposed at both high and clinically relevant concentrations, whereas ddI has yielded positive clastogenic and mutagenic responses only at elevated doses (1). The clastogenic effects of these NRTIs are consistent with their mode of action as DNA chain terminators; however, the mutational spectra data for AZT also suggest that this drug produces point mutations either directly or indirectly by a mechanism(s) not yet established (1). AZT caused vaginal tumors in mice and rats exposed as adults (6), and it induced liver, lung, and female reproductive tumors in mice exposed in utero (7), whereas there are inadequate data in the literature concerning the carcinogenicity of ddI in experimental animals (1). Additionally, all of the currently available NRTIs are positive in one or more in vitro carcinogenicity screening tests (1). In the case of AZT, the mutagenic consequences of exposure in experimental systems and the carcinogenic effects of this drug in rodents are thought to arise primarily as a result of AZT incorporation into cellular DNA and the concomitant termination of DNA replication (1). There is overwhelming evidence that AZT is incorporated into nuclear and mitochondrial DNA in cultured mammalian cells, in animal models, and most importantly in human adults and newborn infants (7–13). However, DNA incorporation measurements and mutagenicity studies of AZT-treated mammalian cells typically have not been conducted in the same samples. A direct comparison between AZT-DNA incorporation and AZT-induced mutagenic effects has therefore been precluded (1).
Our research groups have evaluated the relationships between AZT exposure, AZT incorporation into genomic DNA, and mutagenic response in human B lymphoblastoid cells exposed to AZT in culture. Target sites for mutagenesis included the X-linked hypoxanthine-guanine phosphoribosyltransferase (HPRT) gene (14) and the autosomal thymidine kinase (TK) (15) and adenine phosphoribosyltransferase (APRT) genes (16). In both exposure duration and dose–response experiments, there were significant correlations between AZT-DNA incorporation and AZT-induced mutagenicity at all three loci in the same AZT-exposed cells. Molecular analyses of HPRT, TK, and APRT mutant colonies demonstrated that the majority of AZT-induced mutations were accounted for by large deletions or loss of heterozygosity, consistent with the known mechanism of AZT as a DNA chain terminator (1).
Because some antiretroviral therapies include two NRTIs, the current study was designed to evaluate the genotoxicity and mutagenicity of combined drug exposures as compared with single nucleoside analogue exposures in cultured human cells. Theoretically, a combination of two NRTIs for two differing bases could have at least additive mutagenic effects in human cells. This possibility was addressed by investigating AZT, a nucleoside analogue of thymidine, and ddI, a nucleoside analogue of inosine (which is itself an analogue of adenosine). Here we describe the effects of exposure concentration of each agent alone versus equimolar combinations on cell toxicity, DNA incorporation of AZT, and induction of mutations in the HPRT and TK genes of exposed human lymphoblastoid cells.
Materials and Methods
Cell Culture and Exposures.
TK6 B lymphoblastoid cells were grown in medium in T-flasks (n = 5 per group) as described elsewhere (14, 15). After 3 days of growth in 0, 33, 100, 300, or 900 μM AZT or ddI (dissolved in medium for exposures), cells were washed and plated at a density of 2 or 4 viable cells per well in 96-well U-bottom microtiter dishes in the presence of lethally irradiated feeder cells to measure the cloning efficiency. Ten days after plating, the relative cell survival was obtained by comparing the cloning efficiency in treated cells versus control cells.
Measurement of AZT Incorporation into Cellular DNA.
Genomic DNA was isolated from TK6 cells by using an Oncor nonorganic extraction procedure (Intergen, Purchase, NY), followed by RNase treatment of the extracted DNA. A competitive anti-AZT RIA (AZT-RIA) was used to measure incorporation of AZT into genomic DNA, as previously described (7, 14, 15). Levels of incorporation were expressed as molecules of AZT per 106 nucleotides.
Cell-Cloning Assay for HPRT and TK Mutant Frequencies.
The cell cloning method for determining HPRT mutant frequencies in lymphoblastoid cells has been previously described (14, 15). Briefly, after 3 days of exposure of TK6 cells in medium containing 0, 33, 100, and 300 μM AZT or ddI, or equimolar concentrations of each drug, cells (n = 5 flasks per group) were washed and subcultured in nonselective medium for 7 days to allow phenotypic expression of HPRT mutations. Then cells were plated in microtiter dishes in the absence and presence of selective agent (6-thioguanine) for determining cloning efficiencies and identifying HPRT mutants, respectively, 10 days after plating. HPRT mutant frequency was calculated as the ratio of mean cloning efficiency in selective medium to that in nonselective medium (14).
