Abstract
A major challenge in the study of gene regulation by NF-κB/Rel transcription factors is to understand, at the biological and mechanistic levels, the selective functions of individual Rel family members. To study selectivity, we have examined the NF-κB/Rel protein binding site (Rel site) within the IL-12 p40 promoter. IL-12 is a proinflammatory cytokine expressed by activated macrophages that serves as an essential inducer of T helper 1 cell development. In nuclear extracts from lipopolysaccharideactivated macrophages, the predominant Rel dimers capable of binding the IL-12 p40 Rel site were the p50/p65 and p50/c-Rel heterodimers and p50/p50 homodimer. The two heterodimers bound the site with comparable affinities and exhibited comparable transactivation activities. In striking contrast, p40 mRNA and protein concentrations were reduced dramatically in c-Rel−/− macrophages and only modestly in p65−/− macrophages. Other proinflammatory cytokine mRNAs and proteins were not significantly reduced in c-Rel−/− macrophages. These results reveal that a c-Rel-containing complex is an essential and selective activator of p40 transcription, which may reflect unique regulatory mechanisms or biological functions of IL-12. Furthermore, because selectivity was not observed in vitro or in transient transactivation experiments, these findings suggest that an understanding of the selectivity mechanism may require an analysis of the endogenous p40 locus.
Macrophages are among the first lines of defense in response to pathogenic infection. After the interaction of a pathogen with receptors on the macrophage surface, numerous cellular genes are induced, several of which encode cytokines that stimulate an inflammatory response (1). IL-12 is unique among the proinflammatory cytokines produced by activated macrophages because it serves as an essential inducer of T helper 1 cell development and thus provides a bridge between innate and adaptive immunity (2). T helper 1 cells are required for a cell-mediated immune response, which contributes to the suppression or elimination of numerous intracellular pathogens (2).
Biologically active IL-12 p70 is composed of two subunits, p40 and p35, both of which are induced in macrophages and dendritic cells at the level of transcription (3). In transfection assays, the promoter for the p40 gene is strongly induced by lipopolysaccharide (LPS) and other bacterial products (4–6). Multiple DNA elements contribute to promoter activity, including an element that binds members of the NF-κB/Rel family of transcription factors (4, 6). Rel binding sites have been identified in the promoters for several other proinflammatory cytokine genes expressed in macrophages, including the IL-1β, IL-6, and tumor necrosis factor α (TNF-α) genes (7–10).
Five Rel family members have been identified in mammalian cells: p50, p65 (RelA), c-Rel, p52, and RelB (7). These proteins exist in various homodimeric and heterodimeric complexes that often are maintained in an inactive state by cytoplasmic association with I-κB proteins (7). Cell activation leads to a signal transduction cascade that results in the phosphorylation and degradation of the I-κBs, allowing nuclear translocation and DNA binding of the Rel dimers.
The presence of five Rel family members suggests that each family member and each dimeric complex might regulate unique sets of genes. The strongest evidence for unique functions has been provided by analyses of mice that are deficient for individual family members. Each deficiency results in a different phenotype, suggesting that unique target genes must exist (7). Nevertheless, redundancy and compensation among family members have made it difficult to identify specific target genes and determine the mechanism of family-member selectivity. This analysis is further complicated by the embryonic lethality of p65−/− mice and the inherent challenge of distinguishing direct from indirect effects. Despite these challenges, analyses of Rel-deficient mice have revealed genes that require a specific family member for expression. For example, c-Rel−/− mice do not express the IL-2, A1, and interferon regulatory factor-4 genes in at least some cell types and express the IL-3, granulocyte—macrophage colony-stimulating factor (GM-CSF), inducible nitric-oxide synthase (i-NOS), and TNF-α genes at reduced levels (11–15).
Additional insight into the unique properties of Rel family members has been provided by biochemical studies. For example, although the DNA sequences preferred by the various Rel dimers are similar, differences have been observed that may contribute to family member selectivity (16). Studies of numerous cellular promoters have revealed selective DNA binding and transactivation by specific Rel dimers. However, when dimers with similar sequence preferences were compared, such as the p50/p65 and p50/c-Rel heterodimers, the comparisons usually were performed in the absence of appropriate controls to ensure that equivalent concentrations of the two heterodimers were being analyzed. Thus, the mechanistic basis of family member selectivity remains unresolved, in particular for closely related dimers.
In this study, we have examined the Rel protein interactions at the murine IL-12 p40 promoter and the Rel requirements for p40 transcription. p50/c-Rel and p50/p65 heterodimers were equivalent in their capacity to bind and transactivate the p40 promoter in DNA-binding and transient transactivation assays. However, an analysis of macrophages from Rel mutant mice revealed that a c-Rel-containing complex is essential for activation of the endogenous p40 gene. The implications of these results for elucidating the mechanism and relevance of family member selectivity within the mammalian immune system are discussed.
Materials and Methods
Cells and Transfections.
