Abstract
The Drosophila Genome Project database contains the sequences of two genes, CG8985 and CG13803, which are predicted to code for G protein-coupled receptors. We cloned the cDNAs corresponding to these genes and found that their gene structures had not been correctly annotated. We subsequently expressed the coding regions of the two corrected receptor genes in Chinese hamster ovary cells and found that each of them coded for a receptor that could be activated by low concentrations of Drosophila myosuppressin (EC50,4 × 10–8 M). The insect myosuppressins are decapeptides that generally inhibit insect visceral muscles. Other tested Drosophila neuropeptides did not activate the two receptors. In addition to the two Drosophila myosuppressin receptors, we identified a sequence in the genomic database from the malaria mosquito Anopheles gambiae that also very likely codes for a myosuppressin receptor. To our knowledge, this paper is the first report on the molecular identification of specific insect myosuppressin receptors.
Most insect myosuppressins are decapeptides with the structure X1DVX2HX3FLRFamide (where X1 is pQ, P,T; X2 is D,G,V; X3 is V,S) (1). These neuropeptides have first been isolated from cockroaches and later from locusts, flies, and moths and it can be anticipated that they occur in all insects (1–11). The myosuppressins obtained their name because they have a general inhibitory action on a variety of visceral muscles from insects (1–3, 6–20). Because the insect myosuppressins also block the visceral muscles involved in the passage of food along the alimentary canal (2, 9, 12, 15–17, 19), they have attracted the interest of researchers that are aiming to develop nonpeptide agonists of myosuppressin receptors to use them as insecticides. On several occasions, one compound, benzethonium chloride, has been claimed to be such a myosuppressin receptor agonist (14, 19, 21). This claim was based on binding studies and physiological experiments carried out with isolated insect muscle preparations (14, 19, 21). However, to be sure that benzethonium chloride is specifically acting on the myosuppressin receptor, or to develop more selective and potent myosuppressin agonists, it is necessary to have the cloned and functional myosuppressin receptor available and expressed, for example, in cell lines plated out on microtiter plates for high-throughput screening of chemical libraries. Here we describe the molecular cloning and functional expression in cell lines of two myosuppressin receptors from the fruit fly Drosophila melanogaster. To our knowledge, this article is the first report on the molecular identification of specific myosuppressin receptors from insects.
Materials and Methods
PCR. Total RNA was isolated from adult D. melanogaster (Canton S) by using the TRIzol Reagent (Life Technologies, Grand Island, NY), and further treated with DNase I by using the DNA-free kit (Ambion, Austin, TX). cDNA was synthesized, by using the SMART RACE cDNA Amplification Kit (CLONTECH). From the annotated exons of CG8985 [Drosophila myosupressin receptor (DMSR)-1] and CG13803 (DMSR-2) (www.flybase.org) primers were designed for PCR. For DMSR-1, the sense primer 5′-GGCCAGTGGCAACAATGAAACTGAGC-3′ and the antisense primer 5′-CAGGACACT CAGCAGGCGACTG-3′ (corresponding to positions 3–28 and 1470–1491 in Fig. 1) were used. The 3′ RACE was carried out as a nested PCR with the sense primer 5′-CCTGGACAAGTGGCTGCCGGTG-3′ and the sense nested primer 5′-GGTGCCACGGAGAATCAGCTGTAC-3′ (corresponding to positions 1284–1305 and 1618–1641 in Fig. 1). The 5′ RACE was also nested PCR with the antisense primer 5′-CTGTCCGTCAGGATGTAGTCGTG-3′ and the antisense nested primer 5′-CCAGCATAACTGCCAGGTCGGC-3′ (corresponding to positions 247–269 and 205–226 in Fig. 1). The PCR program was 94°C for 3 min, then a 21-cycle touchdown, 94°C for 30 s, 73°C for 40 s decreasing 0.6°C per cycle, 72°C for 1 min, followed by 25 cycles at 94°C for 30 s, 59°C for 40 s, 72°C for 1 min. For DMSR-2, the sense primer 5′-CTTCATTGACACCATGGTCACG-3′ and the antisense primer 5′-GCTCTCCTGCTACACATTTGTC-3′ (corresponding to positions –13 to +9 and 1446–1476 in Fig. 2) were used in the initial PCR. The 3′ RACE was carried out as a nested PCR with the sense primer 5′-GCACGTTCGCGCTCCTCTTC-3′ and the sense nested primer 5′-CCATCGATCTCGGGCTGACG-3′ (corresponding to positions 1265–1284 and 1427–1446 in Fig. 2). The 5′ RACE was also nested PCR with the antisense primer 5′-AGCTGTAGCTGAGCTGCTCCTC-3′ and the antisense nested primer 5′-CGACAGCCAGACCCGTGAGTATG-3′ (corresponding to positions 274–295 and 177–199 in Fig. 2). The PCR program was 94°C for 3 min, then a 20-cycle touchdown, 94°C for 30 s, 67°C for 45 s decreasing 0.5°C per cycle, 72°C for 2 min, followed by 25 cycles at 94°C for 30 s, 57°C for 45 s, 72°C for 2 min.
