Abstract
Best vitelliform macular dystrophy is a dominantly inherited, early onset, macular degenerative disease that exhibits some histopathologic similarities to age-related macular degeneration. Although the vitelliform lesion is common in the fundus of individuals with Best disease, diagnosis is based on a reduced ratio of the light peak to dark trough in the electrooculogram. Recently, the VMD2 gene on chromosome 11q13, encoding the protein bestrophin, was identified. The function of bestrophin is unknown. To facilitate studies of bestrophin, we produced both rabbit polyclonal and mouse monoclonal antibodies that proved useful for Western blotting, immunoprecipitation, and immunocytochemistry. To characterize bestrophin, we initially probed the retinal pigment epithelium (RPE)-derived cell lines ARPE-19, D407, and RPE-J. All of the cell lines expressed bestrophin mRNA by reverse transcription-PCR, but not on Western blots. Bestrophin in human RPE partitioned in the detergent phase during Triton X-114 extraction and could be modified by biotin in intact cells, indicative of a plasma membrane localization. Immunocytochemical staining of macaque and porcine eyes indicated that bestrophin is localized at the basolateral plasma membrane of RPE cells. When expressed in RPE-J cells by adenovirus-mediated gene transfer, bestrophin again was determined by confocal microscopy and cell surface biotinylation to be a basolateral plasma membrane protein. The basolateral plasma membrane localization of bestrophin suggests the possibility that bestrophin plays a role in generating the altered electrooculogram of individuals with Best disease.
Best vitelliform macular dystrophy (BMD) represents one of a number of single-gene disorders that exhibit symptoms and histopathologies reminiscent of age-related macular degeneration (AMD; refs. 1 and 2). AMD is the leading cause of blindness in the western world and affects nearly 30% of those over the age of 75 (3). BMD is an autosomal dominant disease with a juvenile age of onset (4). Clinically, the disease is characterized by an “egg yolk,” or vitelliform, lesion in the macula, easily visible during fundus examination (1, 5). It is thought that the vitelliform lesion may be caused by the abnormal deposition of lipofuscin in the retinal pigment epithelium (6). Over time, the vitelliform lesion will break up and may appear as a “bull's eye.” A defining characteristic of BMD, however, is a light peak to dark trough ratio of the electrooculogram (EOG) of less than 1.5, without aberrations in the clinical electroretinogram (5). Even otherwise asymptomatic carriers of BMD-associated mutations, as assessed by pedigree, will exhibit an altered EOG (7, 8).
Histopathologically, BMD is poorly characterized with data available from only a small number of donor eyes. In all cases, the disease has been shown to manifest as a generalized retinal pigment epithelium (RPE) abnormality associated with excessive lipofuscin accumulation [although not to the extent seen in Stargardt's disease (6, 9, 10)], regions of geographic RPE atrophy, and deposition of abnormal fibrillar material beneath the RPE, similar to drusen. Occasional breaks in Bruch's membrane with accompanying neovascularization also have been reported, although BMD is not noted for extensive choroidal neovascularization. Many of these features also are found in AMD (11–13).
Understanding the functions of genes mutated in inherited maculopathies will increase our understanding of the physiology of the retina and facilitate the development of treatment strategies for AMD. Recently, Petrukhin et al. (14) identified the gene mutated in Best disease, VMD2, on chromosome 11q13. However, in contrast to the genes mutated in Sorsby's Fundus Dystrophy (TIMP-3; ref. 15), recessive Stargardt's disease (ABCA4; ref. 16), and Doyne's honeycomb retinal dystrophy (EFEMP-1; ref. 17), VMD2 encodes a previously unknown protein designated bestrophin. Although mutations in VMD2 in individuals with AMD are rare, mutations have been reported in up to 1.5% of individuals with AMD (18, 19).
Bestrophin is predicted to be a 585-aa protein with an approximate mass of 68 kDa (14, 20). Based on Northern blotting and in situ hybridization data (14, 20), bestrophin is predominantly expressed in the RPE. Bestrophin shares homology with the Caenorhabditis elegans RFP gene family, named for the presence of a conserved arginine (R), phenylalanine (F), and proline (P) amino acid sequence motif. However, the function of the RFP genes is unknown. Computer-assisted structural analysis predicts that bestrophin is a transmembrane protein with four membrane-spanning α helical domains, although a less likely five-transmembrane domain model recently has been proposed (21). There are no obvious targeting signals present in the amino acid sequence that would provide clues to its subcellular localization, and based on the four-transmembrane domain model, bestrophin has no obvious sites for posttranslational modifications such as N-glycosylation.