The J774 and 293T lines (American Type Culture Collection) were grown in DMEM containing 10% FBS. J774 cells were activated by IFN-γ (10 units/ml) and LPS (10 μg/ml). 293T cells were transfected by using a calcium phosphate procedure (17). Expression plasmids for murine p50, c-Rel, p65, and Flag-tagged c-Rel (fc-Rel) were from Ranjan Sen (Brandeis University, Waltham, MA). Flag-tagged p65 (fp65) was constructed by inserting the Flag epitope sequence at the 5′ end of the murine p65 cDNA in pcDNA3 (Invitrogen). The p40 promoter-luciferase reporter plasmid was described (6).
Peritoneal exudate cells were isolated as described (18). Before performing experiments, cells were harvested and replated at appropriate densities. Fetal liver macrophages were prepared from day 14.5 embryos. Cells (1 × 106/ml) were plated in DMEM with 30% L929-conditioned medium, 20% FBS, 1% l-glutamine, 0.5% sodium pyruvate, and 0.1% β-mercaptoethanol. The cells were cultured for 7 days, with fresh medium added as necessary.
Gel Shift Assays.
Nuclear extracts and gel shift probes were prepared as described (6). The gel shift probe contained the p40 promoter sequence from −142 to −107 (4, 6). Gel shift assays were performed as described (4, 6). Antibodies from Santa Cruz Biotechnology were: p50 (D-17), p65 (A), c-Rel (C), RelB (C-19)X, and p52(K-27)X. Protein-DNA complexes were analyzed on a 5% polyacrylamide/0.4× Tris-borate-EDTA gel.
Immunoprecipitations.
The p50 antibody (4 μg; D-17, Santa Cruz Biotechnologies) was added to nuclear extracts (50 μg) from 293T cells in 100 μl of NETN buffer (100 mM NaCl/1 mM EDTA/20 mM Tris, pH 8/0.5% Nonidet P-40). The solution was incubated on ice for 30 min and at room temperature for 5 min. Protein G-Sepharose beads (25 μl) were added, and the mixture was agitated at 4°C for 1 h. After centrifugation, the supernatant was saved and the pellet was washed four times with RIPA (150 mM NaCl/50 mM Tris, pH 7.5/0.025% deoxycolate Na+/1% Nonidet P-40). Samples were analyzed on a 10% SDS-polyacrylamide gel. Western blots were performed by using anti-flag M2 monoclonal antibody (Kodak) and anti-p50 (H-119)X antibody (Santa Cruz Biotechnologies).
ELISA.
A total of 2 × 106 cells were cultured in 3 ml of DMEM (plus 10% FBS) and activated with LPS/IFN-γ for 24 h. Then, 100 μl of the supernatants and 10-fold serial dilutions were tested by sandwich ELISA. IL-1β was detected in cell extracts. Purified rat anti-mouse IL-12 p40/p70 and IL-6, biotin anti-mouse IL-12 p40/p70 and IL-6, recombinant mouse IL-12 and IL-6, and IL-10 and TNF-α (mono/poly) ELISA kits were from PharMingen. Monoclonal anti-mouse IL-1β, biotin anti-mouse IL-1β, and recombinant murine IL-1β were from R & D Systems.
RNase Protection.
Primary macrophages were cultured in 100-mm tissue culture dishes in 10 ml DMEM containing 10% FBS and were activated with LPS/IFN-γ for 4 h. Total RNA was prepared by using TRI REAGENT (Molecular Research Center, Cincinnati). Then, 1–5 μg of the RNAs were analyzed by using RNase protection kits (PharMingen).
Results
Rel Interactions at the p40 Promoter in Vitro.
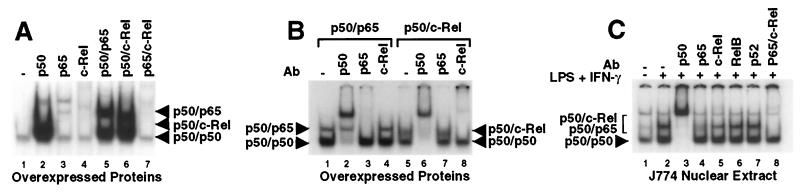
As a first step toward identifying the Rel dimer responsible for p40 activation, we determined which dimers can form stable gel shift complexes with a probe containing the Rel site from the p40 promoter. Homodimers and heterodimers containing the murine p50, p65, and c-Rel proteins were prepared by transfection of 293T cells with expression plasmids. Abundant gel shift complexes were detected with extracts containing overexpressed p50, p50 plus p65, and p50 plus c-Rel (Fig. 1A). The lower complexes correspond to p50 homodimers (see below). The upper complexes in lanes 5 and 6 correspond to p50/p65 and p50/c-Rel heterodimers, respectively. Abundant complexes were not detected when p65 and c-Rel were overexpressed individually or together (lanes 3, 4, and 7).
Figure 1.
Three Rel dimers bind the p40 Rel site in vitro. (A) Gel shift assays were performed with nuclear extracts (4 μg) from 293T cells containing overexpressed Rel proteins as indicated. (B) Gel shift assays were performed with extracts containing overexpressed p50/p65 (lanes 1–4) and p50/c-Rel (lanes 5–8). Specific antibodies were included as indicated. (C) Gel shift assays were performed with extracts (8 μg) from unactivated (lane 1) or LPS/IFN-γ-activated (lanes 2–8) J774 cells.