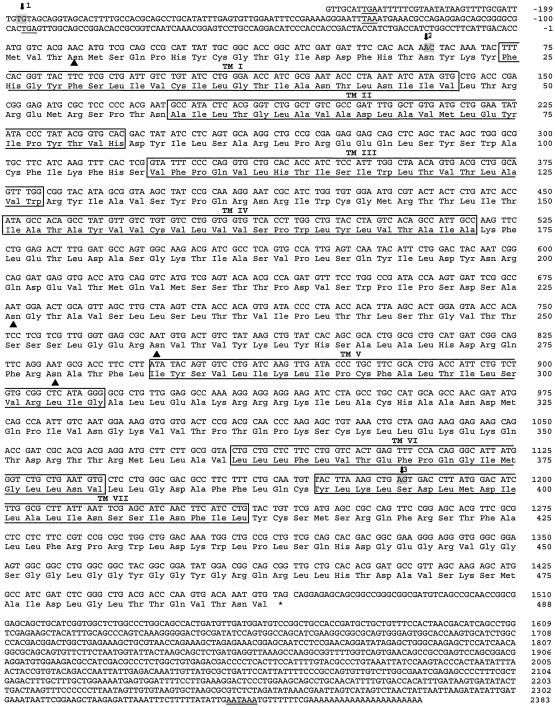
Fig. 1.
cDNA and deduced amino acid sequence of the corrected gene CG8985. The nucleotides are numbered from 5′ to 3′ end and the amino acid residues are numbered, starting with the first ATG (start) codon in the open reading frame. The introns are marked by arrows and numbered 1–4. The two nucleotides, bordering each of the four introns, are highlighted in gray. The stop codons in the 5′ untranslated region are underlined. The putative polyadenylation sites in the 3′ untranslated region are underlined twice. The translation termination codon is marked by an asterisk. The seven transmembrane domains of the receptor protein are boxed and marked TMI–VII. The potential N-glycosylation site in the extracellular N terminus (obeying the NXS/T consensus sequence), and four such sites in the second extracellular loop are marked by filled triangles.
Fig. 2.
cDNA and deduced amino acid sequence of the corrected gene CG13803. Data are displayed in the same way as Fig. 1.
Creation of Stable Cell Lines Expressing the Two Receptors. The sense 5′-ACAATGGCCAGTGGCAACAATGAAACTGAG-3′ and the antisense 5′-CTACAGATTTGTCACCTGCGTGATGTTGGTG-3′ primers (corresponding to nucleotide positions –3 to +27 and 1407–1437 of Fig. 1) were used in a PCR with cDNA from adult flies to amplify the coding region of DMSR-1. This cDNA was also used to amplify DMSR-2 with the primers sense 5′-TTTGAATTCGCCACCATGGTCACGAACATGTCG-3′ and antisense 5′-AAAGAATTCCTACACATTTGTCACTTGGGTCGTCAGC-3′ (the underlined nucleotides correspond to positions 1–18 and 1440–1467 of Fig. 2). The coding sequence of DMSR-1 was cloned into the pCR3.1 vector by using the TA cloning kit from Invitrogen, whereas the coding sequence of DMSR-2 was cloned into the pIRES2-EGFP vector (CLONTECH) by using the EcoRI restriction sites incorporated in the primers. The inserts were checked for the correct sequence. The two plasmids were stably transfected into Chinese hamster ovary (CHO) cells that also stably expressed the human α G protein subunit G-16 (CHO/G-16) (22, 23) by using FuGENE 6 reagent (Roche Diagnostics). The CHO/G-16 cells and the bioluminescence assay are described in refs. 22 and 23. All wells in one bioassay have about the same number of cells.