The method used for measuring HPRT mutant frequencies was used to determine TK mutant frequencies, except that the time allowed for phenotypic expression of TK mutations was 3 days, the selection agent was trifluorothymidine, and plated cells were scored at 28 days after plating (15).
Statistical Analyses.
Statistical significance of the differences in mutant frequency values between control and drug-treated groups were determined by using the Mann–Whitney U statistic. A P value ≤ 0.05 was considered significant.
Results
Effects of AZT, ddI, and AZT–ddI Exposures on Cell Survival.
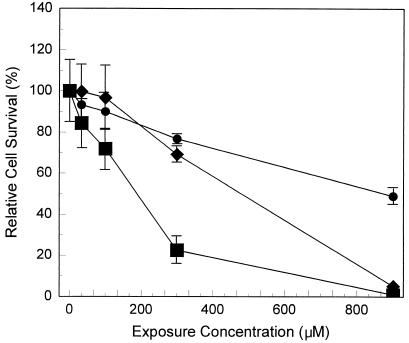
The potential cytotoxic effects of AZT, ddI, and equimolar AZT-ddI treatments were assessed by determining the relative survival of cultured human lymphoblastoid TK6 cells immediately after 3 days of exposure. The cytotoxicity data are shown in Fig. 1, where survival for the unexposed cells is designated as 100%. The 50% inhibitory concentration (IC50) for AZT was about 900 μM, indicating that the sensitivity of TK6 cells to AZT-induced cytotoxicity was similar to that for H9 immortalized human lymphocytes (17), but much less than that for human bone marrow cells (10). ddI was more cytotoxic than AZT in TK6 cells, as demonstrated by the lower IC50 for AZT (≈500 μM). Because very few cells survived exposure to 900 μM ddI, with or without AZT, HPRT and TK mutant frequencies were not measured at this treatment level. Otherwise, the resulting levels of cell survival indicate that there were sufficient viable cells after AZT or AZT-ddI exposure to permit meaningful measurements of HPRT and TK mutant frequencies.
Figure 1.
Effects of concentrations of AZT, ddI, or AZT + ddI on relative survival of TK6 cells. Cultures (n = 5 per group) were exposed to 0, 33, 100, 300, or 900 μM AZT, or ddI, alone or in equimolar combinations for 3 days. Relative cell survivals were ratios of cloning efficiencies in treated versus control samples determined immediately after exposure. ●, AZT; ♦, ddI; ■, AZT + ddI; points, averages; bars, standard errors.
Effects of AZT and AZT-ddI Exposures on DNA Incorporation of AZT.
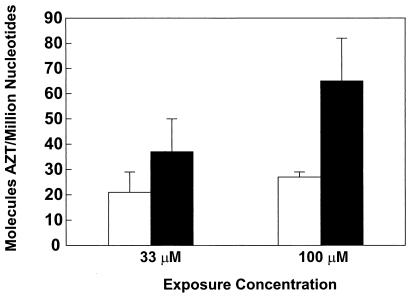
The relationships between exposure concentration and incorporation of AZT into genomic DNA of TK6 cells was assessed after exposure for 3 days to AZT alone, or to equimolar combinations of AZT and ddI (0, 33, 100, or 300 μM). DNA samples from each exposure concentration were coded and analyzed in a blind fashion using a competitive RIA for AZT. The DNA samples yielding the minimal inhibition (representing “zero” value) proved to be from unexposed cells. The levels of AZT incorporation into DNA of cells exposed to 33 μM AZT + 33 μM ddI (37 ± 13 molecules per 106 nucleotides, average ± SE) and 100 μM AZT + 100 μM ddI (65 ± 17 molecules per 106 nucleotides) were significantly increased by 1.8- and 2.4-fold, respectively, compared with those found in cells exposed to 33 μM AZT (21 ± 8 molecules per 106 nucleotides) or 100 μM AZT (27 ± 2 molecules per 106 nucleotides) alone (Fig. 2).
Figure 2.