The effects of Rel antibodies were determined to confirm the identities of the complexes and demonstrate the specificity of the antibodies used for this study. A p50 antibody supershifted both the lower and upper complexes obtained with extracts containing p50 plus p65 and p50 plus c-Rel (Fig. 1B, lanes 2 and 6). (The band comigrating with the p50/p65 complex in lane 2 was not typically observed.) A p65 antibody abolished the upper complex obtained with the p50 plus p65 extract, but had no effect on the complexes obtained with the p50 plus c-Rel extract (Fig. 1B, lanes 3 and 7). A c-Rel antibody elicited the opposite effect (lanes 4 and 8). These results confirm the assignments of the gel shift complexes and the antibody specificities. It is worth noting that the p50/c-Rel dimer consistently yielded a diffuse complex, in contrast to the sharp p50/p65 complex (Fig. 1B, lanes 1 and 5).
To determine which complexes are detectable with nuclear extracts from p40-expressing cells, the J774 macrophage line was used. This line produces substantial quantities of p40 mRNA after LPS activation (18). Gel shift analyses with J774 nuclear extracts from unactivated and activated cells revealed the induction of two complexes (Fig. 1C, lanes 1 and 2). Antibodies to p50 supershifted both complexes (lane 3), suggesting that the lower complex contains p50 homodimers and the upper complex, one or more p50-containing heterodimers. Antibodies to p65 abolished the sharp upper complex, but a weak, diffuse complex reminiscent of the p50/c-Rel heterodimer was retained. This diffuse complex appeared to be abolished by c-Rel antibodies, with the sharp p50/p65 complex retained (lane 5). Simultaneous addition of p65 and c-Rel antibodies abolished both the sharp and diffuse complexes (lane 8). Commercial antibodies to RelB and p52 had no effect on these complexes (lanes 6 and 7). These results demonstrate that three gel shift complexes can be detected with extracts from activated J774 cells: p50/p65 and p50/c-Rel heterodimers and p50/p50 homodimers. Similar results were obtained with extracts from peritoneal macrophages (see Fig. 4C).
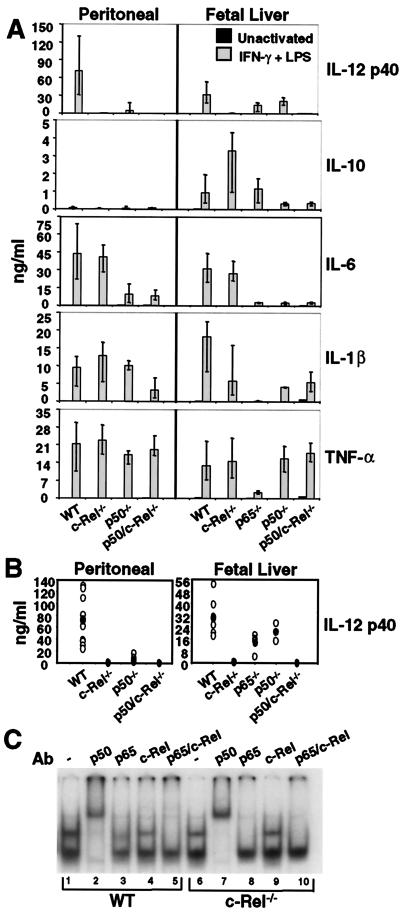
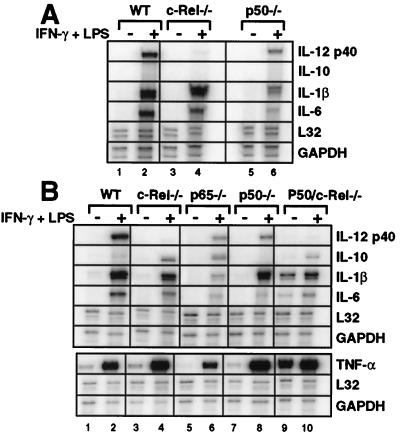
Figure 4.
A selective defect in IL-12 p40 production in c-Rel−/− macrophages. (A) Cytokine expression by peritoneal (Left) or fetal-liver (Right) macrophages was monitored by ELISA. Error bars represent the range of results rather than the standard deviation. (B) The p40 ELISA data from each sample analyzed during this study are shown (○). The ● indicate the average. (C) Gel shift assays were performed with extracts from wild-type (lanes 1–5) and c-Rel−/− (lanes 6–10) peritoneal macrophages activated with LPS/IFN-γ for 4 h.
Comparable DNA-Binding Affinities of p50/c-Rel and p50/p65.