As a first step in understanding the role of bestrophin in macular degenerative disease, we have produced antibodies recognizing the C terminus of bestrophin and used them to probe the cellular and subcellular localization of bestrophin in the eye. Our data indicate that bestrophin is a basolateral plasma membrane protein in situ. Furthermore, although we find bestrophin mRNA in several RPE cell lines, none express bestrophin protein.
Materials and Methods
Cell Culture.
RPE-J cells were maintained in DMEM supplemented with 4% FBS, nonessential amino acids, glutamine, and penicillin/streptomycin at 32°C in a 95% air/5% CO2 environment as previously described (22, 23) and differentiated on Matrigel-coated 1.2-cm diameter Transwell filters as before (23–25). ARPE-19 cells were maintained in DMEM/F12 supplemented with 10% FBS, glutamine, and penicillin/streptomycin at 37°C in a 95% air/5% CO2 environment as previously described (26). D407 cells were maintained in DMEM supplemented with 3% FBS, glutamine, and penicillin/streptomycin at 37°C in a 95% air/5% CO2 environment as previously described (27).
Cloning of Human Bestrophin cDNA and Adenovirus Production.
The primer pair 5′-CCGCTCGAGCCACCATGACCATCACTTACACAAGC-3′ and 5′-GGGGTACCTTAGGAATGTGCTTCATCCCT-3′, containing XhoI and KpnI adapters, was used to amplify a protein-coding region of the bestrophin cDNA from a human retina cDNA library (CLONTECH). The PCR fragment was cloned into the XhoI and KpnI sites of pcDNA3.1(−) (Invitrogen). DNA sequencing was used to verify the absence of PCR-generated errors. The cDNA encoding bestrophin was subcloned into the plasmid pAdlox for production of a type 5, E1-deleted, replication-defective adenovirus vector (AdBest ) by using Cre/Lox recombination according to the method of Hardy et al. (28). Large-scale adenovirus preparation was performed as before (24, 25), using a discontinuous CsCl2 gradient.
Reverse Transcription (RT)-PCR.
Total RNA was isolated by using Trizol (GIBCO) according to the manufacturer's instructions. Total RNA (2 μg) from each cell line was used for RT in a 25 μl total volume. Part of the RT (1 μl) sample was used for a 50-μl PCR reaction. Two sets of primers corresponding to human bestrophin cDNA sequence were used in PCR reactions. Primer set 1 (5′-GGCCAGATCTATGACCATCACTTACACAAGCC-3′ and 5′-CCGCCGCTCGTACTGGTTCCACCAGCGGG-3′) generates a 310-bp long fragment corresponding to the human bestrophin cDNA 5′ coding region. Primer set 2 (5′-GGCCAGATCTATGTACTGGAATAAGCCCGAGC-3′ and 5′-GGCCCTCGAGTTAGGAATGTGCTTCATCCCTG-3′) generates a 773-bp long fragment corresponding to the human bestrophin cDNA 3′ coding region. Both primer sets span multiple exons.
Antibody Production.
Monoclonal antibodies were produced in BALB/c mice immunized with the peptide KDHMDPYWALENRDEAHS coupled to the carrier protein, keyhole limpet hemocyanin by the glutaraldehyde crosslinking method (29). Spleen cells from one mouse were fused with Sp2/0-Ag14 myeloma cells (30) in the presence of 50% polyethylene glycol 4000 containing 10% dimethysulfoxide (31). The cells were plated on 96-well tissue culture plates, in 100 μl of DMEM in complete hypozanthione thymidine medium supplemented with 10% hybridoma cloning factor (Fisher Scientific). The next day, 100 μl of medium plus 4× aminopterin (Sigma) was added to the cultures. Approximately 2 weeks after the fusion, aliquots of media were tested by ELISA assay. Antibody-producing hybridoma cells were cloned by limiting dilution.
Polyclonal antibodies were produced in New Zealand White rabbits immunized with the same antigen used to produce monoclonal antibodies. The polyclonal antibodies used in this study are total IgG fraction purified from rabbit antibestrophin antisera by using protein A Sepharose.
Western Blots.
Western blots were performed as described previously (32) by using alkaline-phosphatase-conjugated secondary antibodies and tetranitroblue tetrazolium/5-bromo-4-chloro-3-indolyl phosphate, or horseradish peroxide conjugated secondary antibodies and ECL+.