Northern Blot Analyses. Northern blots were prepared by using the NorthernMax formaldehyde kit (Ambion) and BrightStar-Plus membranes (Ambion). cDNA probes (nucleotide positions 1284–2216 of Fig. 1 and nucleotide positions 1445–2072 of Fig. 2) were labeled by using the Strip-EZ kit (Ambion). The ribosomal protein-49 probe was generated as described in ref. 24.
Sequence Analyses, Software Programs, and Peptides. DNA sequence comparisons were performed by using the lasergene software package (DNASTAR). For Fig. 4, clustalw was used. The tmhmm v.2.0 prediction server was used for locating transmembrane helices of the proteins (www.cbs.dtu.dk). prism v.3 software (GraphPad, San Diego) was used for plotting of the bioluminescence data, including normalization of the dose–response curves (these curves were plotted as nonlinear regression). Peptides were synthesized by Genemed Synthesis (San Francisco) or Bachem (Bubendorf, Switzerland).
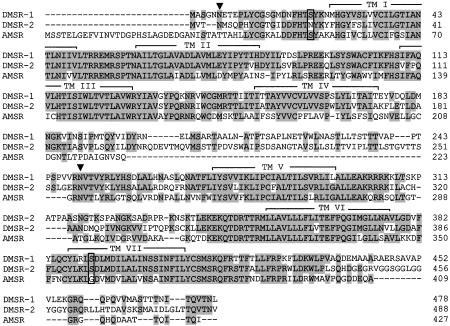
Fig. 4.
Amino acid sequence comparison between the DMSR-1 and -2 encoded, respectively, by the corrected CG8985 and CG13803 genes and the putative myosuppressin receptor from A. gambiae (AMSR; present in the clone with accession no. gb/AAAB01008900.1/). Amino acid residues that are identical between at least two receptors are highlighted in gray. The seven transmembrane domains are indicated by TM I–VII. The two common introns between the three receptor genes are indicated by vertical boxes. The filled triangles indicate common potential N-glycosylation sites. Gaps are introduced to optimize the alignments.
Results
The database from the Drosophila Genome Project consortium contains the sequences of two annotated genes (CG8985 and CG13803) that are supposed to code for two structurally related G protein-coupled receptors (www.flybase.org) (25). We cloned the cDNA of these two genes by using PCR and primers based on the annotated exons. Subsequently, we carried out 3′ and 5′ RACEs to obtain the complete cDNAs. During this process, we found that CG8985 had not been correctly predicted. In the prediction, it contained four exons and three introns, but from the cDNA sequence it became clear that intron 3 was an exon (containing a new “corrected” stop codon). Furthermore, we found two additional exons flanking a large intron at the 5′ end of the predicted gene. The corrected gene CG8985, therefore, has five exons and four introns (Table 1 and Fig. 1). Fig. 1 shows the cloned cDNA of the corrected gene CG8985, which is 3,286 nt long. It contains a putative polyadenylation site at its 3′ end and numerous stop codons preceding the start codon in its 5′ untranslated region. The cDNA codes for a protein of 478 amino acid residues, which contains seven transmembrane domains. The extracellular N terminus contains one potential N-glycosylation site, and four such sites exist in the second extracellular loop (Fig. 1).
Table 1. Intron/exon boundaries of the corrected CG8985 receptor gene.
| Intron | 5′ donor | Intron size, bp | 3′ acceptor | Intron phase | ||
|---|---|---|---|---|---|---|
| 1 | CAA | gtaaatact... | 546 | ...tatccacag | TAA | - |
| 2 | GCA | gtaagtgtc... | 15,252 | ...tatttgcag | GGG | - |
| 3 | AG | gtaatcgac... | 3,657 | ...ttttttcag | C | 2 |
| Ser | Ser | |||||
| 4 | A | gtaagtaaa... | 100 | ...gcccaccag | GT | 1 |
| Ser | Ser |
Comparison of the cDNA of Fig. 1 with the genomic sequence in the database (www.flybase.org) showed 13 nucleotide differences. In four cases, these differences led to different amino acid residues, which, however, were all conserved residues (Table 3, which is published as supporting information on the PNAS web site, www.pnas.org).