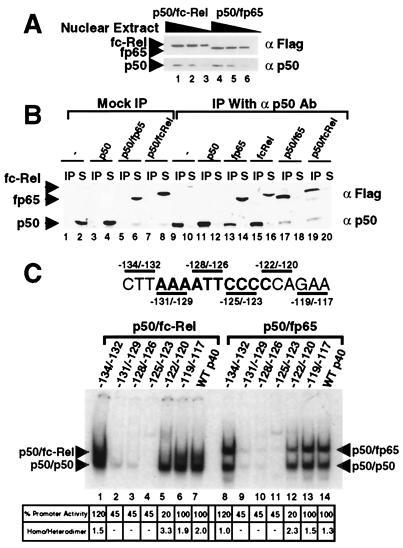
To help identify the Rel activator of p40 transcription, the relative affinities of the p50/p65 and p50/c-Rel heterodimers for the p40 Rel site were determined. For this comparison, extracts containing the heterodimers at comparable concentrations were prepared by overexpressing in 293T cells Flag-tagged versions of c-Rel and p65 (fc-Rel and fp65). Fig. 2A shows Western blot results obtained with three different amounts of a pair of extracts that contain comparable concentrations of fc-Rel and fp65. Next, coimmunoprecipitation experiments were performed to establish that the extracts contained comparable concentrations of the p50/fc-Rel and p50/fp65 heterodimers, not just comparable concentrations of fc-Rel and fp65. Immunoprecipitation with p50 antibodies resulted in the efficient removal of fp65 and fc-Rel (Fig. 2B, lanes 17–20; lanes 1–16 serve as controls), demonstrating that almost all of the fp65 and fc-Rel molecules existed as stable heterodimers with p50.
Figure 2.
p50/p65 and p50/c-Rel bind the p40 Rel site with comparable affinities. (A) 293T-cell extracts were prepared containing similar concentrations of p50/fc-Rel and p50/fp65. Three concentrations of each extract were analyzed by Western blot, using Flag and p50 antibodies. (B) The extracts were analyzed by immunoprecipitation to determine the fraction of fc-Rel and fp65 associated with p50. Control extracts were also analyzed. Immunoprecipitations were performed in the absence (lanes 1–8), and presence (lanes 9–20) of p50 antibody. The immunoprecipitated portion (IP) and 25% of the supernatant (S) were analyzed by Western blot using Flag and p50 antibodies. Extracts containing fp65 were adjusted by dilution so that comparable concentrations of fp65 were present in each reaction. Extracts containing fc-Rel were also adjusted. It is noteworthy the p50 antibody detected and precipitated the 293T-cell endogenous human p50 protein (e.g., lane 2), but did not coprecipitate detectable amounts of fc-Rel and fp65 in the absence of overexpressed murine p50. A likely explanation is that the endogenous human p50 was in fact present at a much lower concentration than the overexpressed murine p50, but yielded a strong signal on the Western blots because the p50 antibody was raised against the human protein and presumably bound the human protein with a higher affinity. (C) Gel shifts were performed with extracts from 293T cells (4 μg) containing p50/fc-Rel (lanes 1–7) or p50/fp65 (lanes 8–14) complexes overexpressed to similar levels. Probes containing the wild-type p40 Rel site (lanes 7 and 14) and various 3-bp mutations (lanes 1–6, 8–13) were analyzed. The effect of each mutation on promoter activity in a transient transfection assay is indicated at the bottom (6). Also indicated is the ratio of p50 homodimer to p50/fp65 or p50/fc-Rel heterodimer.
To compare the relative affinities of the p50/fc-Rel and p50/f65 heterodimers for the p40 Rel site, gel shift experiments were performed with equivalent amounts of each extract, resulting in p50/p65 and p50/c-Rel complexes of comparable abundance (Fig. 2C, compare lanes 7 and 14). As in Fig. 1, the p50/c-Rel complex was diffuse. However, on quantitation by phosphorimager analysis, this complex was found to contain a similar amount of radioactivity as the p50/p65 complex (581 and 562 arbitrary units, respectively). Thus, these results strongly suggest that the affinities of the two heterodimers for the p40 Rel site are comparable.
Correlation Between Promoter Activity and DNA-Binding Activities.
A second test of the functional relevance of the p50/c-Rel, p50/p65, and p50/p50 dimers was a comparison of their abilities to bind a panel of p40 promoter mutants. Dimers that recognize the nucleotides that contribute to promoter function would remain viable candidates for the relevant activator of p40 transcription. Previously, an analysis of a panel of 3- and 6-bp substitution mutants spanning the p40 Rel site revealed that the critical nucleotides for promoter activity in a transient transfection assay extend from −122 to −131 (6). Using extracts from transfected 293T cells and probes containing the 3-bp mutations, gel shift experiments were performed to monitor homodimer and heterodimer binding. The p50/c-Rel and p50/p65 complexes were reduced by the four mutations that disrupt promoter activity (Fig. 2C, lanes 2–5 and 9–12), but not by two mutations that alter flanking sequences (lanes 1, 6, 8, and 13). In contrast, the p50 homodimer complex was consistently unaltered by the −122/−120 mutation, despite the substantial effect of this mutation on promoter activity. The lack of a correlation between p50 homodimer binding and promoter activity supports the hypothesis that the p50 homodimer is unimportant for transcriptional activation.