AZT-DNA incorporation in TK6 cells. Cultures (n = 5 per group) were exposed 0, 33, or 100 μM AZT, alone or in equimolar combinations with ddI for 3 days. After exposure genomic DNA was extracted and assayed by AZT-RIA for incorporation into DNA. AZT, empty bar; AZT + ddI, black bar.
Effects of AZT, ddI, and AZT-ddI Exposures on HPRT and TK Mutant Frequencies.
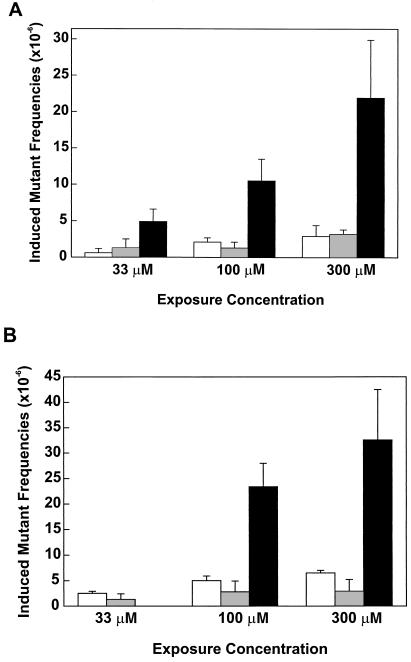
In parallel with the AZT-DNA incorporation studies, mutagenic responses in the HPRT and TK genes were evaluated in the same AZT or AZT-ddI exposed TK6 cells as well as in cells treated for 3 days with only ddI (0, 33, 100, or 300 μM). A cell cloning assay was used to determine HPRT mutant frequencies and the drug-induced mutant frequencies (i.e., the average mutant frequency in treated cells minus the average mutant frequency in controls) as a function of exposure concentration (shown in Fig. 3A). Single drug treatments caused significantly increased HPRT mutant frequencies at 300 μM AZT (P = 0.036) or ddI (P = 0.028), but not at lower exposure concentrations of AZT or ddI. An unexpected and striking finding was that equimolar combinations of AZT and ddI produced significantly elevated HPRT mutant frequencies at all exposure concentrations (P values ranging between 0.018 and 0.004), and that the induced HPRT mutant frequencies in combination treatments were about 3-fold greater than the additive induced mutant frequency values of single treatments with AZT or ddI at exposure concentrations of 33, 100, and 300 μM.
Figure 3.
Comparison of the mutagenic potencies of AZT, ddI, and AZT + ddI, at the HPRT (A) and TK (B) loci in TK6 cells. Cultures (n = 5 per group) were exposed to 0, 33, 100, or 300 μM AZT or ddI, alone or in equimolar combinations for 3 days. A cell cloning assay was used to measure the HPRT or TK mutant frequency. Induced mutant frequencies were obtained by subtracting the mutant frequencies in control groups from those in exposed samples. Average mutant frequencies in control samples ranged from 5.3 ± 1.2 (average ± SD) to 9.0 ± 2.4 × 10−6 for HPRT, and 6.8 ± 0.5 to 15.1 ± 2.4 × 10−6 for TK. AZT, empty bar; ddI, gray bar; AZT + ddI, black bar.
To confirm the observed synergistic mutagenic effects of AZT-ddI coexposures at the HPRT locus, mutant frequencies in the TK gene were measured in TK6 cells exposed to AZT, ddI, or AZT-ddI under the same conditions (Fig. 3B). In AZT-exposed cultures, TK mutant frequencies were significantly increased at exposure concentrations ranging from 33 to 300 μM (P values ranging from 0.004 to 0.008). In contrast, increases in TK mutant frequencies of ddI-exposed cultures were not statistically significant, indicating a lower mutagenic potency for ddI than AZT at the TK locus. Consistent with the HPRT mutant frequency data, the induced TK mutant frequencies in equimolar AZT-ddI combination treatments were 3.0- and 3.5-fold greater than the sum of the mutant frequency values for single treatments with AZT or ddI at exposures of 100 and 300 μM, respectively. In this experiment, the mutagenic effects of 33 μM AZT-ddI were not evaluated.