Immunocytochemistry.
Macaque and porcine eyes were opened posterior to the limbus and fixed in 4% paraformaldehyde in 0.1 M phosphate buffer, pH 7.2, or Dulbecco's PBS containing 1 mM MgCl2 and 0.1 mM CaCl2. Tissues were rinsed in several changes of PBS and processed for paraffin microscopy by using conventional procedures. Sections were deparaffinized with xylene and hydrated through graded ethanols, then subjected to pressurized heat-mediated antigen retrieval in 0.01 M sodium citrate, pH 6.0, at 121°C for 1 min.
Tissue sections were incubated with 6% BSA in 0.1 M phosphate buffer, pH 7.2, for 30 min, then incubated overnight at 4°C with monoclonal antibody E6–6. After washing with PBS, sections were treated with goat anti-mouse IgG conjugated to peroxidase avidin–biotin–peroxidase complex (1:200 dilution; Vector Laboratories) for 1 h. Sections were washed with PBS and then incubated in 0.05% 3,3′-diaminobenzidine and 0.03% hydrogen peroxide in the phosphate buffer. Sections were viewed on a Nikon Microphot2 microscope. Images were acquired by using a SPOT2 charge-coupled device camera and metamorph software (Universal Imaging, Media, PA) and processed in Adobe photoshop 5.0.
Immunofluorescence staining of RPE-J cells was performed as before (24). Cells on Transwell filters were transduced with AdBest at a multiplicity of infection of 3, and moved from 32°C to 40°C. After 48 h, the cells were fixed in −20°C methanol and stained for bestrophin by using monoclonal antibody E6-6. Nuclei were stained with 4′,6-diamidino-2-phenylindole. Cells were examined by using a Leica (Deerfield, IL) TCP-SP confocal microscope with a 60× oil immersion objective lens. Z-series data were acquired in 0.5-μm steps and analyzed by using metamorph software.
RPE Isolation and Triton X-114 (Tx-114) Extraction.
RPE cells were isolated from human donor eyes as follows. The anterior segments were removed and the neural retina was peeled away. RPE cells were brushed from the eyecup in 500 μl of PBS with a camel's hair brush. After two additional washes, the volume was adjusted to 10 ml and the cells were pelleted at 110 × g for 20 min at 4°C. The cell pellet was stored dry at −80°C. Tx-114 phase separation was performed as described (33). Bestrophin was immunoprecipitated from the detergent or aqueous phases, resolved by SDS/PAGE, transferred to poly(vinylidene difluoride), and identified by immunoblot.
Cell Surface Biotinylation.
Cell surface biotinylation assays were performed on freshly isolated human RPE cells. Cells from one pair of eyes were suspended in 1 ml of ice-cold 0.5 mg/ml sulfo-N-hydroxysuccinimide-long chain-biotin (Pierce) in PBS containing 1 mM MgCl2 and 1 mM CaCl2 (PBS-CM). After 30 min, the cell suspension was diluted 10-fold with 50 mM NH4Cl in PBS-CM and pelleted for 20 min at 110 × g. After a second wash in 50 mM NH4Cl in PBS-CM, the pellet was lysed in 1% Triton X-100 in 50 mM Tris, pH 8.0, 150 mM NaCl, and 1 mM EDTA, containing a protease inhibitor mixture. Bestrophin was immunoprecipitated from the lysate by using Pab-125, and Grp78 was immunoprecipitated by using a polyclonal anti-Grp78 antibody (Affinity BioReagents, Golden, CO) as described elsewhere (34). Biotinylated proteins were resolved by SDS/PAGE and visualized by using horseradish peroxidase-conjugated streptavidin and ECL+. The polarity of bestrophin in RPE-J cells transduced with AdBest was determined by using differential cell surface biotinylation as described previously (34).
Results
Production of Antibodies.