Interestingly, the −122/−120 mutation had a relatively small effect on binding by the p50/p65 and p50/c-Rel heterodimers, although it reduced promoter activity to a greater extent than the other mutations in the Rel site (lanes 5 and 12). A reasonable explanation for the discrepancy between binding and promoter activity is that the p50 homodimer preferentially binds this mutant element, thereby suppressing promoter activity in a dominant negative manner. Consistent with this hypothesis, quantitation of the gel shift complexes revealed that the homodimer/heterodimer ratio is enhanced by the −122/−120 mutation (Fig. 2C, Bottom). (The modest 55% reduction in promoter activity by the Rel-site mutations is a unique feature of the transient transfection assay because Rel-site mutations abolished promoter activity in a stable transfection assay; ref. 6.)
Comparable Transactivation Activities of p50/c-Rel and p50/p65.
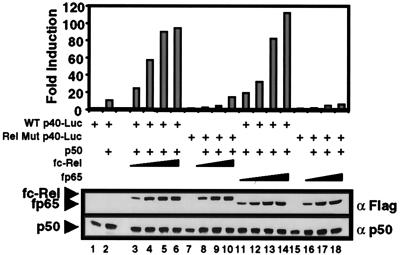
A third common method for identifying the relevant Rel activator of a promoter is to compare the abilities of overexpressed Rel proteins to transactivate a promoter-reporter plasmid. These experiments can be interpreted only if the relative concentrations of overexpressed p65 and c-Rel are known. Therefore, fp65 and fc-Rel were used, with expression levels monitored by Western blot. Overexpression of murine p50 by itself in 293T cells resulted in a 10-fold transactivation of a p40 promoter-luciferase reporter plasmid (Fig. 3, lanes 1 and 2). Inclusion of increasing concentrations of an expression plasmid for either fc-Rel or fp65 gradually enhanced transactivation, leading to maximum transactivation of approximately 100-fold (lanes 3–6 and 11–14). Transactivation depended on the Rel site within the p40 promoter because luciferase activity was greatly diminished when a promoter-reporter plasmid containing a mutation in the Rel site was used (lanes 7–10 and 15–18). Importantly, Western blot analysis using Flag antibodies confirmed that comparable concentrations of fc-Rel and fp65 were expressed (Fig. 3 Lower). Similar results were obtained in three independent experiments (data not shown). These results demonstrate that the p50/fc-Rel and p50/fp65 heterodimers possess similar capacities for transactivation of a transfected p40 promoter.
Figure 3.
Comparable transactivation of a p40 promoter-reporter plasmid by p50/p65 and p50/c-Rel. 293T cells were transfected with 2 μg of a wild-type p40 promoter-luciferase reporter plasmid (ref. 6; lanes 1–6 and 11–14) or a plasmid containing a Rel-site mutation (lanes 7–10 and 15–18). Reporter plasmids were transfected alone (lanes 1, 7, and 15) or with a p50 expression plasmid (0.1 μg; lane 2) and increasing amounts of fc-Rel (2–16 μg; lanes 3–6 and 8–10) or fp65 (0.2–1.6 μg; lanes 11–14 and 16–18) expression plasmids. The fold-induction values were determined by dividing the luciferase activity by the basal activity of the reporter alone (lane 1 for the wild-type and lanes 8 and 16 for the mutant constructs). fc-Rel, fp65, and p50 were monitored by Western blot (Lower). The p50 detected in lanes 1, 7, and 15 corresponds to endogenous human p50.
One potential caveat of using the Flag-tagged proteins is that the tag may alter their activities. To examine this possibility, flag-tagged and untagged p65 and c-Rel were compared. Similar concentrations of p65 and fp65, as determined by Western blot by using p65 antibodies, transactivated the p40 promoter to a similar extent (data not shown). In addition, similar concentrations of c-Rel and fc-Rel, as determined by Western blot by using c-Rel antibodies, yielded similar transactivation results (data not shown).
Selective Defect in IL-12 p40 Production in c-Rel−/− Macrophages.
The above results suggest that both the p50/c-Rel and p50/p65 heterodimers may be capable of activating the endogenous p40 gene. Alternatively, family member specificity may not be manifested in these assays. Rather, specificity may be observed only when the Rel requirements are analyzed in a native chromatin environment. To distinguish between these possibilities, p40 expression was examined in macrophages from mice lacking specific Rel family members.
Thioglycollate-elicited peritoneal macrophages were isolated from wild-type C57BL/6 mice and from c-Rel−/−, p50−/−, and p50/c-Rel−/− mice (19, 20). Cytokine production was then monitored by ELISA 24 h after stimulation with LPS/IFNγ. The results revealed a striking decrease in secreted p40 protein in the c-Rel−/− cells (Fig. 4A, Top Left). In five experiments, p40 protein was reduced between 43- and 1170-fold in activated c-Rel−/− cultures, with an average decrease of 235-fold (Fig. 4B, Left). In contrast, the concentrations of IL-6, IL-1β, and TNF-α were not significantly altered (Fig. 4A, Left). Production of the anti-inflammatory cytokine, IL-10, was low but unaltered in the c-Rel−/− macrophages (Fig. 4A, Left). The unaltered production of these cytokines demonstrates that cell activation proceeded normally, suggesting that the p40 gene is a direct target of a c-Rel-containing complex.