In other experiments, the potentiation in mutagenic response was also apparent as the amount of ddI, compared with AZT, was reduced for AZT-ddI coexposure of cells (only experimental data for the following example are presented here). For example, treatment of TK6 cells with 300 μM AZT + 100 μM ddI for 3 days led to induced TK mutant frequencies that were 2.7-fold greater than the additive mutant frequency values of single treatments with 300 μM AZT (average induced mutant frequency = 6.5 × 10−6) or 100 μM ddI (average induced mutant frequency = 2.8 × 10−6).
Discussion
Mutagenic responses were observed in the HPRT and TK genes of TK6 human B lymphoblastoid cells exposed in culture to AZT, or equimolar combinations of AZT + ddI, at exposure concentrations ranging from 3 to 30 times the peak plasma levels found in some patients (18). Furthermore, the multiplicative synergistic enhancement of the mutagenic responses to the combined drug exposures correlated well with the potentiation of AZT-DNA incorporation by AZT-ddI coexposures, as compared with single drug exposures.
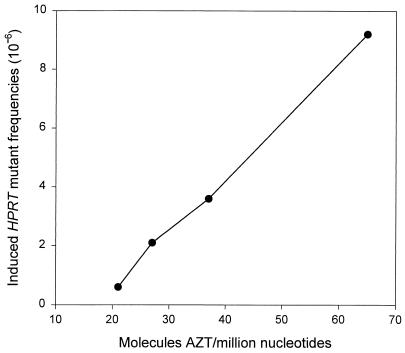
Fig. 4 shows that, in the case of AZT-ddI coexposures, a very smooth curve results when the levels of DNA incorporation of AZT are plotted against induced HPRT mutant frequency values obtained after subtracting the expected contribution in mutagenic response from ddI alone. The near linear shape of this curve supports the hypothesis that the synergistic effect of AZT-ddI coexposure on mutagenesis is due largely to an enhancement of AZT incorporation into DNA.
Figure 4.
The relationship between AZT-DNA incorporation and induced HPRT mutant frequency values obtained following exposure to AZT alone or after subtracting the expected contribution in mutagenic response from ddI alone in the cases where AZT-ddI treatments were combined. The levels of AZT-DNA incorporation were found to have a linear correlation with induced HPRT mutant frequency values following exposure of TK6 cells to 33 μM AZT, 100 μM AZT, 33 μM AZT + 33 μM ddI, and 100 μM AZT + 100 μM ddI.
Still, the synergy in mutation induction with coexposures of AZT-ddI is greater than expected solely by the increase in AZT-DNA incorporation because of the coexposure. This disparity indicates that coexposures of AZT-ddI, compared with ddI alone, lead to a potentiation in the contribution from ddI's mechanisms of mutagenicity, either directly by enhanced DNA incorporation of ddI or indirectly through increased inhibition of DNA polymerases, alterations in nucleotide pools, inhibition of DNA anabolic enzymes, and/or interference with DNA repair processes (1). These findings demonstrate the need for a sensitive assay for investigating (nuclear or mitochondrial) DNA incorporation of ddI, as well as other NRTIs, in complementary cell culture, animal model, and human population studies to define better the potential risks of mutagenicity and/or mitochondrial toxicity arising from antiretroviral drug combinations.
Treatment-related increases in TK mutant frequencies were 1.5- to 2.3-fold greater than HPRT mutant frequencies in cells exposed to AZT + ddI at concentrations of 100 or 300 μM, respectively. The greater mutagenic response in the TK gene than the HPRT gene is likely related to inherent differences in these two loci as mutational targets (19) and the mode of action of AZT and ddI (1). TK6 cells are heterozygous at the TK locus and hemizygous at the HPRT locus. Mutagenic mechanisms that involve homologous interaction, such as gene conversion and mitotic recombination, cannot occur at the X-linked HPRT locus (19). Also, large deletions can be lethal to HPRT mutants because these gross deletions may span the adjacent genes essential for cell survival (19). As DNA chain terminators, AZT and ddI would be expected to induce multilocus deletions (1). Indeed, molecular analyses of mutant colonies indicated that 64%, 84%, and 76% of AZT-induced HPRT (14), TK (15), and APRT (16) mutants, respectively, were due to loss of heterozygosity or large gene deletions. Moreover, loss of heterozygosity is an important mechanism for loss of gene function in recessive genes such as “tumor suppressor” genes, and is a common event in several types of human cancer (20, 21).