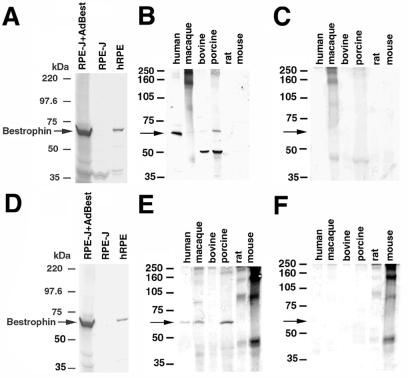
To produce antibodies recognizing bestrophin, two rabbits and four mice were immunized with a peptide corresponding to the C terminus of human bestrophin. Monoclonal antibodies produced by 10 hybridomas that were deemed clonal after three rounds of limiting dilution were characterized. All data in this manuscript were obtained by using monoclonal antibodies from clone E6-6. Antisera from two rabbits also were characterized. All polyclonal antibodies used in this manuscript are from a total IgG fraction derived from the serum of rabbit 125. Accordingly, the polyclonal antibody will be referred to as Pab-125. As shown in Fig. 1 A and D, antibodies were tested by Western blotting against lysates of human RPE, RPE-J cells, and RPE-J cells transduced with the replication-defective adenovirus vector AdBest for bestrophin expression. Both Pab-125 and E6-6 recognized a single band in human RPE lysates and in transduced RPE-J cells of ≈68 kDa. This band was absent in untransduced RPE-J cells. The species specificity of our antibodies was determined by Western blots of RPE or RPE/choroid preparations from human, macaque, bovine, porcine, rat, and mouse eyes. Pab-125 recognized a band of 68 kDa in human, macaque, and porcine eyes, as well as a lower band of ≈50–55 kDa in porcine eyes (Fig. 1B). E6-6 recognized a single band of ≈68 kDa in human, macaque, and porcine tissues (Fig. 1E). Neither antibody recognized bestrophin in lysates of mouse or rat RPE/choroid.
Figure 1.
Specificity of antibestrophin antibodies. The specificity of polyclonal antibody Pab125 (A--C) and monoclonal antibody E6-6 (D–F) was determined by immunoblot. Both antibodies recognize a band of ≈68 kDa, the predicted mass of bestrophin, in human RPE and in RPE-J cells transduced with an adenovirus vector encoding human bestrophin (AdBest) but not in untransduced RPE-J cells (A and D). To determine the crossreactivity of our antibodies with bestrophin in other species, we performed Western blots by using lysates derived from RPE or RPE/choroid preparations from various species (B, C, E, and F). Pab-125 (B) recognized an ≈68kDa band in human-, macaque-, and porcine-derived lysates, as well as an ≈45kDa band in bovine and porcine lysates that was not present in a control blot (C) using only the goat anti-rabbit secondary antibody. Monoclonal antibody E6-6 (E) recognized a single band of ≈68 kDa in human, macaque, and porcine lysates. Extraneous bands were caused by nonspecific reactivity of the secondary goat antimouse antibody (F).
Expression of Bestrophin in RPE-Derived Cell Lines.
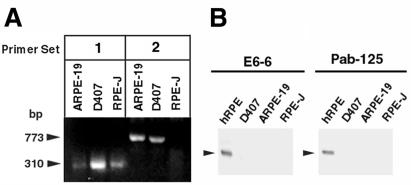
The human-derived RPE cell lines ARPE-19 and D407, as well as RPE-J, were probed by RT-PCR to determine whether they express bestrophin mRNA. Total RNA was isolated from proliferating cultures of the three cell lines. As shown in Fig. 2A, all cell lines were positive by RT-PCR using primer set 1 corresponding to the conserved N-terminal region of bestrophin. Both ARPE-19 and D407 were positive using primer set 2 corresponding to the C terminus of human bestrophin. Western blots of all three cell lines using either Pab-125 or mab E6-6 were negative (Fig. 2B) for protein expression.
Figure 2.
Expression of bestrophin in RPE-derived cell lines. To examine the expression of bestrophin mRNA in RPE-derived cell lines, we probed total RNA derived from RPE-J, ARPE-19, and D407 cells by RT-PCR (A). A portion of the PCR reaction samples (1/5) was resolved on a 2% agarose gel. In PCR reactions using primer set 1, a 310-bp band is visible for all three cell lines. In PCR reactions using primer set 2, a 773-bp band is visible for ARPE-19 and D407, but not RPE-J. This finding is consistent with the lack of immunoreactivity of our antibodies with rat bestrophin, because the antigen used to generate the antibodies is a peptide derived from the C terminus of human bestrophin, which would overlap with the reverse primer in primer set 2. To examine the expression of bestrophin protein (B) in RPE-derived cell lines, we probed 250 μg each of lysates derived from RPE-J, ARPE-19, and D407 cells, and as a positive control, 25 μg of a human RPE lysate, by Western blot with monoclonal antibody E6-6 and Pab-125. Bestrophin was not detected in any of the cell lines. However, because our antibodies do not detect rat bestrophin, we cannot at this time conclude that RPE-J cells do not express bestrophin.