We also cloned the cDNA of gene CG13803 and, again, found that its gene had not been correctly predicted. The annotated gene had four exons and three introns. We found that the predicted intron 2 was, in fact, an exon. Furthermore, we discovered an additional large intron and an exon, both lying in front (5′) of the predicted exon 1. The corrected gene CG13803, therefore, has four exons and three introns (Table 2 and Fig. 2). Fig. 2 shows the cloned cDNA of the corrected gene CG13803, which is 2,618 nt long. The cDNA contains a putative polyadenylation signal at its 3′ end, and several stop codons preceding the start codon in its 5′ untranslated region. It codes for a protein of 488 amino acid residues, which has seven transmembrane domains, one potential N-glycosylation site in its extracellular N terminus, and three such sites in the second extracellular loop (Fig. 2).
Table 2. Intron/exon boundaries of the corrected CG13803 receptor gene.
| Intron | 5′ donor | Intron size, bp | 3′ acceptor | Intron phase | ||
|---|---|---|---|---|---|---|
| 1 | TGT | gtaagttgc... | 14,623 | ...tatttgcag | GTA | - |
| 2 | AA | gtgagtaga... | 429 | ...ccccaacag | C | 2 |
| Asn | Asn | |||||
| 3 | A | gtaagtatt... | 489 | ...tcgtttcag | GT | 1 |
| Ser | Ser |
Comparison of the cDNA of Fig. 2 with the corresponding genomic sequence revealed 37 nucleotide differences. Five of these differences led to changes in amino acid residues, of which four were conserved residues (Table 4, which is published as supporting information on the PNAS web site).
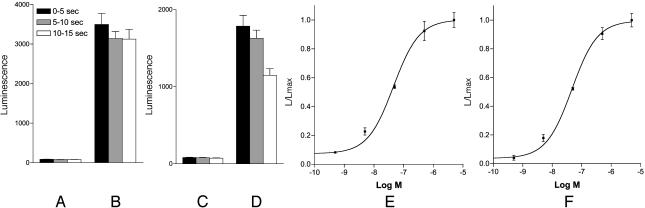
We stably transfected CHO cells with the cDNAs corresponding to the coding regions of either the corrected gene CG8985 or CG13803 and established cloned cell lines, expressing these genes effectively. These cells were also stably expressing the α subunit of the promiscuous G protein, G-16. Two days before the assay, the cells were transiently transfected with DNA, coding for apoaequorin; coelenterazine was added to the culture medium 3 hours before the assay. Addition of receptor ligands and activation of the receptors in these pretreated cells would lead to an inositol 1,4,5-trisphosphate (IP3)/Ca2+-mediated bioluminescence response that could easily be measured and quantified (22, 23, 26–32). We tested a peptide library (consisting of 24 Drosophila or other invertebrate neuropeptides), and seven monoamines on these cells and found that the two receptors were activated by low concentrations of Drosophila myosuppressin (Drome-MS) (5, 11). We also found that both receptors had the same EC50 for Drome-MS (4 × 10–8 M) (Fig. 3 E and F). The other tested peptides and amines (Fig. 3) did not activate the receptors, not even peptides that resembled Drome-MS (TDVDHVLFLRFamide) in their C termini, such as FMRF-amide, Drosophila short neuropeptide F-1 (AQRSPSLRLRF-amide), or perisulfakinin [EQFDDY(SO3H)GHMRFamide]. These results showed that the two receptors are specific for Drome-MS. Another remarkable feature was that the two receptors did not appear to be quickly desensitized. When activated by Drome-MS, they continued to be active for 30 sec or longer (t1/2 ≈ 20 sec for both receptors), which is a kinetics quite different from that of all of the other insect neuropeptide receptors characterized by us so far, where nearly full desensitization occurred within 5 sec (23, 26–32). Finally, we found that the two receptors were not activated by benzethonium chloride (in concentrations up to 10–4 M), which was claimed to be a myosuppressin receptor agonist (14, 19, 21). Concentrations >10–4 M gave bioluminescence responses in both the nontransfected and transfected CHO cells, showing that the actions of benzethonium chloride can be nonspecific.
