Abstract
Human melanoma cells growth-arrest irreversibly and terminally differentiate on treatment with a combination of fibroblast interferon and the protein kinase C activator mezerein. This experimental protocol also results in a loss of tumorigenic potential and profound changes in gene expression. Various cloning and cDNA microarray strategies are being used to determine the complete spectrum of gene expression changes underlying these alterations in human melanoma cells. An efficient approach, Rapid Subtraction Hybridization (RaSH), has been developed that is permitting the identification of genes of potential relevance to cancer growth control and terminal cell differentiation. RaSH cDNA libraries are prepared from double-stranded cDNAs that are enzymatically digested into small fragments, ligated to adapters, and PCR amplified followed by incubation of tester and driver PCR fragments. This subtraction hybridization scheme is technically simple and results in the identification of a high proportion of differentially expressed sequences, including known genes and those not described in current DNA databases. The RaSH approach represents an efficient methodology for identifying and cloning genes displaying differential expression that associate with and potentially regulate complex biological processes.
Keywords: cDNA cloning, reverse Northern blotting, melanoma differentiation associated genes, Northern blotting
Orderly temporal changes in gene expression are major determinants of normal physiological processes, and they represent primary mediators of altered cellular properties that define various disease states (1–3). In these contexts, the ability to elucidate the molecular causes of normal and abnormal cellular changes requires the identification and clarification of function of the spectrum of differentially expressed genes. Attaining this goal continues to represent a major effort in both academic and industrial research laboratories. This task is of high priority and offers promise for discovering target genes regulating specific disease states. Once appropriate genes are recognized, methods such as combinatorial chemistry and high throughput screening can be used to identify and develop new molecules with broad ranges of therapeutic potential.
Several approaches are providing an initial description of potentially important and possibly relevant genes involved in, or associated with, normal processes and specific disease states, including aging, differentiation, development, cancer, cardiovascular disease, and neurodegeneration (1–4). Specific molecular approaches that have proven especially informative in identifying differentially expressed genes include differential RNA display (4, 5), serial analysis of gene expression (6, 7), subtraction hybridization (8–11), reciprocal subtraction differential RNA display (12), representational difference analysis (13), RNA fingerprinting by arbitrarily primed PCR (14), electronic subtraction (15), and combinatorial matrix gene analysis (16). Subtraction hybridization represents a particularly attractive method for isolating genes that are differentially expressed in diverse target cells, without prior knowledge of their functional or biochemical characteristics. A limitation of this scheme can often be attributed to technical difficulties encountered in performing this procedure (11, 17). The traditional subtraction hybridization method involves hybridization of first-strand cDNAs generated from tester mRNAs with mRNAs obtained from drivers. Single-stranded unhybridized cDNAs are then selected by hydroxylapatite column chromatography or biotin-avidin extraction and are used as templates for the second-strand cDNA synthesis. These approaches can only analyze a fraction of the overall changes in gene expression, require large amounts of mRNA, and are lengthy and labor-intensive procedures. To circumvent some of these problems, cDNA libraries in phage plasmid vectors have been used as both testers and drivers leading to successful construction of subtracted cDNA libraries by a number of investigators (8, 18, 19). However, constructing cDNA libraries and preparation of cDNA fragments for hybridization are laborious, sometimes difficult, processes. PCR-based cDNA subtraction considerably accelerates the procedures for cDNA library preparation and provides a new direction for subtraction, although it also involves several tedious steps during or after hybridization (13, 20).
A protocol, Rapid Subtraction Hybridization (RaSH), is presently described and significantly simplifies the process of cDNA subtraction in comparison with other methodologies. Additional advantages of the RaSH scheme include efficiency of subtraction and significant reductions in cost. Proof-of-principle for the RaSH approach is presently provided by using a well established human melanoma cell culture model of terminal differentiation (21, 22). In human melanoma cells, treatment with the combination of fibroblast interferon (IFN-β) + mezerein (MEZ) results in irreversible growth arrest, loss of tumorigenic properties, and terminal cell differentiation (21, 22). These changes in melanoma physiology involve specific temporally regulated modifications in gene expression (22–24). Potentially relevant genes associated with this process have been identified from a temporally spaced subtracted IFN-β + MEZ-treated HO-1 human melanoma cDNA library by using several molecular approaches, including random screening of cDNAs, high throughput microchip cDNA microarrays and random cDNA clonal analysis, and screening by reverse Northern hybridization (8, 23, 24). These approaches provide a molecular snapshot of genes regulated during terminal differentiation, and they have produced a useful database for determining the utility and efficacy of the RaSH approach. In the present study, the RaSH scheme has identified a high proportion of genes (≈45%) displaying differential expression as a function of treatment with IFN-β + MEZ. These include genes previously identified as differentially expressed ESTs (expressed sequence tags) in the HO-1 melanoma system as well as genes not previously identified as differentially expressed and genes without sequence homology to reported genes in DNA databases. In these contexts, the efficacy of the RaSH approach as a strategy for identifying differentially expressed genes, as now applied to the process of cellular differentiation, has been confirmed.
Materials and Methods
Cell Lines and Culture Conditions.
The HO-1 human melanoma cell line was established from a metastatic inguinal node lesion from a 49-year-old female and cultured in DMEM supplemented with 10% (vol/vol) FBS and penicillin/streptomycin (100 units per 100 μg/ml; ref. 21). Cultures were seeded at 1.5 × 106 cells per 10-cm plate, and 24 h later the medium was changed without inducers or with IFN-β (2,000 units/ml), MEZ (10 ng/ml), or IFN-β plus MEZ (2,000 units/ml + 10 ng/ml). For library construction and subtraction, HO-1 cells were untreated or treated with IFN-β plus MEZ (2,000 units/ml + 10 ng/ml) for 2, 4, 8, 16, and 24 h.