The activation of AZT requires functional thymidine kinase, which raised concerns regarding the use of the TK gene to evaluate the mutagenicity of AZT alone or in combination with another NRTI. However, earlier experiments (15, 16) demonstrated that AZT-induced mutant frequencies were quite similar at the TK and APRT loci in human cells exposed to AZT (0, 33, 100, or 300 μM for 3 days) in the same fashion as the current studies. These findings indicate that the involvement of thymidine kinase in AZT anabolism had negligible effect on the mutagenic response to AZT or AZT-ddI at the TK locus, which was in accordance with the fact that AZT triphosphate is formed before any AZT- or AZT-ddI-related mutational events could occur.
Whereas the exposure concentrations of 33 μM and 100 μM AZT or AZT-ddI exceeded peak plasma levels in AZT-treated patients (1), it is noteworthy that the levels of AZT-DNA incorporation that occurred as a result of in vitro exposure of human lymphoblastoid cells (i.e., 21 to 65 molecules of AZT per 106 nucleotides) were similar to or substantially less than the AZT-DNA incorporation values measured (using the same assay) in DNA of lymphocytes from HIV-infected, AZT-treated pregnant women and their newborn infants (12). Among 12 women given 600 mg of AZT per day by mouth for 3 weeks to 9 months of pregnancy, DNA from peripheral blood of lymphocytes from eight individuals contained measurable levels of AZT incorporated into DNA (36 to 215 molecules of AZT per 106 nucleotides). In cord blood samples obtained at delivery from infants of AZT-treated women, lymphocytes from 15 of 22 infants were positive for AZT-DNA incorporation with values ranging from 22 to 452 molecules per 106 nucleotides. Comparison of the AZT-DNA incorporation values obtained in the current in vitro studies and in AZT-treated mother–infant pairs indicates that some humans form more AZT-DNA damage at lower plasma levels of AZT than the DNA damage induced in cells grown at relatively high culture medium concentrations of AZT. These investigations suggest that the levels of AZT-DNA incorporation observed in some humans are sufficiently high to be mutagenic. However, the same studies indicate the existence of interindividual differences (perhaps genetic) that affect levels of DNA incorporation of AZT and probably the risk for mutations (as well as mitochondrial toxicity). Furthermore, the mutagenic response may be enhanced by the use of more than one NRTI.
The important question arising from the results of the current report is whether AZT-ddI or other pairs of NRTIs produce a high degree of potentiation of mutagenicity in transplacentally exposed children who may be genetically predisposed to incorporate these agents into their cellular DNA. Several in vitro studies have indicated that AZT + ddI act synergistically to inhibit HIV viral reverse transcriptase (22, 23), and subsequent clinical studies have demonstrated that this drug combination provides enhanced protection against HIV disease progression and maternal-viral transmission when compared with administration of AZT alone (24). Also, the use of ddI along with AZT is advantageous in cases of acquired drug resistance because it has been demonstrated that mutations in the HIV viral genome leading to AZT resistance do not confer resistance to ddI (25). If, however, some infants prove to be susceptible to AZT-ddI-induced mutation then it will be necessary to establish which susceptibility factors can be used to identify the subset of children who need to be followed long-term.
To date, several investigative groups have attempted unsuccessfully to determine the mechanism(s) underlying this synergism. Mechanisms focusing on alterations in dNTP pools or alterations in the amount of AZT triphosphate formed seem unlikely, given that Palmer and Cox (26) found that the level of AZT phosphorylation was unchanged regardless of whether human lymphoblastoid cells were incubated with AZT alone or in combination with ddI. Instead, they observed a modest increase in the level of ddI-triphosphate following AZT-ddI coexposure of these cells. White and coworkers (27) conducted experiments in some respects analogous to those reported here and found no increase in AZT-DNA incorporation by HIV viral reverse transcriptase when AZT and a second nucleoside analogue (i.e., dideoxyadenosine triphosphate, dideoxycytidine triphosphate, or carbovir) were administered together. Their investigations differed from the present work in several respects, including the fact that they used an extracellular system and evaluated incorporation of AZT by HIV reverse transcriptase instead of DNA polymerase (as was done in the current cell culture studies).