Localization of Bestrophin.
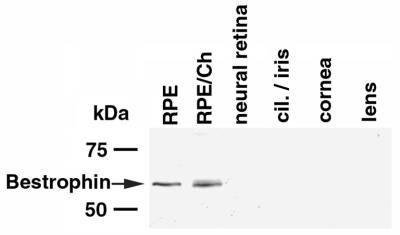
To determine the distribution of bestrophin in the human eye (Fig. 3), Western blot analysis of human RPE, RPE/choroid, neurosensory retina, lens, cornea, and ciliary body/iris was performed by using both Pab-125 and E6-6. Bestrophin was detected only in RPE and RPE/choroid preparations. The intensity of the bestrophin band was similar in both RPE and RPE/choroid samples, despite an 8-fold difference in the amount of protein loaded on the gel.
Figure 3.
Expression of bestrophin in human ocular tissues. To examine the distribution of bestrophin in the human eye we performed Western blots (A) by using monoclonal antibody E6-6 on lysates derived from RPE, RPE/choroid (RPE/Ch), neural retina, ciliary body and iris (cil./iris), cornea, and lens. All were loaded at 400 μg per lane except RPE, which was loaded at 50 μg per lane. As shown in A, bestrophin was detected only in RPE and RPE/choroid preparations. The intensity of the bands is similar even though the protein load in the RPE/Ch lane is 8-fold greater.
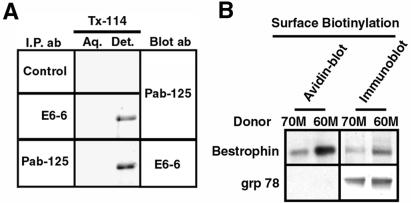
Bestrophin is predicted to be a transmembrane protein (14, 20, 21). To determine whether bestrophin is associated with cellular membranes, three independent assays were used. The first was extraction of human RPE cells with the detergent Tx-114 followed by phase separation into detergent and aqueous phases. Membrane proteins will partition with the detergent phase by using this technique (33). As shown in Fig. 4, the majority of bestrophin was detected in the detergent phase. To determine whether bestrophin is present on the plasma membrane or only in intracellular membranes, surface biotinylation was used to label plasma membrane proteins in isolated human RPE cells. Bestrophin was clearly labeled with biotin, in contrast to the endoplasmic-reticulum-associated protein Grp78, which exhibited no labeling.
Figure 4.
Biochemical localization of bestrophin in human RPE. To test whether bestrophin is a membrane protein we performed Triton X-114 (Tx-114) phase extraction (A) on human RPE cells. Cells were lysed in 1% Tx-114 and bestrophin was immunoprecipitated from either the aqueous or detergent phase by using either Pab-125, E6-6, or a control primary antibody. A strong bestrophin band was present in the detergent phase, indicating that bestrophin is a membrane protein. To determine whether bestrophin is an integral plasma membrane protein, we labeled the surface of human RPE cells with sulfo-N-hydroxysuccinimide-long chain-biotin. After lysis bestrophin was immunoprecipitated with Pab-125 and as a control, the endoplasmic reticulum protein Grp78/bip was immunoprecipitated by using a rabbit anti-Grp78 antibody. Streptavidin blotting of the immunoprecipitates revealed that bestrophin had indeed been biotinylated, in contrast to Grp78, which demonstrates that the cells were intact. To ensure that Grp78 immunoprecipitation worked, we stripped and blotted the membranes with either rabbit anti-Grp78 or Pab-125. Both bestrophin and Grp78 were efficiently immunoprecipitated.