RNA Isolation and Northern Blot Analysis.
Total RNA was isolated by the guanidinium/phenol procedure from untreated or IFN-β-, MEZ-, or IFN-β plus MEZ-treated cells. Northern blotting was performed as described (8–10). Northern blots were quantitated by densitometric analysis with a Molecular Dynamics densitometer. Poly(A)-RNA was purified by using oligo(dT) cellulose columns (GIBCO/BRL).
Primer Designs.
Sequences of oligonucleotides were as follows: XEA-18, TGATCACTCGAGACCAGG; XET-18, TGATCACTCGAGACCTGG; XE-14, CTGATCACTCGAGA; XEA-13, CCAGGTCTCGAG; XET-13, CCTGGTCTCGAG; XDPN-18, CTGATCACTCGAGAGATC; XDPN-14, CTGATCACTCGAGA; XDPN-12, GATCTCTCGAGT. The adapters formed from the two sets of oligonucleotides contain an XhoI recognition site.
Preparation of PCR-Based cDNA Libraries.
Poly(A)-RNA (1 μg) from control cells (driver) or IFN-β + MEZ treated cells (tester) was used for double-stranded cDNAs synthesis by using standard protocols (8, 17, 25). The cDNAs then were digested with EcoRII (Sigma) or DpnII (New England Biolabs) at 37°C for 3 h followed by phenol/chloroform extraction and ethanol precipitation. The digested cDNAs were mixed with primers XE-14/XEA-13/XET-13 (final concentration, 20 μM) or XDPN-14/XDPN-12 (final concentration, 20 μM) in 30 μl of 1× ligation buffer (GIBCO/BRL), heated at 55°C for 1 min, and cooled down to 14°C within 1 h. After adding 3 μl of T4 ligase (5 units/μl) to the mixtures individually, ligation was carried out at 14°C overnight. The mixtures were diluted to 100 μl with 10 mM Tris/1 mM EDTA, pH 7.0, and at least 40 μl of the mixtures was used for PCR amplification. The PCR mixtures were set up as follows: 1 μl of the cDNA mixture, 10 μl 10× PCR buffer, 1 μM dNTPs, 10 μM XEA-18/XET-18 or XDPN-18, and 1 unit of Taq polymerase (GIBCO/BRL). The parameters for PCR were 1 cycle for 5 min at 72°C, followed by 25 cycles for 1 min at 94°C, 1 min at 55°C, and 1 min at 72°C, and 1 cycle for 3 min at 72°C. The PCR products were pooled and purified by using Centricon columns (Amicon). Portions (10 μg) of the tester PCR products were digested with XhoI followed by phenol/chloroform extraction and ethanol precipitation. Two sets of cDNA libraries thus were prepared: the libraries amplified with XEA-18/XET-18 and the libraries amplified with XDPN-18.
Subtraction Hybridization and Generation of Subtracted Libraries.
The tester cDNA (100 ng) was mixed with 3 μg of the driver cDNA in 10 μl of a hybridization solution [0.5 M NaCl/50 mM Tris, pH 7.5/0.2% SDS/40% (vol/vol) formamide] and, after boiling for 5 min, incubated at 42°C for 48 h. The hybridization mixture was phenol/chloroform extracted, ethanol precipitated, and dissolved in 20 μl of 10 mM Tris/1 mM EDTA, pH 7.0. Part (1 μl) of the mixture was ligated with 1 μg of XhoI-digested, calf intestinal phosphatase-treated pCRII plasmids overnight at 14°C and transformed into Shot-1 bacteria.
Colony Screening.
Bacterial colonies were picked randomly and PCR amplified. The PCR products were blotted onto filters, and reverse Northern blotting was performed to identify cDNAs displaying differential expression in control and differentiated cells (12, 24). The results were confirmed by Northern blotting. The sequences of these clones were determined by using automated cycle sequencing at the DNA facility of Columbia University.
Results and Discussion
Melanoma Cell Culture Model System for Defining the Molecular Determinants of Growth Control and Differentiation.
Abnormalities in differentiation are common properties in many diverse cancers (26, 27). Treatment of human melanoma cells with the combination of IFN-β + MEZ results in a rapid cessation of growth, an induction of melanogenesis, production of profound morphological changes including the development of dendrite-like processes, changes in cell surface antigens, and alterations in gene expression (8, 9, 21–24, 26, 28). Kinetic studies designed to define the temporal relationship between inducer treatment and induction of terminal differentiation in HO-1 human melanoma cells demonstrate that the first 24 h of inducer treatment is critical for irreversibly committing the majority of treated cells to terminal differentiation (21, 22).
