Abstract
Antimicrobial peptides constitute an important component of the mammalian innate immune response. Several types of antimicrobial peptides, including the β-defensins, are produced at epithelial surfaces in response to infectious threats. Here we show that a class of small molecules, including l-isoleucine and several of its analogs, can specifically induce epithelial β-defensin expression. This induction is transcriptional in nature and involves activation of the NF-κB/rel family of trans-activating factors. We hypothesize that these substances represent unique markers for the presence of pathogens and are recognized by innate immune pattern recognition receptors. Isoleucine or its analogs ultimately may have clinical utility as novel immunostimulants that could bolster the barrier defenses of mucosal surfaces.
Antimicrobial peptides represent an important component of the innate immune defense of organisms ranging from plants to insects to humans. They are broad-spectrum, surface-active agents that kill microbes by forming pores in their membranes. The defensins are a subclass of antimicrobial peptides. In mammals defensins are present in neutrophil granules where they are necessary for the nonoxidative killing of phagocytized microbes (1). It is now well established that defensins also are produced at virtually all mammalian epithelial surfaces, including those of the skin, airway, gut, and urogenital tracts (2, 3). Expression of some epithelial defensins is constitutive and contributes to a noninflammatory antimicrobial barrier at the epithelial surface. Other defensins are inducible and are highly expressed at sites of inflammation or infection (2, 4, 5). Impairment of defensin function leads to susceptibility to infection of the airway in cystic fibrosis (6) and to enhanced Salmonella infection in the mouse intestinal tract (7). In addition to their direct antimicrobial activities, β-defensins are chemotactic for memory T cells and dendritic cells, suggesting that they play an important role in the integration of the innate and acquired immune responses (8).
The molecular mechanisms underlying epithelial defensin induction remain largely unexplored. Pattern recognition receptors (9) likely play a critical role in this process, as has been shown in the case of CD-14-mediated induction of β-defensins by bacterial lipopolysaccharide (10). Whole heat-killed bacteria and fungi induce human β-defensin-2 in human keratinocytes but the molecular basis of this response is not yet known (5). Inflammatory cytokines such as tumor necrosis factor α and IL-1β also have been shown to induce β-defensins (11–13). Because pharmacological induction of defensins at epithelial barriers may have therapeutic utility, we initiated a search for novel molecules that could induce epithelial defensin production by using cell-based assays. We found that l-isoleucine and its analogs are highly specific β-defensin inducers in epithelial cells.
Materials and Methods
Cell Culture.
Madin–Darby bovine kidney (MDBK) epithelial cells were obtained from the American Type Culture Collection and maintained in MEM (GIBCO/BRL) supplemented with 10% horse serum and 1% MEM nonessential amino acids (GIBCO/BRL) in a humidified 37°C incubator containing 5% CO2. The medium was supplemented with G418 (0.4 mg/ml) to maintain the stably transformed MDBK cell line.
Cell-Based Assay Using Reverse Transcription–PCR and Quantitative PCR.
For stimulation, cells were plated at a concentration of 5 × 105 cells per well in 6-well dishes for 24 h. For initial experiments the MEM then was replaced by bronchial epithelial growth medium (Clonetics) and cells were incubated in the presence of test substances for 24 h. For subsequent experiments evaluating dose–response relationships, structure-activity relationships, and NF-κβ stimulation MEM was replaced by bronchial epithelial growth/labeling medium lacking isoleucine (Clonetics). Total RNA was extracted with Trizol solution, as recommended by the manufacturer (Life Technologies, Gaithersburg, MD). After DNase (GIBCO/BRL) treatment of 1 μg of total RNA, reverse transcription was performed. PCR was carried out by using Perkin–Elmer Ampli Taq DNA polymerase under the following cycling conditions: 1 min at 95°C, 1 min at 52°C, and 1 min at 72°C for 30 cycles and a final 15-min extension at 72°C. The sequence of the biotin- and ruthenium-conjugated primers (Baron Technologies, Milford, CT) used to quantify bovine enteric β-defensin (EBD) transcripts were designed based on the sequence of the highly related lingual antimicrobial peptide cDNA. The sequences of these primers were as follows: forward, 5′-CTC TTC CTG GTC CTG TCT-3′, and reverse, 5′-CTT CTT TTA CTT CCT CCT GCA GCA-3′. The conjugated bovine tubulin primers were 5′-GTT CCC AAA GAT GTC AAT GCT GCC-3′ and 5′-ATG CTG CAA GGC TGA AAG GAA TGG-3′. The PCR products were quantified by using the Perkin–Elmer quantitative PCR (Perkin-Elmer System 5000) instrument, and the values obtained for the EBD products were normalized with those corresponding to tubulin.
Plasmids.
PCR was performed on bovine genomic DNA (CLONTECH) by using primers specific for the β-defensin promoter region (nucleotides 1–882). The primers contained engineered restriction sites for MluI and BglII, and their sequences were as follows: 5′-GCC CGC ACG CGT ATT ACT TTC CTT CCA AGG AAT AAG CAT C-3′ and 5′-GGC GCC AGA TCT GGC GTC CCG AGC TCT TCG GCT GAT GCT GGA-3′. The DNA product then was cloned into the TA cloning vector (Invitrogen) before sequencing. Finally, the enteric β-defensin promoter region was cloned into the pGL2 basic vector (Promega) carrying the firefly luciferase gene by using standard procedures.