Fig. 3.
Bioluminescence responses of nontransfected CHO/G-16 cells and of CHO/G-16 cells transfected with DNA coding for the coding region of either the corrected gene CG8985 or CG13803. The vertical bars represent SEM, which are sometimes lower than the symbols (filled squares) used. In these cases, only the symbols are given. (A and C) Bioluminescence responses of CHO/G-16 cells after addition of 5 × 10–7 M Drome-MS. (B) Bioluminescence response of CHO/G-16/CG8985 cells after addition of 5 × 10–7M Drome-MS. (D) Bioluminescence response of CHO/G-16/CG13803 cells after addition of 5 × 10–7M Drome-MS. (E) Dose–response curve of the bioluminescence responses of CHO/G-16/CG 8985 cells induced by Drome-MS. The responses 0–5, 5–10, and 10–15 sec after addition of the peptide were added and counted as one response. (F) A similar curve for the CHO/G-16/CG13803 cells. The following peptides did not activate the two receptors (tested up to 10–6or 10–5M): crustacean cardioactive peptide; capa-1, -2, and -3; corazonin; Drosophila adipokinetic hormone; Drosophila tachykinin-3; Drosophila short neuropeptide F-1; Drosophila ecdysis triggering hormones-1 and -2; Drosophila pigment dispersing hormone; Drosophila pyrokinin-2; drostatins-A4 -B2, and -C; FMRFamide; Heliothis zea hypertrehalosaemic neuropeptide; hug-γ; leucopyrokinin; leucokinin-III; perisulfakinin; and proctolin. For peptide structures see refs. 1, 10, and 11. The following amines did not activate the two receptors (tested up to 10–5M): adrenaline; dopamine; histamine; noradrenaline; octopamine; serotonin; and tyramine.
The two Drome-MS receptors (DMSR-1 and -2) strongly resemble each other (65% overall amino acid residue identity; 71% identity in the transmembrane region) (Fig. 4). A comparison of the two receptors with the other known Drosophila neuropeptide receptors or other proteins from the GenBank database revealed no proteins with significant structural similarities. However, the genomic database from the recently sequenced malaria mosquito Anopheles gambiae (33), contained a gene sequence coding for a putative G protein-coupled receptor that strongly resembled DMSR-1 and -2, both with respect to amino acid sequence (55–58% overall amino acid residue identities; 65–67% identical residues in the transmembrane region) and gene structure (two shared introns between the three genes with identical intron phasings) (Fig. 4). All of these data strongly suggest that the Anopheles receptor is a myosuppressin receptor. Whether Anopheles also has a second myosuppressin receptor could not be confirmed at present.
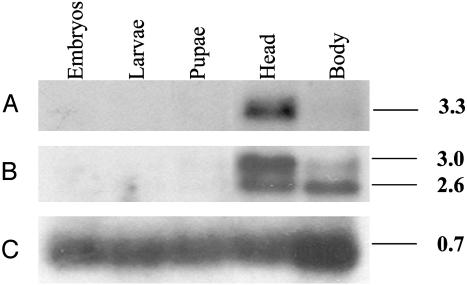
Northern blots revealed that the two Drome-MS receptors were only very weakly expressed in embryos, larvae, and pupae. In adult flies, DMSR-1 was strongly expressed in the head, but virtually absent in the body (thorax/abdomen) (Fig. 5A), whereas DMSR-2 was present in both adult head and body (Fig. 5B). The DMSR-2 mRNA displayed a double band, of which the smaller one corresponded to the size of the cloned cDNA (Fig. 5B).
Fig. 5.
Northern blots of mRNA isolated from various developmental stages from Drosophila. The sizes of the transcripts are given at the right (in kb). Each lane contained ≈5 μg of mRNA from either embryos (0–24 h), mixed first- to third-instar larvae, pupae, and heads or bodies (thorax + abdomen) from adult flies. (A) The lanes were hybridized with a cDNA probe, corresponding to CG8985. (B) The Northern blot from A was stripped and subsequently hybridized with a cDNA probe, corresponding to CG13803. (C) The Northern blot from B was stripped and subsequently hybridized with a cDNA probe, coding for ribosomal protein 49. This blot gives the loading efficiency of each lane.