To define the spectrum of gene expression changes occurring during commitment to differentiation and maintenance of terminal differentiation in human melanoma cells, a modified subtraction hybridization technique was used (8). Because differentiation is a temporal process, our approach to producing subtracted libraries involved the isolation of temporally spaced mRNAs (encompassing the first 24 h of treatment) from IFN-β + MEZ and control untreated cells and subtracting control (driver) from differentiation inducer-treated cells (tester; ref. 8). Initial screening of this differentiation inducer-treated HO-1 subtracted library included 70 random clones, of which 23 clones (≈33%) displayed differential expression as a consequence of treatment with the inducing agents, and seven of these differentially expressed clones initially represented sequences not reported in then current DNA databases (8, 26). The initially represented melanoma differentiation associated (mda) genes included mda-2, which is homologous to the germ-cell-specific transcription repressor Tctex-1 (29, 30); mda-4, a member of the human IFN-inducible gene family associated with control of tumorigenicity in a model of human melanoma (31); mda-6, the universal cyclin-dependent kinase inhibitor p21 (8, 32, 33); mda-7, a ubiquitous cancer growth-suppressor gene (34, 35); mda-9, a differentiation-associated and IFN γ-inducible gene (36, 37); and mda-1 and mda-5 (8), still without representation in the DNA database and without a defined function. By using the same subtracted library and screening of 1,000 immobilized cDNAs on glass chips, an additional 112 cDNAs (26 known and 11 previously unknown; ≈11%) displaying elevated expression as a function of differentiation in HO-1 cells have been identified (23). Moreover, screening of an additional 400 random cDNA clones from the subtracted library by reverse Northern hybridization identified an additional 65 differentially expressed cDNAs (30 known and 26 previously unknown; ≈16%), some of which had also been identified in the first library screen or by the high-density cDNA microarray approach (24). These analyses provide baseline data for specific gene expression changes occurring during the processes of induction and maintenance of differentiation in human melanoma cells.
RaSH Protocol and Its Application for Identifying Genes Differentially Expressed During HO-1 Differentiation.
Subtraction hybridization provides a general methodology for directly selecting for unique cDNA species and removing common expressed sequences between cellular genomes (8–11). The application of the original approach often was not straightforward, and success required a high degree of technical competence (11). The ability to use subtraction hybridization for identifying differentially expressed genes has been improved by the development of PCR-based subtraction hybridization approaches (13, 20). However, this innovation is not trouble-free, because this approach is laborious and often ineffective in identifying differentially expressed genes. To simplify the subtraction hybridization approach and make this methodology more amenable to diverse research laboratories, we have developed a simple, efficient, and affordable rapid subtraction hybridization approach, RaSH.
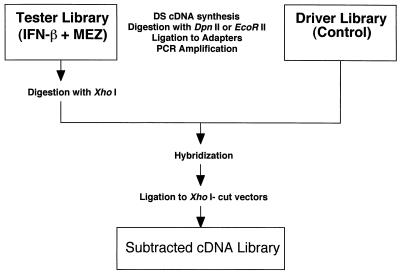
A schematic of the RaSH approach is shown in Fig. 1. For the present study, pooled RNAs (2, 4, 8, 16, and 24 h) extracted from control and IFN-β + MEZ-treated cells were used in the RaSH protocol to identify genes differentially expressed in the inducer-treated cells. The cDNA libraries were constructed by synthesizing double-stranded cDNAs, digesting the cDNAs into small fragments, ligating the fragments to adapters, and amplifying with PCR. Purifying the PCR products by means of Centricon columns produces reproducible results. Experiments were performed by using the 4-bp restriction enzyme DpnII and the 4.5-bp restriction enzyme EcoRII to compare the effects of cDNA fragment size on redundancy of subtracted cDNA species. Compared with the average size of 256 bp generated by DpnII, EcoRII digested cDNAs into fragments of 512 bp on average, which reduced the redundancy of large-gene species while decreasing the representation of small-gene species in the RaSH cDNA libraries.
Figure 1.
Schematic outline of the RaSH protocol. This scheme involves construction of tester (IFN-β + MEZ) and driver (control) HO-1 libraries, followed by digestion of only the tester library with XhoI. After hybridization, differentially expressed sequences are cloned into XhoI-digested vectors, resulting in a subtracted cDNA library enriched for mda genes displaying elevated expression. By using the control HO-1 library as the tester and the IFN-β + MEZ library as the driver, RaSH can also be used to produce a subtracted cDNA library enriched for genes down-regulated during terminal differentiation.
Subtraction hybridization during the RaSH approach is performed by incubating the tester and driver PCR fragments without the need for any additional PCR amplification steps. Instead of using the PCR-based subtraction approach, which can result in amplification bias, RaSH uses a different strategy based on mass-driven subtraction by altering the ratio of input tester to driver. In the RaSH scheme, subtracted cDNAs are selected simply by matching the ends of the cDNA fragments to the ends of the plasmid vectors during ligation, from which subtracted libraries are constructed. This simple step of subtraction makes RaSH different from any other cDNA subtraction protocol.
The RaSH approach incorporates a reverse Northern analysis, which offers the ability to rapidly pretest large numbers of cDNA clones for differential expression (12, 24). The reverse Northern step permits the first tier identification and elimination of possible false positive clones. This procedure can, theoretically, also be used for subsequent rounds of RaSH clone identification by using previously identified RaSH sequences to eliminate redundant clonal selection. Moreover, it is possible to scale up this approach and use high-throughput screening platforms containing RaSH-derived clones for monitoring temporal and distinct patterns of gene expression of multiple cDNAs within one assay. Once identified as a potential positive clone, confirmation of differential expression can be verified by Northern blotting analyses (12, 24).
To construct subtracted cDNA libraries, tester cDNAs were digested with DpnII or EcoRII, ligated to DpnII- or EcoRII-end adapters, amplified with specific primers, digested with XhoI (which recognizes the XhoI site in the primers), and subtracted with driver cDNAs treated in an identical manner without enzymatic digestion (Fig. 1). In this manner, two cDNA libraries (Dpn-sLib and Eco-sLib) were constructed.