Stable Transfections and Reporter Gene Assays.
MDBK cells were cotransfected with the newly engineered pGL2 plasmid (described above) and the LXSN vector (14), carrying the G418 resistance gene, in a ratio of 1:5 (LXSN/pGL2) by using Fugene 6 transfection kit (Roche Molecular Biochemicals), following the instructions recommended by the manufacturer. The cells were maintained in MEM, with 10% horse serum and 1% MEM nonessential amino acids, and supplemented with G418 (0.4 mg/ml) for the selection of a stable cell line.
For stimulation, the transfected cells were plated at a concentration of 2 × 104 cells/well into a 96-well dish (Packard) and maintained overnight at 37°C. The cells were washed with PBS and incubated in bronchial epithelial growth/labeling medium lacking isoleucine (Clonetics) as described above for the PCR-based assay. The cells then were exposed to the test substances for 16–24 h. Luciferase activity was detected by using the chemiluminescent reporter assay system, LucLite (Packard). The luciferase substrate was added to the culture medium, and luminescence was measured with a Top Count (Packard) instrument.
Extraction and Purification.
An autolysed yeast extract (Saccharomyces cerevisiae) (Food Ingredients Specialities, Solon, OH) at a concentration of 400 mg/ml in water was brought to 90% ethanol. The resulting suspension was clarified by centrifugation. The supernatant was adjusted to 50% acetone, and the resulting precipitate was collected by centrifugation and lyophilized.
The material was loaded on a Kromasil C18 HPLC column (250 × 10 mm) equilibrated with 15 mM heptaflourobutyric acid (Aldrich). Elution was performed with a linear gradient of 0–40% acetonitrile (J.T. Baker) in the same buffer over 90 min at a flow rate of 6 ml/min. Data were collected at 280 nm. Fractions were collected automatically, concentrated under vacuum, and reconstituted in deionized water.
HPLC analysis with Ortho-Phthaldialdehyde (OPA) Derivatization.
OPA reagent solution (Sigma) was formulated for precolumn derivatization of amines and amino acids. Briefly, 10 μl of the sample (1–10 μg) was added to 10 μl of OPA reagent. The mixture then was analyzed by HPLC equipped with a fluorescence detector. The mixture was loaded on a reversed-phase column, Kromasil C18 (250 × 4.6 mm), equilibrated with 10% acetonitrile in 0.1% trifluoroacetic acid (TFA) (J.T. Baker). Elution was carried out by using a linear gradient of acetonitrile from 18% to 63% of acetonitrile in 0.1% TFA over 30 min at a flow rate of 1 ml/min and a temperature of 45°C.
HPLC Analysis with Evaporative Light Scattering Detection (ELSD).
HPLC analysis monitored with an ELSD detector, was performed on a Kromasil C18 column (250 × 4.6 mm) equilibrated with 15 mM heptaflourobutyric acid and a linear gradient of acetonitrile, from 0% to 38%, over 40 min, at a flow rate of 1 ml/min.
1-Fluoro-2.4-Dinitrophenyl-5-Alanine Amide (FDAA) Derivatization.
Amino acids (l- and d-isomers) and FDAA (Sigma-Aldrich) were used for the synthesis of diastereomers as described (15). HPLC separations were carried out on an ODS-AQ C18 reversed-phase column (100 × 4.6 mm; YMC, Wilmington, DE) (100 × 4.6 mm) reversed-phase column equilibrated in 0.1% trifluoroacetic acid at a temperature of 45°C. Elution was performed with a linear gradient of 20–70% acetonitrile over 6 min at 2 ml/min. Absorbance was monitored at 340 nm.
NMR Analysis.
Proton and carbon spectra were obtained with a 5-mm dual probe on a Bruker AC-400 at 400 MHz and 100 MHz. Proton and carbon chemical shifts in CD3OD were reported relative to tetramethylsilane with residual signal of CD3OD serving as a secondary reference at 3.3 ppm for proton and 49.15 ppm for carbon spectra.
Electrophoretic Mobility Shift Assay.