Discussion
In this paper we have cloned and characterized two Drosophila receptors that are specific for Drome-MS and that do not react with other Drosophila neuropeptides, not even with peptides that have C-terminal structures resembling Drome-MS, such as the sulfakinins, the FMRFamides, or peptides belonging to the neuropeptide-F family (Fig. 3). Interestingly, the two receptors are not activated by benzethonium chloride, which was claimed to be a myosuppressin receptor agonist (14, 19, 21), but whose actions, therefore, must be quite different.
The two Drome-MS receptors have an EC50 for Drome-MS of 4 × 10–8 M. This value compares well with the concentrations of Drome-MS needed in pharmacological or physiological experiments to inhibit Drosophila heartbeat (EC50, 10–7 M) or contractions of the crop (10–6 M) (20), and with the concentrations of other insect myosuppressins that were used to inhibit the blowfly crop (4 × 10–8 M) (19), the cockroach foreand hindgut (between 10–10 M and 3 × 10–8 M) (9, 14), the locust heart (EC50, 5 × 10–8 M) (13), or the salivary gland from Rodnius prolixus (between 5 × 10–9 and 5 × 10–7 M) (18). These data, therefore, suggest that the two cloned Drome-MS receptors are the physiologically relevant myosuppressin receptors.
In addition to the two Drome-MS receptors characterized in this paper, we have earlier identified the first insect FMRFamide receptor in Drosophila (28). This receptor is activated by very low concentrations of Drosophila FMRFamides (EC50,9 × 10–10 M), but also by higher concentrations of Drome-MS (EC50, 2 × 10–7 M) and Drosophila short neuropeptide F-1 (EC50, 9 × 10–8 M). These findings were recently confirmed by another research group (34). We have previously considered the activation of the Drosophila FMRFamide receptor by the other two peptides as cross reactions caused by the similarities of their C-terminal peptide structures (28). However, the FMRFamide receptor's EC50 value for Drome-MS is only five times higher than the EC50 values found for the two Drome-MS receptors identified in the present study. This finding would imply that the Drosophila FMRFamide receptor, under certain conditions, possibly also might function as a third Drome-MS receptor. It also implies that high (“pharmacological”) concentrations of Drome-MS (e.g., 10–6 M, see above) could effect the Drosophila FMRFamide receptor instead of the presumed Drome-MS receptor. However, similarly to the two Drome-MS receptors, the Drosophila FMRFamide receptor is not activated by benzethonium chloride (G.C., unpublished observations).
Northern blots showed that DMSR-2 is expressed in both the head and body (thorax and abdomen) of adult flies (Fig. 5B). These results agree very well with the actions of the myosuppressins on numerous visceral muscles present in both the body and head of insects (see above). However, the very strong expression of both receptors in the head (compare, for example, the loading efficiencies in the last two lanes of Fig. 5C), also suggests the presence of myosuppressin receptors in the brain. In fact, anatomical studies have shown that myosuppressin-like material was abundant in various parts of the insect brain (19, 20, 35–37). These two findings together, then, suggest a so far unknown role of the myosuppressins in the insect central nervous system.
It is interesting that two transcripts were observed with DMSR-2 (Fig. 5B), of which the smaller one corresponded to the cloned receptor (Fig. 2), whereas the somewhat larger transcripts probably corresponded to a mRNA species, having an alternative polyadenylation signal, lying ≈400 bp downstream from our cloned polyadenylation signal. This longer transcript has not been cloned by us, but can be deduced from the genomic sequence in the database (www.flybase.org). It is also interesting that the smaller transcript was mainly present in the body, whereas the longer transcript was mainly in the head (Fig. 5). This could mean that nerve cells predominantly produce the larger transcript and visceral muscle cells produce the smaller one.