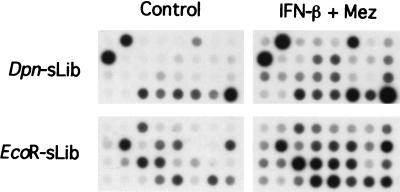
Colonies from the two libraries were isolated randomly and the PCR-amplified products were used for reverse Northern analysis. This analysis resulted in the identification of ≈50% differentially expressed clones. The results of the screening of 32 representative clones from each library are shown in Fig. 2 indicating differential expression in the majority of clones from both libraries when one compares control and differentiation-inducer treated HO-1 cells. Clones from both libraries were sequenced and compared with previously identified genes deposited in GenBank and European Molecular Biology Organization databases. Both libraries resulted in the identification of known and unidentified gene sequences. Although the majority of clones contained single-gene inserts, as with other cloning strategies, some colonies were isolated that contained more than one insert (usually two) ligated in tandem.
Figure 2.
Reverse Northern blot analysis of differentially expressed sequence tags identified by RaSH. PCR-amplified products from bacterial clones of both RaSH-derived subtracted libraries, Dpn-sLib and EcoR-sLib, were dot-blotted onto nylon membranes and were probed with [32P]cDNA reverse-transcribed from RNA samples of control or IFN-β + MEZ-treated HO-1 cells.
The reverse Northern analysis results were confirmed with Northern blots by using both known and unidentified sequences from each library (Tables 1 and 2). This analysis resulted in a high degree of concordance (≈89%) between positive signals identified by using the reverse Northern screen and true differential expression as indicated by Northern blotting (Figs. 2 and 3). It should be noted, however, that because equal amounts of mRNA at 2, 4, 8, 16, and 24 h after treatment of HO-1 cells with the differentiation inducers were pooled and used for poly(A)-RNA selection and tester cDNA synthesis, the results of reverse Northern blots in the current experiments should be interpreted as a comparison between the expression level of control and the average level of the differentiation-induced cells at different time points. If a single time point had been used, the proportion and types of differentially expressed genes identified would, in all likelihood, be different unless they displayed elevated expression over a broad time period (22–24). Defined temporal expression kinetics seem to be the rule rather than the exception during a 72-h evaluation of the expression of a subset of microarrayed subtracted HO-1 cDNAs with temporally spaced RNAs isolated from HO-1 cells treated with IFN-β + MEZ (23). This effect is also apparent after inspection of the various mda genes identified by using RaSH as indicated by temporal kinetics of expression over a 72-h time course, 2, 4, 8, 24, and 72 h (Fig. 3). For example, genes such as mda-D-57, mda-D-55, mda-E-64, mda-E-61, and mda-D-42 are inducible within 2 or 4 h exposure to IFN-β + MEZ, whereas other mda genes, such as mda-D-33, mda-D-47, mda-D-66, mda-D-56, mda-D-27, mda-E-47, and mda-D-34, display low or no induction within this time frame (Fig. 3). In fact, all of the mda genes analyzed exhibit defined temporal kinetics confirming similar results obtained by using high-throughput microarray analyses (23).
Table 1.
Clones isolated with RaSH with the restriction enzyme DpnII
| Nomenclature* | Identity† | Mda type‡ |
|---|---|---|
| mda-D-25 | HLA cw6 | Type I |
| mda-D-26 | Novel, human homolog of Mus musculus small GTP-binding protein | Type I |
| mda-D-27, D-49, D-53, D-58, D-69 | Fibronectin | Type IV |
| mda-D-28 | Vascular endothelial growth factor (VEGF) (165) | Type IV |
| mda-D-33 | Cig5 (IFN-responsive RNA) | Type I |
| mda-D-34, D-36, D-46 | IL-11 | Type IV |
| mda-D-35 | Prolactin receptor-associated protein (PRA) | Type II |
| mda-D-39 | IL-1 | Type II |
| mda-D-40 | Plasminogen activator inhibitor type II | Type II |
| mda-D-41 | G-binding protein I | Type I |
| mda-D-42, D-54 | LIF | Type II |
| mda-D-45 | KIAA0439 from human brain | Type II |
| mda-D-48, D-52 | α5 integrin | Type II |
| mda-D-50 | Mac-2 binding protein 90 Kd product | Type I |
| mda-D-51, D-61 | Urokinase-type plasminogen activator receptor | Type II |
| mda-D-70 | Monocyte activation antigen | Type I |
| mda-D-55 | 2′-5′ oligoadenylate synthetase | Type I |
| mda-D-56 | Mn superoxide dismutase | Type III |
| mda-D-57, D-59, D-62 | 1-8U from the IFN-inducible gene family | Type I |
| mda-D-65 | ICAM-1 (intercellular adhesion molecule 1) | Type I |
| mda-D-68 | snRNP protein B | Type I |
| mda-D-32, D-47, D-63, D-66 | Novel | Type II |
Clones are designated as melanoma differentiation associated (mda) and the D designation refers to the fact that the restriction enzyme DpnII was used for isolation by the RaSH approach.
Sequences were searched against various DNA databases to determine sequence identities. Novel, no similar sequence reported in current DNA databases.
Type I mda gene, inducible by IFN-β and IFN-β + MEZ; Type II mda gene, inducible by MEZ and IFN-β + MEZ; Type III mda gene, inducible by IFN-β, MEZ and IFN-β + MEZ; Type IV mda gene, inducible predominantly by IFN-β + MEZ.