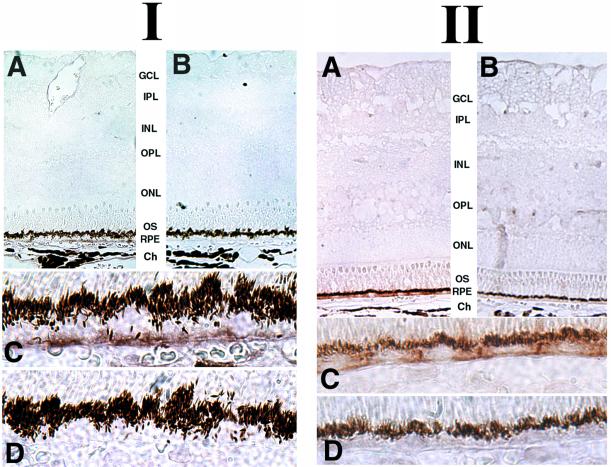
Having determined that bestrophin is a plasma membrane protein, its localization in the cell was confirmed by using immunocytochemistry. Macaque and porcine eyes were probed (Fig. 5) with mab E6-6 by using standard horseradish peroxide detection to avoid problems arising from the overwhelming autofluorescence associated with RPE-lipofuscin granules even in normal eyes (35, 36). On examination of sections at low magnification, bestrophin staining was observed only in the RPE layer of eyes in each of the species studied. Staining was absent from control sections in which the antibody was preincubated with the peptide antigen. Inspection of the sections at higher magnifications revealed that bestrophin staining was primarily associated with the basolateral plasma membrane of the RPE cells.
Figure 5.
Immunohistochemical localization of bestrophin in macaque and porcine RPE. Macaque (I) or porcine (II) eyes fixed by immersion in 4% paraformaldehyde were stained for bestrophin by using monoclonal antibody E6-6 (A and C), or to control for specificity with monoclonal antibody E6-6 preincubated with the peptide antigen (B and D) as described in Methods. Inspection of sections at low magnification (A and B) revealed that bestrophin expression is restricted to the RPE in both macaque and porcine eyes. Inspection at higher magnification (C and D) revealed that bestrophin is concentrated along the basal surface and the lateral borders of the cells.
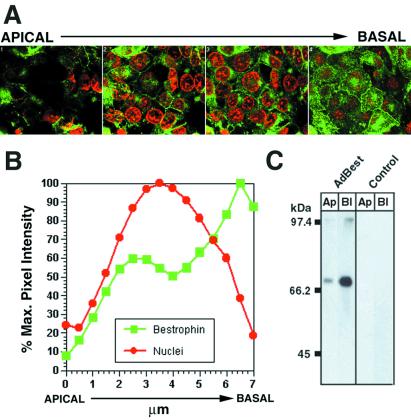
To further confirm the localization of bestrophin, human bestrophin was expressed in polarized RPE-J cells by adenovirus-mediated gene transfer. RPE-J cells transduced with AdBest were examined by using immunofluorescence and confocal microscopy (Fig. 6A). In every section series examined, maximal staining was of the basal surface and lateral borders of the cells. Some intracellular staining of vesicular and perinuclear compartments also was noted, the degree of which correlated with the level of bestrophin expression in the cells and is most likely because of overexpression of the protein. Analysis of the pixel intensity of bestrophin staining (Fig. 6B) with respect to nuclear staining in the various planes of each series clearly indicated that bestrophin is basolateral polarized. Cell surface biotinylation also was used as a biochemical marker of polarity. As shown in Fig. 6C, the polarity of bestrophin in RPE-J cells transduced with AdBest at a multiplicity of infection of 3 (Fig. 6C) was distinctly basolateral, in agreement with immunofluorescence microscopy experiments.
Figure 6.
Localization of bestrophin in RPE-J cells. Confluent polarized monolayers of RPE-J cells were transduced with AdBest as described in Methods. Cells were fixed in methanol at −20°C and stained with monoclonal antibody E6-6 and goat anti-mouse IgG conjugated to FITC (green). Nuclei were stained with 4′,6-diamidino-2-phenylindole (red). Confocal microscopy was used to produce a section series with planes spaced ≈0.5 μm apart. A montage is shown (A) that depicts the average fluorescence in four adjacent images, covering a 2-μm depth of the cells. Inspection of the planes revealed staining along the basal surface and lateral borders as well as in a perinuclear compartment and intracellular vesicular structures. Analysis of average pixel intensities (B) in each plane indicated a pattern of bestrophin immunoreactivity consistent with its localization in the basolateral plasma membrane of the cells. This basolateral localization in RPE-J cells transduced with AdBest was reinforced by the greater streptavidin reactivity of bestrophin immunoprecipitates from cells biotinylated selectively on either their apical or basolateral surface (C).
Discussion
Bestrophin is the protein altered in BMD (14), an inherited retinal degenerative disease similar to AMD. BMD is diagnosed by its unique electrophysiologic symptoms, a depressed EOG (light peak to dark trough ratio <1.5) in the absence of an altered electroretinogram (1, 2). Our understanding of bestrophin function, how mutations in bestrophin cause BMD, and how bestrophin may contribute to the transepithelial potential of RPE cells, requires knowledge of its tissue-specific expression patterns and its localization within the cell. In this study, we have used a variety of biochemical and immunocytochemical means to identify the localization of bestrophin.