The identification in this paper of two specific myosuppressin receptors in Drosophila opens the possibility of finding similar receptors in other insects or arthropods. That this is a realistic option is shown by our discovery of a probable myosuppressin receptor in the malaria mosquito A. gambiae (Fig. 4). The identification of myosuppressin receptors in other model insects or other arthropods will certainly contribute to our understanding of the endocrinology and physiology of these animals. Furthermore, the availability of myosuppressin receptors in recombinant cell lines (grown in 96- or 384-well plates) will also make it possible to screen large chemical libraries for agonists, which could be used as leads to develop specific and environmentally safe insecticides.
Supplementary Material
Acknowledgments
We thank Drs. S. Rees and J. Stables (Glaxo Wellcome, Stevenage, U.K.) for supplying cell line CHO/G-16, Birgitte Paulsen for typing the manuscript, and Lundbeck Foundation and Fabrikant Vilhelm Pedersen og Hustrus Mindelegat (Manufacturer Vilhelm Pedersen and Wife Memorial Legacy; this support was granted on recommendation from the Novo Nordisk Foundation) for financial support.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: CHO, Chinese hamster ovary; Drome-MS, Drosophila myosuppressin; DMSR-1 and -2, Drome-MS receptor 1 and 2; G-16, G protein 16.
References
- 1.Nässel, D. R. (2002) Prog. Neurobiol. 68, 1–84. [DOI] [PubMed] [Google Scholar]
- 2.Holman, G. M., Cook, B. J. & Nachman, R. J. (1986) Comp. Biochem. Physiol. C 85, 329–333. [DOI] [PubMed] [Google Scholar]
- 3.Robb, S., Packman, L. C. & Evans, P. D. (1989) Biochem. Biophys. Res. Commun. 160, 850–856. [DOI] [PubMed] [Google Scholar]
- 4.Kingan, T. G., Teplow, D. B., Phillips, J. M., Riehm, J. P., Rao, K. R., Hildebrand, J. G., Homberg, U., Kammer, A. E., Jardine, I., Griffin, P. R., et al. (1990) Peptides 11, 849–856. [DOI] [PubMed] [Google Scholar]
- 5.Nichols, R. (1992) J. Mol. Neurosci. 3, 213–218. [DOI] [PubMed] [Google Scholar]
- 6.Fonagy, A., Schoofs, L., Proost, P., Van Damme, J., Bueds, H. & De Loof, A. (1992) Comp. Biochem. Physiol. C 102, 239–245. [DOI] [PubMed] [Google Scholar]
- 7.Schoofs, L., Holman, G. M., Paemen, L., Veelaert, D., Amelinckx, M. & De Loof, A. (1993) Peptides 14, 409–421. [DOI] [PubMed] [Google Scholar]
- 8.Peef, N. M., Orchard, I. & Lange, A. B. (1994) Peptides 15, 387–392. [DOI] [PubMed] [Google Scholar]
- 9.Predel, R., Rapus, J. & Eckert, M. (2001) Peptides 22, 199–208. [DOI] [PubMed] [Google Scholar]
- 10.Gäde, G., Hoffmann, K. H. & Spring, J. H. (1997) Physiol. Rev. 77, 963–1032. [DOI] [PubMed] [Google Scholar]
- 11.Vanden Broeck, J. (2001) Peptides 22, 241–254. [DOI] [PubMed] [Google Scholar]
- 12.Cook, B. J. & Wagner, R. M. (1991) Comp. Biochem. Physiol. C 99, 95–99. [DOI] [PubMed] [Google Scholar]
- 13.Robb, S. & Evans, P. D. (1994) J. Exp. Biol. 197, 437–442. [DOI] [PubMed] [Google Scholar]
- 14.Nachman, R. J., Olender, E. H., Roberts, V. A., Holman, G. M. & Yamamoto, D. (1996) Peptides 17, 313–320. [DOI] [PubMed] [Google Scholar]
- 15.Lange, A. B. & Orchard, I. (1998) Peptides 19, 459–467. [DOI] [PubMed] [Google Scholar]
- 16.Fuse, M. & Orchard, I. (1998) Regul. Pept. 77, 163–168. [DOI] [PubMed] [Google Scholar]