Table 2.
Clones isolated with RaSH with the restriction enzyme EcoRII
| Nomenclature* | Identity† | Mda type‡ |
|---|---|---|
| mda-E-28, E-34, E-38, E-41, E-45, E-49, E-50 | Fibronectin | Type IV |
| mda-E-31 | 2′-5′ oligoadenylate synthetase | Type I |
| mda-E-32, E-57, E-58 | LIF | Type II |
| mda-E-35 | Transporter 1, ABC | Type I |
| mda-E-36, E-42, E-52 | HLA-B | Type I |
| mda-E-39 | KIAA0180 | Type II |
| mda-E-40 | Vimentin | Type I |
| mda-E-44 | Tumor necrosis factor-α | Type II |
| mda-E-46, E-55 | Calcium-activated potassium channel | Type IV |
| mda-E-47 | Ninjurin 1 | Type IV |
| mda-E-51 | HMG-Y protein isoform | Type I |
| mda-E-54 | Proteasome 26S subunit | Type IV |
| mda-E-61 | Human GADD34 | Type II |
| mda-E-68 | mda-6 (p21, Waf1, CIP1, SDI1) | Type IV |
| mda-E-43, E-63, E-64 | Novel | Type I |
Clones are designated as melanoma differentiation-associated (mda) and the E designation refers to the fact that the restriction enzyme EcoRII was used for isolation by the RaSH approach.
Sequences were searched against various DNA databases to determine sequence identities. Novel, no similar sequence reported in current DNA databases.
Type I mda gene, inducible by IFN-β and IFN-β + MEZ; Type II mda gene, inducible by MEZ and IFN-β + MEZ; Type III mda gene, inducible by IFN-β, MEZ and IFN-β + MEZ; Type IV mda gene, inducible predominantly by IFN-β + MEZ.
Figure 3.
Differential expression of representative types of mda genes identified by RaSH and reverse Northern blotting. Northern blots of total RNA were prepared from HO-1 cells untreated (control; lanes 1 and 7) or treated for 2 h (lane 2), 4 h (lane 3), 8 h (lane 4), 24 h (lanes 5 and 10), or 72 h (lane 6) with IFN-β + MEZ, 24 h with IFN-β (lane 8), or 24 h with MEZ (lane 9). Membranes were probed with radiolabeled (32P) labeled mda sequence tags, identified by RaSH in the Dpn-sLib and EcoR-sLib. Equal loading of samples was confirmed by hybridization with a 32P-labeled glyceraldehyde-3-phosphate dehydrogenase cDNA probe.
The genes identified by using RaSH in the melanoma differentiation model can be classified into four mda gene subtypes based on their pattern of induction, as confirmed by Northern blotting (8). These include, type I mda genes (up-regulated by IFN-β and IFN-β + MEZ), type II mda genes (up-regulated by MEZ and IFN-β + MEZ), type III mda genes (up-regulated by IFN-β, MEZ and IFN-β + MEZ), and type IV mda genes (up-regulated primarily by IFN-β + MEZ; ref. 8; Fig. 3). Excluding redundant gene identification, in the initial RaSH analyses (≈10% of the subtracted libraries) with the enzymes DpnII and EcoRII a total of 17 type I, 14 type II, 1 type III, and 7 type IV mda genes were identified based on Northern blotting analyses (Tables 1 and 2). Examples of the four types of mda genes initially identified by reverse Northern blotting and then classified into specific mda subtypes by Northern blotting are shown in Fig. 3. By using the enzyme DpnII in the RaSH approach, a comparable number of type I and type II genes were identified, 10 and 11 respectively, whereas only 1 type III and 3 type IV genes were identified (Table 1). With the enzyme EcoRII in the RaSH approach, eight type I genes and only four type II genes were identified, whereas no type III and five type IV genes were cloned (Table 2). As found in previous screenings of temporally spaced IFN-β + MEZ subtracted HO-1 cDNA libraries, several categories of known genes were identified, including IFN-inducible genes (HLA, Cig-5, 1–8U, GBP I), MEZ-inducible genes (prolactin receptor-associated protein), a differentiation factor, leukemia inhibitory factor (LIF), genes involved in growth inhibition or apoptosis (mda-6, HuGADD34), and cytoskeleton and extracellular matrix genes (fibronectin, integrin α5; Tables 1 and 2). In addition, seven sequences without representation in current DNA databases also were identified by using RaSH. Of the 25 distinct genes identified from the Dpn-sLib and the 17 distinct genes cloned from the EcoR-sLib, only one type I (2′-5′ oligoadenylate synthetase), one type II (LIF), and one type IV (fibronectin) gene were common to both libraries (Tables 1 and 2).
It is well established that the induction of many IFN-stimulated genes occur through activation of the JAK/STAT signaling pathway (38, 39), and MEZ can induce activation of specific protein kinase C (PKC) subtypes (40). Based on the relative proportion of mda gene subtypes identified from subtracted libraries by using RaSH and other gene identification strategies (23, 24), it is probable that the ability of IFN-β + MEZ to induce terminal differentiation may involve the combined activation of both of these pathways. Further studies to address the role of each respective pathway, i.e., JAK/STAT signaling and PKC activation, in induction of growth arrest and terminal differentiation in human melanoma cells by IFN-β + MEZ should prove informative.