In the context of these earlier studies, the findings of the current work are surprising and raise the possibility that the mechanism(s) of AZT-ddI-induced potentiation in therapeutic response and host cell mutagenic response may lie with one or more cellular factors acting independently from those previously investigated. For example, alterations in post-DNA integration events, including modulation of DNA repair mechanisms and/or pathways leading to apoptosis, could lead to the survival of host cells with increased levels of AZT incorporation in DNA and thus the results reported here. An experimental basis for hypothesizing a role for one or both of these systems has already been established (28, 29). Nonetheless, results of the current study suggest that the synergistic mutagenic effects of AZT + ddI in cultured human cells, as well as the synergistic antiviral effects of this drug combination in patients, are probably related in part to enhanced incorporation of AZT into both cellular and proviral DNA by coexposure to these two NRTIs. Elucidation of the factors leading to this increase in DNA incorporation of AZT and the concomitant synergism of this drug combination offer the possibility that these unknown factors can be manipulated to enhance viral susceptibility and/or decrease host sensitivity.
Acknowledgments
We gratefully acknowledge the use of the services of the Wadsworth Center Media and Glassware Support Service Group for preparing media components. In addition, we appreciate the considerate contributions of the journal's reviewers. This work was supported, in part, by National Institutes of Health Grant RO1HD33648 (to V.E.W.) from the National Institute of Child Health and Human Development, the National Cancer Institute, and Office of AIDS Research.
Abbreviations
- AZT
zidovudine
- ddI
didanosine
- NRTIs
nucleoside reverse transcriptase inhibitors
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.220203197.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.220203197
References
- 1.IARC Monographs on the Evaluation of Carcinogenic Risks to Humans, Some Antiviral and Antineoplastic Drugs, and Other Pharmaceutical Agents (2000) International Agency for Research on Cancer Scientific Publication No. 76 (Lyon), pp. 35–42, 73–127, 153–173.
- 2.Blanche S, Tardieu M, Rustin P, Slama A, Barret B, Firtion G, Ciraru-Vigneron N, Lacroix C, Ropuzioux C, Mandelbrot L, et al. Lancet. 1999;354:1084–1089. doi: 10.1016/S0140-6736(99)07219-0. [DOI] [PubMed] [Google Scholar]
- 3.Fischl M A, Richman D D, Grieco M H, Gottlieb M S, Volberding P A, Laskin O L, Leedom J M, Groopman J E, Mildvan D, Schooley R T, et al. N Engl J Med. 1987;317:185–191. doi: 10.1056/NEJM198707233170401. [DOI] [PubMed] [Google Scholar]
- 4.Richman D D, Fischl M A, Grieco M H, Gottlieb M S, Volberding P A, Laskin O L, Leedom J M, Groopman J E, Mildvan D, Hirsch M S, et al. N Eng J Med. 1987;317:192–197. doi: 10.1056/NEJM198707233170402. [DOI] [PubMed] [Google Scholar]
- 5.Yarchoan R, Mitsuya H, Thomas R V, Pluda J M, Hartman N R, Perno C F, Marczk K S, Allain J P, Johns D G, Broder S. Science. 1989;245:412–415. doi: 10.1126/science.2502840. [DOI] [PubMed] [Google Scholar]
- 6.Ayers K M, Clive D, Tucker W E, Jr, Hajian G, Miranda P. Fundam Appl Toxicol. 1996;32:148–158. doi: 10.1006/faat.1996.0118. [DOI] [PubMed] [Google Scholar]
- 7.Olivero O A, Anderson L M, Diwan B A, Haines D C, Harbaugh S W, Moskal T J, Jones A B, Rice J M, Riggs C W, Logsdon D, et al. J Natl Cancer Inst. 1997;89:1602–1608. doi: 10.1093/jnci/89.21.1602. [DOI] [PubMed] [Google Scholar]