- 17.Duttlinger, A., Berry, K. & Nichols, R. (2002) Peptides 23, 1953–1957. [DOI] [PubMed] [Google Scholar]
- 18.Orchard, I. & Te Brugge, V. T. (2002) Peptides 23, 693–700. [DOI] [PubMed] [Google Scholar]
- 19.Richer, S., Stoffolano, J. G., Yin, C. M. & Nichols, R. (2000) J. Comp. Neurol. 421, 136–142. [PubMed] [Google Scholar]
- 20.Nichols, R. (2003) Annu. Rev. Entomol. 48, 485–503. [DOI] [PubMed] [Google Scholar]
- 21.Lange, A. B., Orchard, I., Wang, Z. & Nachman, R. J. (1995) Proc. Natl. Acad. Sci. USA 92, 9250–9253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stables, J., Green, A., Marshall, F., Fraser, N., Knight, E., Sautel, M., Milligan, G., Lee, M. & Rees, S. (1997) Anal. Biochem. 252, 115–126. [DOI] [PubMed] [Google Scholar]
- 23.Staubli, F., Jørgensen, T. J. D., Cazzamali, G., Williamson, M., Lenz, C., Søndergaard, L., Roepstorff, P. & Grimmelikhuijzen, C. J. P. (2002) Proc. Natl. Acad. Sci. USA 99, 3446–3451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hauser, F., Nothacker, H.-P. & Grimmelikhuijzen, C. J. P. (1997) J. Biol. Chem. 272, 1002–1010. [DOI] [PubMed] [Google Scholar]
- 25.Hewes, R. S. & Taghert, P. H. (2001) Genome Res. 11, 1126–1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lenz, C., Williamson, M., Hansen, G. N. & Grimmelikhuijzen, C. J. P. (2001) Biochem. Biophys. Res. Commun. 286, 1117–1122. [DOI] [PubMed] [Google Scholar]
- 27.Secher, T., Lenz, C., Cazzamali, G., Sørensen, G., Williamson, M., Hansen, G. N., Svane, P. & Grimmelikhuijzen, C. J. P. (2001) J. Biol. Chem. 276, 47052–47060. [DOI] [PubMed] [Google Scholar]
- 28.Cazzamali, G. & Grimmelikhuijzen, C. J. P. (2002) Proc. Natl. Acad. Sci. USA 99, 12073–12078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cazzamali, G., Saxild, N. P. E. & Grimmelikhuijzen, C. J. P. (2002) Biochem. Biophys. Res. Commun. 298, 31–36. [DOI] [PubMed] [Google Scholar]
- 30.Iversen, A., Cazzamali, G., Williamson, M., Hauser, F. & Grimmelikhuijzen, C. J. P. (2002) Biochem. Biophys. Res. Commun. 299, 628–633. [DOI] [PubMed] [Google Scholar]
- 31.Iversen, A., Cazzamali, G., Williamson, M., Hauser, F. & Grimmelikhuijzen, C. J. P. (2002) Biochem. Biophys. Res. Commun. 299, 924–931. [DOI] [PubMed] [Google Scholar]
- 32.Cazzamali, G., Hauser, F., Kobberup, S., Williamson, M. & Grimmelikhuijzen, C. J. P. (2003) Biochem. Biophys. Res. Commun. 303, 146–152. [DOI] [PubMed] [Google Scholar]
- 33.Holt, R. A., Subramanian, G. M., Halpern, A., Sutton, G. G., Charlab, R., Nusskern, D. R., Wincker, P., Clark, A. G., Ribeiro, J. M. C., Wides, R., et al. (2002) Science 298, 129–149.12364791 [Google Scholar]
- 34.Meeusen, T., Mertens, I., Clynen, E., Baggerman, G., Nichols, R., Nachman, R. J., Huybrechts, R., De Loof, A. & Schoofs, L. (2002) Proc. Natl. Acad. Sci. USA 99, 15363–15368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Meola, S. M., Wright, M. S., Holman, G. M. & Thompson, J. M. (1991) Neurochem. Res. 16, 543–549. [DOI] [PubMed] [Google Scholar]
- 36.McCormick, J. & Nichols, R. (1993) J. Comp. Neurol. 338, 278–288. [DOI] [PubMed] [Google Scholar]
- 37.Lu, D., Lee, K. Y., Horodyski, F. M. & Witten, J. L. (2002) J. Comp. Neurol. 446, 377–396. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.