MDBK cells were grown to 80% confluency and were serum-deprived during the treatment with isoleucine (Aldrich) at a concentration of 25 μg/ml, followed by preparation of nuclear extracts. Nuclear extracts were made as described (16). Complementary oligonucleotides representing portions of the mouse Ig κ gene containing the NF-κB site (upper strand 5′-CCC CAG AGG GGA CTT TCC GAG AGG CTC-3′; lower strand 5′-GGG GAG CCT CTC GGA AAG TCC CCT CTG-3′) and bovine enteric β-defensin promoter, EBD 800 (upper strand 5′-GGG AGC CAG CGT GGA ATT CCT CCC AGA ACC TGG-3′; lower strand 5′-CCC CCA GGT TCT GGG AGG AAT TCC ACG CTG GCT-3′) were annealed and 3′ end-labeled with [α-32P]dCTP with Klenow polymerase via standard procedures. Binding reactions were performed by preincubating 10 μg of nuclear extract protein in 20 mM Hepes (pH 7.9), 60 mM KCl, 1 mM MgCl2, 0.1 mM EDTA, 10% glycerol, 0.5 mM DTT, and 2 μg of poly(dI-dC) on ice for 10 min, followed by addition of the double-stranded 32P-labeled probe and a second incubation at room temperature for 20 min. Samples were loaded directly onto nondenaturing 6% polyacrylamide gels (29:1 acrylamide to bisacrylamide) prepared in 45 mM Tris borate/45 mM boric acid/0.1 mM EDTA (0.5× TBE). Competitions were performed by using unlabeled double-stranded oligonucleotides. The specific NF-κB site competitor contained the mouse Ig κ sequence listed above. The nonspecific competitor consisted of a portion of the human haptoglobin promoter (17) containing a C/EBP binding site (5′-CCC GAT CCA AGT GTG AAG CAA GAG CG-3′ and 5′-GGG CGC TCT TGC TTC ACA CTT GGA TC-3′). The mutant NF-κB competitor probe was identical to the NF-κB probe except for a single base pair mutation that is known to prevent NF-κB binding (18) in the second position of the NF-κB consensus recognition sequence (5′-CCC CAG AGG CGA CTT TCC GAG AGG CTC-3′ and (5′-GGG GAG CCT CTC GGA AAG TCG CCT CTG-3′). Electrophoresis was performed at room temperature for 2–2.5 h at approximately 170 V. The gels then were dried and exposed to Kodak MS film with the appropriate intensifying screens.
Results
Isolation and Characterization of Defensin-Inducing Activity.
We suspected that microbial products or extracts might contain novel substances able to stimulate innate host defense mechanisms. Therefore, we chose to screen these products in a cell-based assay in which we monitored mRNA levels of the epithelially expressed EBD (19). Among several preparations, a food grade autolysate of the yeast S. cerevisiae was found to be positive. To purify and identify the molecule(s) responsible for stimulating the cells to produce defensins, we first subjected the crude yeast extract to a two-step precipitation with ethanol and acetone. The resulting precipitate elicited the same level of defensin induction as the crude extract. As shown in Table 1, this purification step effected about a 10-fold enrichment of the active substance(s). The active precipitate, consisting primarily of amino acids and salts, then was further fractionated by HPLC on a reversed-phase column under acidic conditions, as described in Materials and Methods. The HPLC fractions, eluted with a linear gradient of acetonitrile, then were tested by using the cell-based induction assay. The structures of the different constituents of the most active fractions were determined by NMR and confirmed by amino acid analysis and ortho-pthaldialdehyde derivatization (data not shown). We tested for the presence of molecules other than amino acids in the precipitate by HPLC with evaporative light scattering detection (data not shown). Taken together, these data showed that the active fractions contained only the amino acid isoleucine as their common component.
Table 1.
Purification of the epithelial defensin inducer from yeast extract
| Weight, mg | Specific activity, unit/mg | Total units | Yield, % | |
|---|---|---|---|---|
| Yeast extract | 50 × 103 | 1 | 5 × 104 | 100 |
| Acetone precipitate | 2.7 × 103 | 10.67 | 2.88 × 104 | 58 |
| Isoleucine | 490 | 50.80 | 2.49 × 104 | 50 |
The defensin-inducing activity was purified as described in Materials and Methods. A unit is defined as the amount of substance that effects peak stimulation of defensin expression.
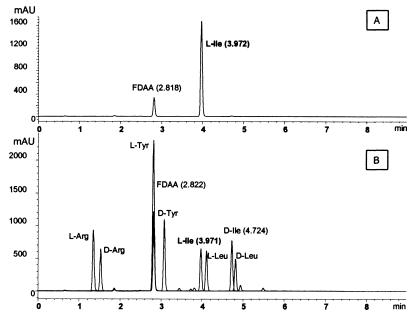
To elucidate the chiral configuration of the purified isoleucine, the fractions containing isoleucine were chemically derivatized by using FDAA. The diastereomers generated then were separated by HPLC on a reversed-phase column. The chromatograms revealed that the fractions contained only the l-enantiomers of the amino acids (data not shown). The chromatogram (Fig. 1) obtained after FDAA derivatization of an active fraction containing only isoleucine demonstrated that it contained only the l-enantiomer. We conclude that l-isoleucine is the substance responsible for the observed activation of the β-defensin gene. The presence of activating concentrations of isoleucine in the yeast autolysate is likely due to the highly digested nature of the autolysate and is therefore serendipitous rather than reflective of a yeast-specific pattern.
Figure 1.
Configuration analysis of the active substance after derivatization with FDAA. (A) Reversed-phase HPLC separation of isoleucine purified from the yeast extract and subsequently derivatized. Comparison to the standards in B indicates that the purified isoleucine consists entirely of the l-enantiomer. (B) HPLC separations on a reversed-phase column for l- and d-diastereomers (l/d-aa-DNP-l-Ala-NH2) of the standard amino acids Arg, Tyr, Ile, and Leu.
Isoleucine Is a Specific Inducer of Epithelial Defensin Expression.