As with previous subtraction hybridization approaches (8, 23, 24), clonal redundancy was apparent in both RaSH libraries (Tables 1 and 2). As examples, fibronectin (≈8 kb) was isolated five times from the Dpn-sLib and seven times from the EcoR-sLib, and LIF (≈7.6 kb) was isolated twice from the Dpn-sLib and three times from the EcoR-sLib. In contrast, redundancy in the isolation of smaller cDNAs, including α5-integrin (two isolates; 4.1 kb) and the IFN-responsive gene 1-8U (three isolates; 0.8 kb) occurred only in the Dpn-sLib, and these sequences were not detected in the EcoR-sLib. Size may not be the only factor resulting in differential abundance of clones identified in libraries prepared with the two different enzymes, because IL-11 was isolated three times from the Dpn-sLib, ≈6.9 kb in size and was not present in the 10% of EcoR-sLib-evaluated clones. It is apparent, however, from this limited analysis of the two RaSH-derived libraries that using EcoRII to digest cDNAs may result in reduction of sequence redundancy compared with DpnII. However, the use of DpnII in the RaSH approach may facilitate the isolation of smaller sized cDNAs moreso than the use of the enzyme EcoRII.
The RaSH approach successfully identified genes previously recognized as those differentially expressed during induction of terminal differentiation in human melanoma cells (8, 22–24). This grouping includes genes identified during the initial random screening of the subtracted cDNA library [mda-6 (p21) and vimentin], identified by using high-density microchip cDNA arrays (fibronectin, HLA-B, Mn superoxide dismutase, and 2′-5′ oligoadenylate synthetase), and identified by random clonal isolation and screening by reverse Northern hybridization (HLA-B). Moreover, previous studies have demonstrated changes in the expression of intercellular adhesion molecule 1 (28) and α5-integrin (22) during induction of differentiation in HO-1 cells, and both of these gene changes were identified by using the RaSH approach. In addition, RaSH has identified sequences not detected by using the previous protocols (Tables 1 and 2). In contrast, genes, such as the transcription factor ISGF-3 and mitochondrial-associated genes that were identified 11 and 52 times, respectively, by using microarrays of subtracted cDNA clones (23) were not identified by using the RaSH approach. The reason for the differences in the types of genes identified by using the dissimilar methodologies is not known. One possibility is that all of the approaches, including RaSH, have not evaluated the entire spectrum of cDNAs present in the subtracted libraries that have been generated. In the case of the high-density cDNA microarray analysis, only 1,000 of the ≈10,000 clones in the subtracted cDNA library were evaluated (23). Similarly, by using the RaSH approach, only 10% of either the Dpn-sLib or the EcoR-sLib have been evaluated. Alternatively, it is possible that specific cloning and identification biases are present in the different approaches, thereby resulting in only a subset of genes being identified with any specific subtraction approach. This preferential gene identification may be true with respect to the microchip array approach, because only random cDNAs of ≥500 bp could be formatted for evaluation on chips (23), whereas smaller subtracted cDNAs were used for the reverse Northern approach (24).
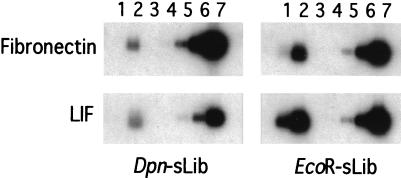
Efficiency of subtraction in RaSH was determined by comparing the percentage of specific gene fragments present in the PCR-amplified cDNA library versus those in the subtracted library. The fibronectin and LIF fragments were used for this purpose because of their large cDNA size, ≈8 and ≈7.6 kb, respectively, which resulted in redundant copies of these genes in both subtracted libraries (Dpn-sLib and EcoR-sLib; Tables 1 and 2). The relative amounts of each of these cDNA fragments were estimated by Southern blotting of the PCR cDNA libraries and in comparison with different quantities of the cDNA fragments (Fig. 4). Enrichment was then calculated as the percentage of the fragment in the PCR libraries divided by the percentage of the clones in the subtracted library. Enrichment of LIF was 544-fold in the Dpn-sLib and 96-fold in the EcoR-sLib, whereas enrichment of fibronectin was 75-fold in the Dpn-sLib and 6-fold in the EcoR-sLib. The representation of cDNA species was reduced in the EcoRII PCR-amplified library, because larger cDNA fragments were produced by EcoRII digestion. The presence of fewer cDNA species in this library might explain the “lower efficiency” in relative copy number. The present data also indicate that PCR amplification in the RaSH procedure does not significantly alter the proportion of expressed genes. Based on this consideration, it is possible that RaSH could be performed without a PCR-amplification step, and this possibility is currently being tested. However, it is also probable that low-abundance messages may be lost if PCR amplification is not used in the RaSH approach.
Figure 4.
Determination of the amounts of fibronectin and LIF fragments in PCR cDNA libraries. Relative amount of fibronectin and LIF fragments in PCR libraries was determined by comparison of the signal intensity of fibronectin and LIF in specified cDNA libraries (Dpn-sLib and EcoR-sLib) with the signal intensity of defined amounts of the cDNA fragments by Southern blot hybridization. Lane 1, 100 ng of PCR-cDNA library from untreated cells; lane 2, 100 ng of PCR-cDNA library from IFN-β + MEZ treated cells; lanes 3 to 7, increasing amounts of cDNA fragments of fibronectin and LIF (0.001, 0.01, 0.1, 1, and 10 ng, respectively). PhosphorImager (Molecular Dynamics) scanning determined hybridized signal intensity.