Our biochemical data indicate that bestrophin will partition into the detergent phase when extracted with Tx-114. This is a property unique to integral and peripheral membrane proteins (33). The amino acid sequence of bestrophin predicts that it is an integral membrane protein (14, 20, 21, 37), a finding confirmed by our cell surface biotinylation experiments. Because the biotin does not have access to the interior of the cell, the only way that bestrophin could be labeled would be if a portion of it were exposed to the extracellular environment. The most likely model for bestrophin topology would place two loops of 21 aa (present between transmembrane domains 1 and 2) and 17 aa (present between transmembrane domains 3 and 4) outside of the cell (14, 37). These domains contain two lysine residues that would be available for modification by sulfo-N-hydroxysuccinimide-long chain-biotin. There are no predicted modifications of these extracellular loops that would interfere with the possibility of biotinylation. A second model recently has been proposed for bestrophin that predicts five transmembrane domains (21). According to this model, the C terminal domain of bestrophin would be extracellular and would contain two potential N-linked glycosylation sites. Neither Pab-125, nor E6-6 detects exogenous bestrophin in 4% paraformaldehyde-fixed RPE-J cells without permeabilization (data not shown). Furthermore, bestrophin is detected on Western blots at the mass predicted by the primary amino acid sequence. If bestrophin is a glycoprotein, we would predict that the N-glycans would change the mobility of bestrophin in SDS/PAGE gels or result in a more diffuse band than we detect. Thus, we currently favor the four-transmembrane domain model.
Finding bestrophin on the plasma membrane by biochemical means is not sufficient to conclude that the protein is in fact a resident plasma membrane protein. Furthermore, if bestrophin is a resident plasma membrane protein, determining its polarity is critical to formulating models of bestrophin function. To address this question, we localized bestrophin by immunocytochemistry in macaque and porcine eyes. As predicted by the in situ hybridization experiments of others (14), and in agreement with our Western blot data (see Figs. 3 and 5), bestrophin immunoreactivity was confined to the RPE. When sections in several orientations were inspected at higher magnifications, intense staining of the basal and lateral surfaces of RPE cells was observed, consistent with basolateral plasma membrane localization. To further examine the polarized localization of bestrophin, we expressed the protein in polarized monolayers of the rat RPE-derived cell line, RPE-J. Both cell surface labeling and confocal microscopy experiments confirmed a basolateral localization (Fig. 6).
We examined the expression of bestrophin in three commonly used RPE cell lines: ARPE-19, D407, and RPE-J. Bestrophin mRNA was detected in all three by RT-PCR. However, consistent with a lack of homology in the C terminus of mouse and human bestrophin (data not shown), we could not detect a PCR product from RPE-J cells by using primer set 2, recognizing the nucleic acid sequence at the C terminus of human bestrophin. Western blot analysis of ARPE-19, D407, and RPE-J lysates were negative, indicating that neither D407 nor ARPE-19 cells produce bestrophin. Because our antibodies do not recognize rat bestrophin, we cannot conclude that RPE-J cells do not produce the protein. However, these data do indicate that bestrophin mRNA is synthesized by all three RPE cell lines. The mRNA is either untranslated, or the protein is present in quantities below our limit of detection. A similar phenomenon has been reported for RPE-65 (38), which is transcribed but not translated in ARPE-19 cells (39). Interestingly, RPE-65 protein is made by primary fetal human RPE cultures. However, although these cultures are positive for bestrophin mRNA on Northern blots, they do not produce the protein until after many months in culture (personal communication, D. Bok).
In summary, we have produced monoclonal and polyclonal antibodies that recognize bestrophin, the protein product of the VMD2 gene. By using these antibodies, we have demonstrated that bestrophin is a basolateral plasma membrane protein in vivo, placing it in a location consistent with a role in the effects on the EOG characteristic of BMD.
Acknowledgments
We thank Dr. Cindy Duchala of the Cleveland Clinic Foundation hybridoma core for assistance with antibody production, Dr. Judy Drazba for assistance with confocal microscopy, Dr. Ignacio Rodriguez for macaque eyes, and Dr. Neal Peachy for reading the manuscript. Wen Li provided excellent technical assistance. We are also grateful to the Cleveland Eye Bank. This work was supported by National Institutes of Health Grant R01-EY13160 (to A.D.M.), a Kirchgessner foundation research grant (to A.D.M.), and funds from the Foundation Fighting Blindness and the Cleveland Clinic Foundation.