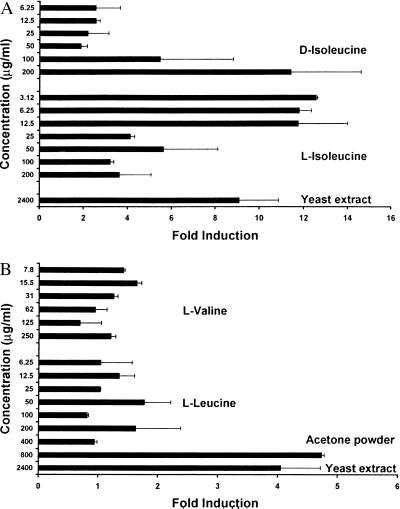
We measured the relative expression of defensin after the MDBK cells were stimulated with the l- and d-enantiomers of isoleucine over a range of concentrations between 3.12 μg/ml and 200 μg/ml. l-isoleucine induced the expression of the β-defensin by 10- to 12-fold with peak activity between 3.12 μg/ml and 12.5 μg/ml (see Fig. 2A). d-isoleucine was substantially less potent than its l-enantiomer, requiring a concentration of 200 μg/ml to produce a similar level of induction to that observed with approximately 3 μg/ml l-isoleucine in this experiment (Fig. 2A), supporting the specificity of l-isoleucine and suggesting the involvement of a chiral receptor.
Figure 2.
Relative expression of EBD after treatment of the MDBK cells with branched chain amino acids. After stimulation with the test substances, total RNA was extracted from the cells. Reverse transcription–PCR was performed on 1 μg of total RNA, and the PCR products were quantified by quantitative PCR. The values obtained for the EBD products were normalized with those corresponding to tubulin. (A) Treatment with l- and d-isoleucine. (B) Treatment with l-leucine and l-valine.
The results shown for l-isoleucine are representative of many experiments in which we observed reduced or no activation at higher isoleucine concentrations. Many experiments testing defensin induction with isoleucine yielded “bell-shaped” dose-response curves. This type of dose response is commonly observed with biological response modifiers and likely represents homeostatic down modulation of the response by the cell in the face of a chronic strong stimulus. Peak activating concentrations of isoleucine in these biological assays often varied by 1–2 dilutions from experiment to experiment, with typical peak activity in the range of 6.25 μg/ml to 25 μg/ml. The lower defensin expression levels observed at higher isoleucine concentrations were not caused by cellular toxicity because the cells remained viable based on morphology and trypan blue exclusion under these conditions (data not shown). The other members of the branched-chain amino acid family, valine and leucine, do not promote the activation of the defensin gene in the concentration range where isoleucine exhibits its effect (Fig. 2B). These results demonstrate that isoleucine is a highly specific inducer of β-defensin expression. Additional evidence for this specificity came from tests of all of the remaining naturally occurring amino acids, which were observed to have no effect under the conditions tested (data not shown).
Structure-Activity Relationship Between Isoleucine and Epithelial Defensin Induction.
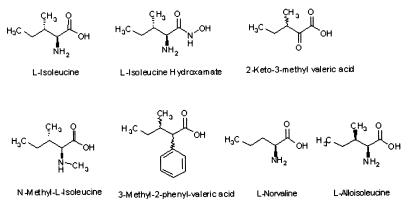
To further elucidate the structure-function relationships between isoleucine and epithelial defensin induction, we tested a variety of analogs of isoleucine in a range of concentrations between 1.56 μg/ml and 400 μg/ml by measuring the expression of luciferase in the defensin promoter/reporter gene assay. The results of these studies are shown in Table 2. The structures of the tested analogs are shown in Fig. 3. In these experiments peak activity was centered on a concentration of 25 μg/ml isoleucine, and we therefore report the activities of the analogs at this concentration. Several modifications led to molecules of similar potency to l-isoleucine. These include l-isoleucine hydroxamate in which the carboxylic acid group is modified and dl,2-keto-3-methylvaleric acid where the amine group is substituted by a keto function. When the amine group is replaced by a phenyl group (3-methyl, 2-phenylvalerate) or blocked by a methyl group (N-methyl-l-isoleucine) biological activity is lost, suggesting that the amino substituent is important in binding and/or steric interactions leading to activation of the defensin gene. Modifications of the alkyl chain affect the biological activity of isoleucine as well. The C3 enantiomer (l-alloisoleucine) and the straight chain analog (l-norvaline) of isoleucine are both inactive. These results, together with those presented above for valine and leucine, show the importance of the primary amine group, the configuration of the alkyl chain, and the two chiral centers of the molecule. Further, they demonstrate that isoleucine is a specific inducer of epithelial defensins and strongly suggest that it acts via a chiral binding site, likely in a polypeptide receptor or enzyme. The observation that isoleucine stimulates defensin promoter-driven luciferase production strongly supports the idea that the regulation of defensin mRNA expression described above is primarily transcriptional in nature.
Table 2.