The differences between RaSH and other PCR-based subtraction protocols are readily apparent. Except for the use of PCR for cDNA amplification, the experimental approaches are distinct because RaSH includes different primer designs, subtraction approaches, and subtracted sequence selection. Although both cDNA populations from control and differentiated HO-1 cells are amplified by using the same primers, only the tester cDNA fragments are digested with XhoI, which recognizes the internal XhoI site in the primer. Therefore, the cDNA fragments from the differentiated cells have clonable XhoI sticky-ends, which distinguishes them from those of driver cDNAs. Only one round of hybridization is necessary in the RaSH protocol because the subtraction stringency can be adjusted by simply changing the ratio of tester and driver during subtraction. Unlike other PCR-based protocols, no PCR amplification is used in RaSH during subtraction. The absence of these additional steps also significantly simplifies the subtraction procedures. The use of EcoRII (4.5-bp cutter) as opposed to DpnII in producing RaSH subtracted libraries can be used to reduce redundant gene identification. However, the application of either restriction enzyme in the RaSH approach results in the efficient identification of both similar and distinct genes. It should also be emphasized that the efficiency of the RaSH approach in identifying differentially expressed genes is superior to previous approaches, i.e., 45% versus ≈20% for the combination of random cDNA clone isolation, high-throughput screening of microarrayed subtracted cDNAs, and random cDNA isolation and analysis by reverse Northern blotting (8, 23, 24).
In conclusion, we have developed a rapid and simple cDNA subtraction protocol, RaSH, and, presently applied, this procedure to isolate mda genes. The originality of this method is reflected in the manner in which cDNA fragments are prepared, the steps used in subtraction, and the method of selecting subtracted sequences. The mda genes isolated by using RaSH represent both known and previously unidentified sequences of potential relevance to growth control and differentiation. Based on the ease of performance and the efficiency of differential gene isolation embodied in the RaSH approach, this methodology should find wide application for facilitating the identification of relevant genes associated with and potentially causative of important biological phenomena.
Acknowledgments
The present research was supported in part by National Institutes of Health Grants CA35675 and NS31492, the Samuel Waxman Cancer Research Foundation, and the Chernow Endowment. P.B.F. is the Michael and Stella Chernow Urological Cancer Research Scientist in the Departments of Pathology and Urology.
Abbreviations
- MEZ
mezerein
- RaSH
Rapid Subtraction Hybridization
- LIF
leukemia inhibitory factor
- mda
melanoma differentiation associated
Footnotes
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. BF023602, BF023603, BF023604, BF023605, BF023606, BF023607, and BF023608).
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.220431297.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.220431297
References
- 1.Schena M, Shalon D, Heller R, Chai A, Brown P O, Davis R W. Proc Natl Acad Sci USA. 1996;93:10614–10619. doi: 10.1073/pnas.93.20.10614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Iyer V R, Eisen M B, Ross D T, Schuler G, Moore T, Lee J C F, Trent J M, Staudt L M, Hudson J, Jr, Boguski M S, et al. Science. 1999;283:83–87. doi: 10.1126/science.283.5398.83. [DOI] [PubMed] [Google Scholar]
- 3.Fambrough D, McClure K, Kazlauskas A, Lander E S. Cell. 1999;97:727–741. doi: 10.1016/s0092-8674(00)80785-0. [DOI] [PubMed] [Google Scholar]
- 4.Liang P, Pardee A B. Science. 1992;257:967–971. doi: 10.1126/science.1354393. [DOI] [PubMed] [Google Scholar]
- 5.Shen R, Su Z, Olsson C A, Fisher P B. Proc Natl Acad Sci USA. 1995;92:6778–6782. doi: 10.1073/pnas.92.15.6778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Velculescu V E, Zhang L, Vogelstein B, Kinzler K W. Science. 1995;270:484–487. doi: 10.1126/science.270.5235.484. [DOI] [PubMed] [Google Scholar]
- 7.Zhang L, Zhou W, Velculescu V E, Kern S E, Hruban R H, Hamilton S R, Vogelstein B, Kinzler K W. Science. 1997;276:1268–1272. doi: 10.1126/science.276.5316.1268. [DOI] [PubMed] [Google Scholar]
- 8.Jiang H, Fisher P B. Mol Cell Differ. 1993;1:285–299. [Google Scholar]
- 9.Jiang H, Lin J J, Su Z-Z, Goldstein N I, Fisher P B. Oncogene. 1995;11:2477–2486. [PubMed] [Google Scholar]