Consistent with the hypothesis that a p50/c-Rel heterodimer is the relevant activator of p40 transcription, p40 concentrations were reduced 15-fold in p50−/− peritoneal macrophages (Fig. 4A, Left)., In p50/c-Rel−/− macrophages, p40 protein was undetectable (Fig. 4A, Left). In both of these strains, the other cytokines were unaffected or were reduced to a modest extent. The substantial expression of p40 in the p50−/− cells may be a reflection of redundancy between p50 and p52 (A.H., unpublished data).
To monitor the effect of the c-Rel deficiency on the expression of Rel dimers, gel shift experiments were performed with the p40 probe and extracts from activated wild-type and c-Rel−/− macrophages. Abundant p50/p50, p50/p65, and p50/c-Rel complexes were detected in wild-type extracts (Fig. 4C, lanes 1–5), closely resembling the results obtained in J774 cells (Fig. 1). In c-Rel−/− extracts, the p50/p50 and p50/p65 complexes were detected (lanes 6–10). However, the diffuse p50/c-Rel complex was absent, as is most apparent from an examination of the results in the presence of p65 antibodies (compare lanes 3 and 8).
To examine further the possibility that the p40 promoter is a direct target of a c-Rel-containing complex, it was necessary to analyze p40 production in p65−/− macrophages. This experiment could not be performed with peritoneal macrophages because disruption of the p65 gene results in embryonic lethality (21). We therefore used macrophages differentiated in vitro from day-14.5 fetal liver cells (22). Activation of wild-type cells with LPS/IFNγ yielded p40, IL-6, IL-1β, and TNF-α at concentrations comparable to those observed with the peritoneal macrophages (Fig. 4A, WT).
Fetal liver-derived macrophages from c-Rel−/− mice produced p40 at greatly reduced concentrations (44-fold; Fig. 4 A and B, Right). The most notable effect of the c-Rel deficiency on the other cytokines was the 3- to 4-fold enhancement of IL-10. Although IL-10 has been reported to suppress IL-12 p40 expression (23), the reduction in p40 expression in the c-Rel−/− cells appears to be independent of the IL-10 enhancement. First, IL-10 was not detected in supernatants from c-Rel−/− peritoneal macrophages, despite the reduction in p40 expression. Second, addition of IL-10 antibodies to the cultured macrophages had no effect on p40 production (data not shown). Third, IL-10 suppresses all of the proinflammatory cytokines shown here (24), yet IL-6 and TNF-α were unaffected in the c-Rel−/− cells.
In macrophages prepared from p65−/− fetal livers, the concentration of p40 protein was reduced by only 2-fold (Fig. 4 A and B). Even this minor reduction probably overestimates the effect on p40 expression because, when the supernatants were collected for the ELISA, a large fraction of the p65−/− macrophages appeared to be apoptotic under a light microscope. Therefore, the reduced production of p40 and other cytokines may result, at least in part, from cell death (that is presumably mediated by TNF-α). These results support the hypothesis that a c-Rel-containing complex is an essential activator of p40 transcription.
Analysis of Cytokine mRNAs.
To determine whether the ELISA results reflect differences in steady-state mRNAs, total RNA was prepared from wild-type and mutant peritoneal and fetal liver-derived macrophages before and after activation with LPS/IFN-γ for 4 h. A comparison of RNase protection assay results revealed a close correlation with the ELISA results (Fig. 5). IL-12 p40 mRNA was reduced to nearly undetectable levels in the c-Rel−/− cells, whereas IL-1β, IL-6, and TNFα mRNAs were unaffected. The most significant difference between the ELISA and RNase protection results was that, in p65−/− macrophages from fetal liver, p40 mRNA was consistently reduced to a greater extent (approximately 10-fold) than was p40 protein (Fig. 5B). The reason for this difference is unknown, but because the mRNAs for almost all of the genes monitored are substantially reduced in the apoptotic p65−/− macrophages, this reduction may be because of a general effect of the p65 deficiency on cell growth and viability.
Figure 5.
Steady-state mRNA levels in wild-type and Rel-deficient macrophages. Total RNA was isolated from peritoneal (A) and fetal liver-derived (B) macrophages before and 4 h after activation with LPS/IFN-γ. Specific mRNA levels were determined by RNase protection. The data shown correspond only to the cytokine mRNAs analyzed by ELISA in Fig. 4.
Discussion
This study revealed much greater selectivity in the Rel-protein requirements for expression of a proinflammatory cytokine gene than was anticipated. The biological significance of the c-Rel requirement for p40 expression remains to be determined. Although p40 transcription is regulated by many of the same inducers and antiinflammatory agents as other proinflammatory cytokine genes, it is uniquely regulated by a number of agents, whose mechanisms of action may depend on c-Rel (2, 3, 25). IFN-γ is a prominent example of an agent that enhances p40 expression, with relatively little effect on other proinflammatory cytokines (3, 25). However, c-Rel is not required solely for IFN-γ responsiveness because the c-Rel requirement was observed on LPS stimulation in the absence of IFN-γ (data not shown). Another unique feature of p40 induction is its requirement for new protein synthesis (20, 24). Furthermore, p40 is expressed in immature cells of the myeloid lineage as well as in mature macrophages, whereas TNF-α and IL-1β are expressed only in mature macrophages (26). The c-Rel requirement may be related to one of these unique features of p40 regulation or to many others that have been reported (25).