Structure-activity relationship between isoleucine and epithelial defensin induction
| Treatment, 25 μg/ml | Fold induction |
|---|---|
| No treatment | 1.0 ± 0.17 |
| l-isoleucine | 5.0 ± 0.17 |
| l-alloisoleucine | 1.3 ± 0.23 |
| l-norvaline | 2.4 ± 0.16 |
| 3-Methyl-2-phenyl-valeric acid | 1.4 ± 0.13 |
| N-methyl-l-isoleucine | 1.8 ± 0.17 |
| d,l-2-keto-3-methyl-valeric acid | 4.5 ± 0.05 |
| l-isoleucine hydroxamate | 6.1 ± 0.71 |
The compounds were tested by using the defensin promoter/reporter gene assay. All compounds were tested over a range of 1.56 to 400 μg/ml. The reported induction ratios are those observed at the peak isoleucine activating concentration of 25 μg/ml in these experiments.
Figure 3.
Structures of compounds tested in structure-activity studies. Defensin-inducing activities are listed in Table 2.
Isoleucine Induces NF-κB/rel Activities.
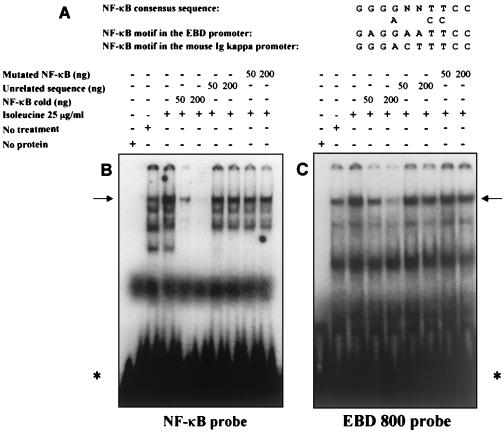
The NF-κB/rel family of transcription factors is critical for the expression of a wide variety of immune response genes and has been hypothesized to also control mammalian β-defensin expression (10, 11). We therefore examined the effect of isoleucine treatment on the binding activity of the NF-κB/rel family of transcription factors to elucidate the cell signaling events involved in isoleucine-mediated defensin induction. We made nuclear extracts from isoleucine-treated and untreated MDBK cells and used electrophoretic mobility shift assays to measure factor binding to a probe containing a known NF-κB recognition sequence. As shown in Fig. 4B, multiple species bind to the NF-κB probe and one major species, indicated by an arrow, is induced in response to isoleucine treatment. The sequence specificity of these interactions was demonstrated by using competition experiments. Addition of an excess of unlabeled NF-κB probe abolished binding but a similar excess of a probe of unrelated sequence did not. We next tested an unlabeled competitor probe containing a single base pair mutation known to prevent interaction of NF-κB with its recognition sequence. This mutated probe is no longer able to compete for binding of the factors recognizing the labeled NF-κB probe. These results demonstrate that isoleucine induces specific DNA binding proteins that recognize the NF-κB consensus sequence.
Figure 4.
Isoleucine induces the binding activity of an NF-κB protein. Transfected MDBK cells were serum-deprived during treatment with isoleucine at a concentration of 25 μg/ml. Nuclear extracts were made as described in Materials and Methods. (A) A comparison of the positive control mouse Ig κ and the EBD promoter NF-κB motifs are compared with the NF-κB consensus binding sequence. Factor binding to the 32P-labeled oligonucleotide containing the NK-κB consensus motif (B) and an NF-κB motif from the EBD promoter (C) was analyzed by gel shift and autoradiography. DNA-binding reactions were carried out in 20 μl containing 10 μg of nuclear protein extract. Inducible NF-κB/rel complexes are indicated by arrows. * indicate the free probe.
A computer analysis did not reveal any sites that perfectly matched the NF-κB consensus binding sequence within the portions of the EBD promoter that are represented in the reporter construct. However, visual inspection revealed several potential NF-κB binding motifs that contained one or two mismatches with the consensus sequence. A comparison of one of these sequences to the NF-κB consensus recognition sequence (20), shown in Fig. 4A, shows the similarity of this putative NF-κB site to the consensus. To test the ability of this putative site to bind NF-κB/rel species we used a labeled probe containing the EBD promoter sequence in electrophoretic mobility shift assay experiments. As shown in Fig. 4C, this probe yields a protein–DNA complex that is induced by isoleucine treatment and is of similar mobility to the major species produced by using the NF-κB site containing probe. The sequence specificity of this interaction was demonstrated via competition with the NF-κB binding probe as specific competitor and separately with the control probe containing an unrelated sequence. The results in Fig. 4C show that binding of the major isoleucine-induced protein is abolished by an excess of the unlabeled probe containing an authentic NF-κB binding site. In contrast, a similar excess of the unlabeled probe of unrelated sequence did not effectively compete for binding. The mutated NF-κB competitor probe was not able to compete for proteins binding to the labeled EBD promoter probe. These results, taken together, demonstrate that the EBD promoter contains an authentic recognition sequence for NF-κB/rel factors. This idea is further supported by the fact that the single mismatch in the EBD promoter NF-κB binding sequence, an A in place of a G in the second position, is also present in a known p65 homodimer binding site in the human granulocyte- colony stimulating factor promoter (21).