- 10.Su Z-Z, Shi Y, Fisher P B. Proc Natl Acad Sci USA. 1997;94:9125–9130. doi: 10.1073/pnas.94.17.9125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sagerstrom C G, Sun B I, Sive H L. Annu Rev Biochem. 1997;66:751–783. doi: 10.1146/annurev.biochem.66.1.751. [DOI] [PubMed] [Google Scholar]
- 12.Kang D-C, LaFrance R, Su Z-Z, Fisher P B. Proc Natl Acad Sci USA. 1998;95:13788–13793. doi: 10.1073/pnas.95.23.13788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hubank M, Schatz D G. Nucleic Acids Res. 1994;22:5640–5648. doi: 10.1093/nar/22.25.5640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McClelland M, Mathieu-Daude F, Welsh J. Trends Genet. 1995;11:242–246. doi: 10.1016/s0168-9525(00)89058-7. [DOI] [PubMed] [Google Scholar]
- 15.Adams M D, Kerlavage A R, Fields C, Venter J C. Nat Genet. 1993;4:256–267. doi: 10.1038/ng0793-256. [DOI] [PubMed] [Google Scholar]
- 16.Schena M, Shalon D, Davis R W, Brown P O. Science. 1995;270:467–470. doi: 10.1126/science.270.5235.467. [DOI] [PubMed] [Google Scholar]
- 17.Reddy P G, Su Z-Z, Fisher P B. In: Chromosomes and Genetic Analysis: Methods in Molecular Genetics. Adolph K W, editor. Vol. 1. Orlando, FL: Academic; 1993. pp. 68–102. [Google Scholar]
- 18.Rubenstein J L R, Brice A E J, Ciaranello R D, Denney D, Porteus M H, Usdin T B. Nucleic Acids Res. 1990;18:4833–4842. doi: 10.1093/nar/18.16.4833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Herfort M R, Garber A T. BioTechniques. 1991;11:598–603. [PubMed] [Google Scholar]
- 20.Diatchenko L, Lau Y-F C, Campbell A P, Chenchik A, Moqadam F, Huang B, Lukyanov K, Gurskaya N, Sverdlov E, Siebert D. Proc Natl Acad Sci USA. 1996;93:6025–6030. doi: 10.1073/pnas.93.12.6025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fisher P B, Prignoli D R, Hermo H, Jr, Weinstein I B, Pestka S. J Interferon Res. 1985;5:11–22. doi: 10.1089/jir.1985.5.11. [DOI] [PubMed] [Google Scholar]
- 22.Jiang H, Su Z-Z, Boyd J, Fisher P B. Mol Cell Differ. 1993;1:41–46. [Google Scholar]
- 23.Huang F, Adelman J, Jiang H, Goldstein N I, Fisher P B. Oncogene. 1999;18:3546–3552. doi: 10.1038/sj.onc.1202715. [DOI] [PubMed] [Google Scholar]
- 24.Huang F, Adelman J, Jiang H, Goldstein N I, Fisher P B. Gene. 1999;236:125–131. doi: 10.1016/s0378-1119(99)00244-9. [DOI] [PubMed] [Google Scholar]
- 25.Gubler U, Hoffman B J. Gene. 1983;25:263–269. doi: 10.1016/0378-1119(83)90230-5. [DOI] [PubMed] [Google Scholar]
- 26.Jiang H, Lin J, Fisher P B. Mol Cell Differ. 1994;2:221–239. [Google Scholar]
- 27.Waxman S, editor. Differentiation Therapy. Rome: Serono Symposium; 1995. [Google Scholar]
- 28.Guarini L, Graham G M, Jiang H, Ferrone S, Zucker S, Fisher P B. Pigm Cell Res Suppl. 1992;2:123–131. doi: 10.1111/j.1600-0749.1990.tb00361.x. [DOI] [PubMed] [Google Scholar]
- 29.Roux A F, Rommens J, McDowell C, Anson-Cartwright L, Bell S, Schappert K, Fishman G A, Musarella M. Hum Mol Genet. 1994;3:257–263. doi: 10.1093/hmg/3.2.257. [DOI] [PubMed] [Google Scholar]
- 30.O'Neill M J, Artzt K. Development (Cambridge, UK) 1995;12:561–568. doi: 10.1242/dev.121.2.561. [DOI] [PubMed] [Google Scholar]
- 31.DeYoung K L, Ray M E, Su Y A, Anzick S L, Johnstone R W, Trapani J A, Meltzer P S, Trent J M. Oncogene. 1997;15:453–457. doi: 10.1038/sj.onc.1201206. [DOI] [PubMed] [Google Scholar]
- 32.Xiong Y, Hannon G J, Zhang H, Casso D, Kobayashi R, Beach D. Nature (London) 1993;366:701–704. doi: 10.1038/366701a0. [DOI] [PubMed] [Google Scholar]
- 33.Jiang H, Lin J, Su Z-Z, Herlyn M, Kerbel R S, Weissman B E, Welch D R, Fisher P B. Oncogene. 1995;10:1855–1864. [PubMed] [Google Scholar]
- 34.Jiang H, Su Z-Z, Lin J J, Goldstein N I, Young C S H, Fisher P B. Proc Natl Acad Sci USA. 1996;93:9160–9165. doi: 10.1073/pnas.93.17.9160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Su Z-Z, Madireddi M T, Lin J J, Young C S H, Kitada S, Reed J C, Goldstein N I, Fisher P B. Proc Natl Acad Sci USA. 1998;95:14400–14405. doi: 10.1073/pnas.95.24.14400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lin J J, Jiang H, Fisher P B. Mol Cell Differ. 1996;4:317–333. [Google Scholar]
- 37.Lin J J, Jiang H, Fisher P B. Gene. 1998;207:105–110. doi: 10.1016/s0378-1119(97)00562-3. [DOI] [PubMed] [Google Scholar]
- 38.Darnell J E, Jr, Kerr I M, Stark G R. Science. 1994;264:1415–1421. doi: 10.1126/science.8197455. [DOI] [PubMed] [Google Scholar]
- 39.Stark G R, Kerr I M, Williams B R, Silverman R H, Schreiber R D. Annu Rev Biochem. 1998;67:227–264. doi: 10.1146/annurev.biochem.67.1.227. [DOI] [PubMed] [Google Scholar]
- 40.Geiges D, Meyer T, Marte B, Vanek M, Weissgerber G, Stabel S, Pfeilschifter J, Fabbro D, Huwiler A. Biochem Pharmacol. 1997;53:865–875. doi: 10.1016/s0006-2952(96)00885-4. [DOI] [PubMed] [Google Scholar]