- 8.Darnowski J W, Goulette F A. Biochem Pharmacol. 1994;48:1797–1805. doi: 10.1016/0006-2952(94)90466-9. [DOI] [PubMed] [Google Scholar]
- 9.Olivero O A, Beland F A, Poirier M C. Int J Oncol. 1994;4:499–504. doi: 10.3892/ijo.4.1.49. [DOI] [PubMed] [Google Scholar]
- 10.Sommadossi J, Carlisle R, Zhou Z. Mol Pharmacol. 1989;36:9–14. [PubMed] [Google Scholar]
- 11.Vazquez-Padua M A, Starnes M C, Cheng Y C. Cancer Commun. 1990;2:55–62. doi: 10.3727/095535490820874740. [DOI] [PubMed] [Google Scholar]
- 12.Olivero O A, Shearer G M, Chougnet C A, Kovacs A A S, Landay A L, Baker R, Stek A M, Khoury M M, Proia L A, Kessler H A, et al. AIDS. 1999;13:919–925. doi: 10.1097/00002030-199905280-00007. [DOI] [PubMed] [Google Scholar]
- 13.Poirier M C, Patterson T A, Slikker W, Jr, Olivero O A. J Acquired Immune Defic Syndr. 2000;22:477–483. doi: 10.1097/00126334-199912150-00008. [DOI] [PubMed] [Google Scholar]
- 14.Sussman H E, Olivero O A, Meng Q, Pietras S M, Poirier M C, O'Neill J P, Finette B A, Bauer M J, Walker V E. Mutat Res. 1999;429:249–259. doi: 10.1016/s0027-5107(99)00111-6. [DOI] [PubMed] [Google Scholar]
- 15.Meng Q, Su T, Olivero O A, Poirier M C, Shi X, Ding X, Walker V E. Toxicol Sci. 2000;54:323–329. doi: 10.1093/toxsci/54.2.322. [DOI] [PubMed] [Google Scholar]
- 16.Meng Q, Grosovsky A J, Shi X, Walker V E. Mutagenesis. 2000;15:405–410. doi: 10.1093/mutage/15.5.405. [DOI] [PubMed] [Google Scholar]
- 17.Furman P A, Fyfe J A, St. Clair M H, Weinhold K, Rideout J L, Freeman G A, Lehrman S N, Bolognesi D P, Broder S, Mitsuya H, et al. Proc Natl Acad Sci USA. 1986;83:8333–8337. doi: 10.1073/pnas.83.21.8333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dudley M N. J Infect Dis. 1995;171:S99–S112. doi: 10.1093/infdis/171.supplement_2.s99. [DOI] [PubMed] [Google Scholar]
- 19.Liber H L, Yandell D W, Little J B. Mutat Res. 1989;216:7–17. doi: 10.1016/0165-1161(89)90018-6. [DOI] [PubMed] [Google Scholar]
- 20.Hansen M F, Koufos A, Gallie B L, Phillips R A, Fodstad O, Brogger A, Gedde-Dahl T, Cavenee W K. Proc Natl Acad Sci USA. 1985;82:6216–6220. doi: 10.1073/pnas.82.18.6216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fearon E R, Vogelstein B. Cell. 1990;61:759–767. doi: 10.1016/0092-8674(90)90186-i. [DOI] [PubMed] [Google Scholar]
- 22.Schinazi R F, Sommadossi J P, Saalmann V, Cannon D L, Xie M Y, Hart G C, Smith G A, Hahn E F. Antimicrob Agents Chemother. 1990;34:1061–1067. doi: 10.1128/aac.34.6.1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dornsife R E, St. Clair M H, Huang A T, Panella T J, Koszalka G W, Burns C L, Averett D E. Antimicrob Agents Chemother. 1991;35:322–328. doi: 10.1128/aac.35.2.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Graham N M, Hoover D R, Park L P, Stein D S, Phair J P, Mellors J W, Detels R, Saah A J. Ann Intern Med. 1996;124:1031–1038. doi: 10.7326/0003-4819-124-12-199606150-00002. [DOI] [PubMed] [Google Scholar]
- 25.Ren J, Esnouf R M, Hopkins A L, Jones E Y, Kirby I, Keeling J, Ross C K, Larder B A, Stuart D I, Stammers D K. Proc Natl Acad Sci USA. 1998;95:9518–9523. doi: 10.1073/pnas.95.16.9518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Palmer S, Cox S. Antiviral Chem Chemother. 1994;5:403–409. [Google Scholar]
- 27.White E L, Parker W B, Ross L J, Shannon W M. Antiviral Res. 1993;22:295–308. doi: 10.1016/0166-3542(93)90039-l. [DOI] [PubMed] [Google Scholar]
- 28.Fink D, Aebi D, Howell S B. Clin Cancer Res. 1998;4:1–6. [PubMed] [Google Scholar]
- 29.Morris S, Chen J J, Domon O E, McGarrity L J, Bishop M E, Manjanatha M G, Casciano D A. Mutat Res. 1998;405:41–56. doi: 10.1016/s0027-5107(98)00126-2. [DOI] [PubMed] [Google Scholar]