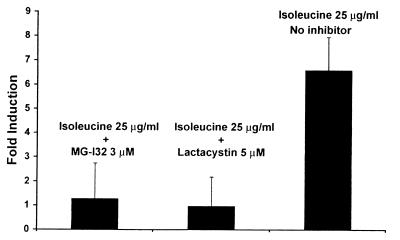
We next tested the effect of pharmacological inhibitors of NF-κB activation on the ability of isoleucine to induce defensin promoter activation in cultured epithelial cells. Proteasome activity has been shown to be necessary for the signal-induced activation of NF-κB (22). The proteasome functions to degrade I-κB, a cellular inhibitor of NF-κB nuclear translocation and activation that interacts with the p65 subunit of NF-κB in the cytoplasm (22, 23). Several small molecule proteasome inhibitors have been shown to block NF-κB activation in cultured cells (23). We tested the effect of two structurally diverse proteasome inhibitors, lactacystin (24) and MG-132 (22), on isoleucine induction of the EBD promoter-luciferase construct. The results, shown in Fig. 5, demonstrate that both proteasome inhibitors completely abolish isoleucine-mediated defensin promoter induction. At the tested concentrations both inhibitors used in Fig. 5 had no effect on cell morphology or viability as measured by trypan blue exclusion (data not shown). These results further support a role for NF-κB or closely related species in isoleucine induction of defensin expression.
Figure 5.
NF-KB activation stimulated by isoleucine is abolished by proteasome inhibitors. The transfected MDBK cells were treated with the proteasome inhibitors MG-132 (3 μM) or lactacystin (5 μM) starting 30 min before exposure to 25 μg/ml isoleucine. Both inhibitors completely blocked isoleucine activation of the defensin promoter.
Discussion
Antimicrobial peptides, including the defensins, recently have been recognized as important mediators of mucosal host defense (25). As part of the innate rather than the acquired immune system their induction is controlled by a system of “pattern recogniton” receptors (26) through which host cells recognize and respond to molecules that signal the presence of potentially harmful microbes. Examples of these epitopes include lipopolysaccharide (27), β-glucans (28), formyl-methionynl-leucyl-phenylalanine (29), some mannose-containing carbohydrates (30), peptidoglycan (31), and bacterial DNA (32). In the case of defensins, existing evidence demonstrates that lipopolysaccharide and heat-killed bacteria or fungi can provoke epithelial cells to produce more of these antimicrobial peptides. IL-1 is also a strong inducer (12, 13, 33), a finding consistent with IL-1's role as a host-defense regulating cytokine acting through the IL-1R1/Toll superfamily of innate immune response receptors (34). Here we have shown that in addition to these known inducers a class of small molecules, including l-isoleucine and several of its analogs, also can specifically induce the production of epithelial defensins. These results suggest that the presence of free isoleucine or similar compounds at micromolar levels may serve as a previously unappreciated “pattern,” signaling the presence of invading microbes to epithelial cells.
The question of why micromolar levels of free isoleucine or similar compounds should serve as a recognizable marker of bacterial presence or invasion remains open. It is intriguing that an essential amino acid acts as a defensin inducer. Because isoleucine cannot be synthesized by the host, free isoleucine must either arise from an external source or result from the degradation of host proteins. One possible explanation for isoleucine's emergence as a microbial marker is that bacterial proteases, in the process of degrading host proteins, may release free isoleucine in amounts that are substantially in excess of those found in the uninfected state. Perhaps more likely is the hypothesis that isoleucine, or a very similar compound, is directly secreted by bacteria as a metabolite or as an intercellular communication molecule. Indeed, there is precedent for the use of amino acids and fatty acid derivatives as intercellular bacterial messengers (35, 36). It also has been reported that one class of low molecular weight organic compounds, the alkylkamines, are recognized by the γδ T cell receptor and can promote the expansion of the γδ receptor-bearing cell population (37). Such alkylamines are produced in vitro by a number of pathogenic bacteria including Salmonella typhimurium, Listeria monocytogenes, and Yersinia enterocolitica. The antigenic effects of the alkylamines are observed at high micromolar to low millimolar concentrations, similar to our observations with isoleucine induction of epithelial β-defensin expression.
Our data strongly suggest that isoleucine and its active analogs mediate their effects through a chiral receptor or enzyme. Because isoleucine is readily transported into cells it is possible that receptor binding takes place intracellularly. Although the nature of the receptor remains unknown, we have demonstrated that one feature of the cellular signal produced by isoleucine exposure is the activation of the NF-κB/rel family of transcription factors. NF-κB is part of an ancient, highly conserved signaling pathway that typically is involved in host defense and immune response. The role of NF-κB in the induction of antimicrobial peptides in insects is well established (38). In mammals NF-κB also is involved in many immune type responses, including IL-2 induction on T cell activation (39) and granulocyte-macrophage colony stimulating factor induction (40). NF-κB consensus sequences have been reported in the bovine tracheal antimicrobial peptide defensin promoter as well as the human β-defensin-2 promoter (41, 42). In the case of human β-defensin-2 a functional role for NF-κB in induction by IL-1-α and invading bacteria has been demonstrated in human colon cell lines (33).