It is most intriguing to consider the possibility that the c-Rel requirement may be related to IL-12's unique biological role as an inducer of T helper 1 cell differentiation (2, 3). This function would be reminiscent of the selective functions of Rel proteins in Drosophila, where the expression of antifungal and antibacterial peptides exhibit different Rel-protein requirements (27, 28). In some respects, the IL-12 p40 gene in mammals is analogous to the antifungal and antibacterial genes in Drosophila because its expression is required for an effective immune response against a specific subset of pathogens (in particular, intracellular pathogens). The data therefore suggest a potential parallel between Drosophila and mammals; in these distantly related organisms, unusually strong requirements for specific Rel family members appear to be associated with the induction of pathogen-specific genes.
The c-Rel requirement for p40 transcription is most likely because of an essential interaction between a c-Rel-containing complex (presumably a p50/c-Rel dimer) and the Rel site within the p40 promoter. This hypothesis is based on the following reasoning: First, the in vitro DNA-binding studies provided evidence that the only abundant dimers in activated macrophages capable of binding the p40 Rel site are p50/p50, p50/p65, and p50/c-Rel. The p50 homodimer is unlikely to be an essential activator because p50 lacks a transactivation domain and does not recognize all of the nucleotides that are important for promoter function (i.e., −122; see Fig. 2C). The p50/p65 dimer can transactivate the promoter in transfection assays, but p40 mRNA and protein remain quite abundant in p65−/− macrophages, despite the apoptotic phenotype of these cells. In contrast, p40 mRNA and protein are reduced to nearly undetectable levels in c-Rel−/− macrophages.
If a p50/c-Rel dimer is truly required for the activity of the Rel site within the p40 promoter, the mechanistic basis of selectivity may be difficult to elucidate. A likely explanation for the lack of selectivity in the DNA-binding assays is that differential binding affinities are not responsible for the c-Rel requirement. Furthermore, the c-Rel requirement does not appear to be because of differences in the intrinsic transactivation activities of p50/p65 and p50/c-Rel. Although selective transactivation of the p40 promoter by c-Rel was observed in a previous study (4), the p65 and c-Rel expression levels were not compared. Most likely, it will be necessary to study the promoter in its native chromosomal environment to elucidate the mechanistic basis of selectivity. c-Rel may carry out a selective interaction with another DNA-binding protein, coactivator complex, or general transcription factor that is required for promoter activity, but only when the promoter is in a native chromosomal context.
Although the current data suggest an essential role for a c-Rel-containing complex at the p40 promoter, they do not rule out the possibility that p50/p65 and p50/c-Rel complexes are equally competent for activation of the endogenous promoter, with an essential c-Rel interaction occurring elsewhere in the locus. The current data also do not rule out the possibility that the c-Rel requirement for p40 transcription is indirect. Although this possibility cannot be excluded until the mechanistic basis of the c-Rel requirement is fully understood, it seems unlikely, given the highly specific effect of the c-Rel deficiency.
In conclusion, the selective defect in IL-12 p40 transcription in c-Rel−/− macrophages provides a rare opportunity to examine the mechanistic basis of Rel family member selectivity. By introducing c-Rel mutants and c-Rel/p65 chimeras into the c-Rel−/− macrophages, it should be possible to determine the domains of c-Rel that are responsible for the selectivity, leading to the proteins that interact with the selectivity domains. Future analyses of the properties of p40 regulation also may reveal the biological significance of the selectivity.
Acknowledgments
We thank Ranjan Sen for Rel expression plasmids. This work was supported by U.S. Public Health Service Grants R01-AI42549 (to D.B.) and T32-AI07323 (to S.S.). S.T.S. is an Investigator with the Howard Hughes Medical Institute.