In this study we report that isoleucine induces NF-κB/rel species in MDBK cells and the presence of an NF-κB recognition site in an isoleucine-inducible defensin promoter. The functional importance of NF-κB/rel species in isoleucine-mediated defensin induction is supported by the observed abolition of induction by pharmacologic inhibitors of NF-κB activation. The exact molecular nature of the induced NF-κB/rel species involved in isoleucine induction remains to be determined. Further work defining the upstream regulators of NF-κB/rel activation by isoleucine ultimately may help to elucidate the nature of the proximal pattern recognition receptor that interacts with isoleucine. These findings extend the known mechanisms host epithelial cells use to recognize pathogenic microbes. Isoleucine or its analogs also may have clinical utility as a class of immunostimulants that could bolster the barrier defenses of mucosal surfaces by using an approach similar to that suggested by Bevins for the surface of the airway (43).
Acknowledgments
We thank Joshua Turse for his technical assistance. We are very grateful to Dr. Jamila Louahed, Dr. Stephen Jones, and Dr. William Kinney for helpful discussions and critical reading of the manuscript.
Abbreviations
- EBD
enteric β-defensin
- MDBK
Madin–Darby bovine-kidney
- FDAA
1-fluoro-2.4-dinitrophenyl-5-alanine amide
Footnotes
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.220424597.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.220424597
References
- 1.Ganz T, Lehrer R I. Pharmacol Ther. 1995;66:191–205. doi: 10.1016/0163-7258(94)00076-f. [DOI] [PubMed] [Google Scholar]
- 2.Stolzenberg E D, Anderson G M, Ackerman M R, Whitlock R H, Zasloff M. Proc Natl Acad Sci USA. 1997;94:8686–8690. doi: 10.1073/pnas.94.16.8686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhao C, Wang I, Lehrer R I. FEBS Lett. 1996;396:319–322. doi: 10.1016/0014-5793(96)01123-4. [DOI] [PubMed] [Google Scholar]
- 4.Schonwetter B S, Stolzenberg E D, Zasloff M A. Science. 1995;267:1645–1648. doi: 10.1126/science.7886453. [DOI] [PubMed] [Google Scholar]
- 5.Harder J, Bartels J, Christophers E, Schroder J M. Nature (London) 1997;26:861. doi: 10.1038/43088. [DOI] [PubMed] [Google Scholar]
- 6.Goldman M J, Anderson G M, Stolzenberg E D, Kari U P, Zasloff M, Wilson J M. Cell. 1997;88:553–560. doi: 10.1016/s0092-8674(00)81895-4. [DOI] [PubMed] [Google Scholar]
- 7.Wilson C L, Ouellette A J, Satchell D P, Ayabe T, Lopez-Boado Y S, Stratman J L, Hultgren S J, Matrisian L M, Parks W C. Science. 1999;286:113–117. doi: 10.1126/science.286.5437.113. [DOI] [PubMed] [Google Scholar]
- 8.Yang D, Chertov O, Bykovskaia S N, Chen Q, Buffo M J, Shogan J, Anderson M, Schroeder J M, Wang J M, Howard O M, Oppenheim J J. Science. 1999;286:525–538. doi: 10.1126/science.286.5439.525. [DOI] [PubMed] [Google Scholar]
- 9.Janeway C A., Jr Immunol Today. 1992;13:11–16. doi: 10.1016/0167-5699(92)90198-G. [DOI] [PubMed] [Google Scholar]
- 10.Diamond G, Russell J P, Bevins C L. Proc Natl Acad Sci USA. 1996;93:5156–5160. doi: 10.1073/pnas.93.10.5156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Russell J P, Diamond G, Tarver A P, Scanlin T F, Bevins C L. Infect Immun. 1996;64:1565–1568. doi: 10.1128/iai.64.5.1565-1568.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Singh P K, Jia H P, Wiles K, Hesselberth J, Liu L, Conway B D, Greenberg E P, Valores E V, Welsh M J, Ganz T, et al. Proc Natl Acad Sci USA. 1998;95:14961–14966. doi: 10.1073/pnas.95.25.14961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mathews M, Jia H P, Guthmiller J M, Losh G, Graham S, Johnson G K, Tack B F, McCray P B., Jr Infect Immun. 1999;67:2740–2745. doi: 10.1128/iai.67.6.2740-2745.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miller A D, Rosman G J. BioTechniques. 1989;7:980–990. [PMC free article] [PubMed] [Google Scholar]
- 15.Mafrey P. Carlsberg Res Commun. 1984;49:591–596. [Google Scholar]
- 16.Lee K A W, Bindereif A, Green M R. Gene Anal Tech. 1988;5:22–31. doi: 10.1016/0735-0651(88)90023-4. [DOI] [PubMed] [Google Scholar]