Abbreviations
- LPS
lipopolysaccharide
- fc-Rel
Flag-tagged c-Rel
- fp65
Flag-tagged p65
- TNF-α
tumor necrosis factor α
Footnotes
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.230436397.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.230436397
References
- 1.Sweet M J, Hume D A. J Leukocyte Biol. 1996;60:8–26. doi: 10.1002/jlb.60.1.8. [DOI] [PubMed] [Google Scholar]
- 2.Gately M K, Renzetti L M, Magram J, Stern A S, Adorini L, Gubler U, Presky D H. Annu Rev Immunol. 1998;16:495–521. doi: 10.1146/annurev.immunol.16.1.495. [DOI] [PubMed] [Google Scholar]
- 3.Trinchieri G. Adv Immunol. 1998;70:83–243. doi: 10.1016/s0065-2776(08)60387-9. [DOI] [PubMed] [Google Scholar]
- 4.Murphy T L, Cleveland M G, Kulesza P, Magram J, Murphy K M. Mol Cell Biol. 1995;15:5258–5267. doi: 10.1128/mcb.15.10.5258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ma X, Chow J M, Gri G, Carra G, Gerosa F, Wolf S F, Dzialo R, Trinchieri G. J Exp Med. 1996;183:147–157. doi: 10.1084/jem.183.1.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Plevy S E, Gemberling J H, Hsu S, Dorner A J, Smale S T. Mol Cell Biol. 1997;17:4572–4588. doi: 10.1128/mcb.17.8.4572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ghosh S, May M J, Kopp E B. Annu Rev Immunol. 1998;16:225–260. doi: 10.1146/annurev.immunol.16.1.225. [DOI] [PubMed] [Google Scholar]
- 8.Hiscott J, Marois J, Garoufalis J, D'Addario M, Roulston A, Kwan I, Pepin N, Lacoste J, Nguyen H, Bensi G, Fenton M. Mol Cell Biol. 1993;13:6231–6240. doi: 10.1128/mcb.13.10.6231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Libermann T A, Baltimore D. Mol Cell Biol. 1990;10:2327–2334. doi: 10.1128/mcb.10.5.2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Collart M A, Baeuerle P, Vassalli P. Mol Cell Biol. 1990;10:1498–1506. doi: 10.1128/mcb.10.4.1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Köntgen F, Grumont R J, Strasser A, Metcalf D, Li R, Tarlinton D, Gerondakis S. Genes Dev. 1995;9:1965–1977. doi: 10.1101/gad.9.16.1965. [DOI] [PubMed] [Google Scholar]
- 12.Gerondakis S, Strasser A, Metcalf D, Grigoriadis G, Scheerlinck J Y, Grumont R J. Proc Natl Acad Sci USA. 1996;93:3405–4509. doi: 10.1073/pnas.93.8.3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grigoriadis G, Zhan Y, Grumont R J, Metcalf D, Handman E, Cheers C, Gerondakis S. EMBO J. 1996;15:7099–7107. [PMC free article] [PubMed] [Google Scholar]
- 14.Grumont R J, Rourke I J, Gerondakis S. Genes Dev. 1999;13:400–411. doi: 10.1101/gad.13.4.400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grumont R J, Gerondakis S. J Exp Med. 2000;191:1281–1291. doi: 10.1084/jem.191.8.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kunsch C, Ruben S M, Rosen C A. Mol Cell Biol. 1992;12:4412–4421. doi: 10.1128/mcb.12.10.4412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ausubel F M, Brent R E, Kingston R, Moore D D, Seidman J G, Smith J A, Struhl K. Current Protocols in Molecular Biology. New York: Wiley; 1994. [Google Scholar]
- 18.Weinmann A S, Plevy S E, Smale S T. Immunity. 1999;11:665–675. doi: 10.1016/s1074-7613(00)80141-7. [DOI] [PubMed] [Google Scholar]
- 19.Sha W C, Liou H-C, Tuomanen E I, Baltimore D. Cell. 1995;80:321–330. doi: 10.1016/0092-8674(95)90415-8. [DOI] [PubMed] [Google Scholar]
- 20.Tumang J R, Owyang A, Andjelic S, Jin Z, Hardy R R, Liou M L, Liou H-C. Eur J Immunol. 1998;28:4299–4312. doi: 10.1002/(SICI)1521-4141(199812)28:12<4299::AID-IMMU4299>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 21.Beg A A, Sha W C, Bronson R T, Ghosh S, Baltimore D. Nature (London) 1995;376:167–170. doi: 10.1038/376167a0. [DOI] [PubMed] [Google Scholar]
- 22.Beg A A, Baltimore D. Science. 1996;274:782–784. doi: 10.1126/science.274.5288.782. [DOI] [PubMed] [Google Scholar]
- 23.Aste-Amezaga M, Ma X, Sartori A, Trinchieri G. J Immunol. 1998;160:5936–5944. [PubMed] [Google Scholar]
- 24.Fiorentino D F, Zlotnik A, Mosmann T R, Howard M, O'Garra A. J Immunol. 1991;147:3815–3822. [PubMed] [Google Scholar]
- 25.Sutterwala F S, Mosser D M. J Leukocyte Biol. 1999;65:543–551. [PubMed] [Google Scholar]
- 26.Kubin M, Chow J M, Trinchieri G. Blood. 1994;83:1847–1855. [PubMed] [Google Scholar]
- 27.Hedengren M, Åsling B, Dushay M S, Ando I, Ekengren S, Wihlborg M, Hultmark D. Mol Cell. 1999;4:827–837. doi: 10.1016/s1097-2765(00)80392-5. [DOI] [PubMed] [Google Scholar]
- 28.Rutschmann S, Jung A C, Hetru C, Reichhart J-M, Hoffmann J A, Ferrandon D. Immunity. 2000;12:569–580. doi: 10.1016/s1074-7613(00)80208-3. [DOI] [PubMed] [Google Scholar]