- 17.Oliviero S, Cortese R. EMBO J. 1989;8:1145–1151. doi: 10.1002/j.1460-2075.1989.tb03485.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lenardo M J, Baltimore D. Cell. 1989;58:227–229. doi: 10.1016/0092-8674(89)90833-7. [DOI] [PubMed] [Google Scholar]
- 19.Tarver A P, Clark D P, Diamond G, Russell J P, Erdjument-Bromage H, Tempst P, Cohen K S, Jones D E, Sweeney R W, Wines M, et al. Infect Immun. 1998;66:1045–1056. doi: 10.1128/iai.66.3.1045-1056.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bauerle P A. Biochim Biophys Acta. 1991;1072:63–80. doi: 10.1016/0304-419x(91)90007-8. [DOI] [PubMed] [Google Scholar]
- 21.Dunn S M, Coles L S, Lang R K, Gerondakis S, Vadas M A, Shannon M F. Blood. 1994;83:2469–2479. [PubMed] [Google Scholar]
- 22.Palombella V J, Rando O J, Goldberg A L, Maniatis T. Cell. 1994;78:773–785. doi: 10.1016/s0092-8674(94)90482-0. [DOI] [PubMed] [Google Scholar]
- 23.Grisham M B, Palombella V J, Elliott P J, Conner E M, Brand S, Wong H L, Pien C, Mazzola L M, Destree A, Parent L, Adams J. Methods Enzymol. 1999;300:345–363. doi: 10.1016/s0076-6879(99)00140-8. [DOI] [PubMed] [Google Scholar]
- 24.Fenteany G, Standaert R F, Lane W S, Choi S, Corey E J, Schreiber S L. Science. 1995;268:726–731. doi: 10.1126/science.7732382. [DOI] [PubMed] [Google Scholar]
- 25.Huttner K M, Bevins C L. Pediatr Res. 1999;45:785–794. doi: 10.1203/00006450-199906000-00001. [DOI] [PubMed] [Google Scholar]
- 26.Medzhitov R, Janeway C A., Jr Cell. 1997;91:295–298. doi: 10.1016/s0092-8674(00)80412-2. [DOI] [PubMed] [Google Scholar]
- 27.Ingalls R R, Heine H, Lien E, Yoshimurs A, Golenbock D. Infect Dis Clin North Am. 1999;13:341–353. doi: 10.1016/s0891-5520(05)70078-7. [DOI] [PubMed] [Google Scholar]
- 28.Czop J K, Kay J. J Exp Med. 1991;173:1511–1520. doi: 10.1084/jem.173.6.1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Panaro M A, Mitolo V. Immunopharmacol Immunotoxicol. 1999;21:397–419. doi: 10.3109/08923979909007117. [DOI] [PubMed] [Google Scholar]
- 30.Fraser I P, Koziel H, Ezekowitz R A. Semin Immunol. 1998;10:363–372. doi: 10.1006/smim.1998.0141. [DOI] [PubMed] [Google Scholar]
- 31.Jin Y, Gupta D, Dziarski R. J Infect Dis. 1998;177:1629–1638. doi: 10.1086/515318. [DOI] [PubMed] [Google Scholar]
- 32.Heeg K, Sparwasser T, Lipford G B, Hacker H, Zimmermann S, Wagner H. Eur J Clin Microbiol Infect Dis. 1998;17:464–469. doi: 10.1007/BF01691128. [DOI] [PubMed] [Google Scholar]
- 33.O'Neil D A, Porter E M, Elewaut D, Anderson G M, Eckman L, Ganz T, Kagnoff M F. J Immunol. 1999;163:6718–6724. [PubMed] [Google Scholar]
- 34.O'Neill L A J, Greene C. J Leukocyte Biol. 1998;63:650–657. [PubMed] [Google Scholar]
- 35.Downard J, Toal D. Mol Microbiol. 1995;16:171–175. doi: 10.1111/j.1365-2958.1995.tb02290.x. [DOI] [PubMed] [Google Scholar]
- 36.Flavier A B, Clough S J, Schell M A, Denny T P. Mol Microbiol. 1997;26:251–259. doi: 10.1046/j.1365-2958.1997.5661945.x. [DOI] [PubMed] [Google Scholar]
- 37.Bukowski J F, Morita C T, Brenner M B. Immunity. 1999;11:57–65. doi: 10.1016/s1074-7613(00)80081-3. [DOI] [PubMed] [Google Scholar]
- 38.Hoffmann J A, Kafatos F C, Janeway C A, Ezekowitz R A B. Science. 1999;284:1313–1318. doi: 10.1126/science.284.5418.1313. [DOI] [PubMed] [Google Scholar]
- 39.Jain J, Loh C, Rao A. Curr Opin Immunol. 1995;7:333–342. doi: 10.1016/0952-7915(95)80107-3. [DOI] [PubMed] [Google Scholar]
- 40.Schreck R, Bauerle P A. Mol Cell Biol. 1990;10:1281–1286. doi: 10.1128/mcb.10.3.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Diamond G, Jones D E, Bevins C L. Proc Natl Acad Sci USA. 1993;90:4596–4600. doi: 10.1073/pnas.90.10.4596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu L, Wang L, Jia H P, Zhao C, Heng H H Q, Schutte B C, McCray P B, Jr, Ganz T. Gene. 1998;222:237–244. doi: 10.1016/s0378-1119(98)00480-6. [DOI] [PubMed] [Google Scholar]
- 43.Bevins C L. Am J Respir Cell Mol Biol. 1999;20:861–863. doi: 10.1165/ajrcmb.20.5.f149. [DOI] [PubMed] [Google Scholar]