Abbreviations
- BMD
Best vitelliform macular dystrophy
- EOG
electrooculogram
- Tx-114
Triton X-114
- AMD
age-related macular degeneration
- RPE
retinal pigment epithelium
- RT-PCR
reverse transcription–PCR
Footnotes
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.220402097.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.220402097
References
- 1.Gass D J M. In: Stereoscopic Atlas of Macular Diseases, Diagnosis and Treatment. Gass D J M, editor. Vol. 1. St. Louis: Mosby; 1997. pp. 303–313. [Google Scholar]
- 2.Marmor M F, Small K. In: The Retinal Pigment Epithelium. Wolfensberger T J, Marmor M F, editors. New York: Oxford Univ. Press; 1998. pp. 330–333. [Google Scholar]
- 3.Leibowitz H M, Krueger D E, Maunder L R, Milton R C, Kini M M, Kahn H A, Nickerson R J, Pool J, Colton T L, Ganley J P, et al. Surv Ophthalmol. 1980;24:335–610. [PubMed] [Google Scholar]
- 4.Godel V, Chaine G, Regenbogen L, Coscas G. Acta Ophthalmol Suppl. 1986;175:1–31. [PubMed] [Google Scholar]
- 5.Cross H E, Bard L. Am J Ophthalmol. 1974;77:46–50. doi: 10.1016/0002-9394(74)90603-5. [DOI] [PubMed] [Google Scholar]
- 6.Weingeist T A, Kobrin J L, Watzke R C. Arch Ophthalmol. 1982;100:1108–1114. doi: 10.1001/archopht.1982.01030040086016. [DOI] [PubMed] [Google Scholar]
- 7.Maloney W F, Robertson D M, Duboff S M. Arch Ophthalmol. 1977;95:979–983. doi: 10.1001/archopht.1977.04450060065003. [DOI] [PubMed] [Google Scholar]
- 8.Bard L A, Cross H E. Trans Am Acad Ophthalmol Otolaryngol. 1975;79:OP865–OP873. [PubMed] [Google Scholar]
- 9.O'Gorman S, Flaherty W A, Fishman G A, Berson E L. Arch Ophthalmol. 1988;106:1261–1268. doi: 10.1001/archopht.1988.01060140421045. [DOI] [PubMed] [Google Scholar]
- 10.Frangieh G T, Green W R, Fine S L. Arch Ophthalmol. 1982;100:1115–1121. doi: 10.1001/archopht.1982.01030040093017. [DOI] [PubMed] [Google Scholar]
- 11.Green W R, Enger C. Ophthalmology. 1993;100:1519–1535. doi: 10.1016/s0161-6420(93)31466-1. [DOI] [PubMed] [Google Scholar]
- 12.Curcio C A, Medeiros N E, Millican C L. Invest Ophthalmol Vis Sci. 1998;39:1085–1096. [PubMed] [Google Scholar]
- 13.Bird A C, Bressler N M, Bressler S B, Chisholm I H, Coscas G, Davis M D, de Jong P T, Klaver C C, Klein B E, Klein R, et al. Surv Ophthalmol. 1995;39:367–374. doi: 10.1016/s0039-6257(05)80092-x. [DOI] [PubMed] [Google Scholar]
- 14.Petrukhin K, Koisti M J, Bakall B, Li W, Xie G, Marknell T, Sandgren O, Forsman K, Holmgren G, Andreasson S, et al. Nat Genet. 1998;19:241–247. doi: 10.1038/915. [DOI] [PubMed] [Google Scholar]
- 15.Weber B H, Vogt G, Pruett R C, Stohr H, Felbor U. Nat Genet. 1994;8:352–356. doi: 10.1038/ng1294-352. [DOI] [PubMed] [Google Scholar]
- 16.Allikmets R, Singh N, Sun H, Shroyer N F, Hutchinson A, Chidambaram A, Gerrard B, Baird L, Stauffer D, Peiffer A, et al. Nat Genet. 1997;15:236–246. doi: 10.1038/ng0397-236. [DOI] [PubMed] [Google Scholar]
- 17.Stone E M, Lotery A J, Munier F L, Heon E, Piguet B, Guymer R H, Vandenburgh K, Cousin P, Nishimura D, Swiderski R E, et al. Nat Genet. 1999;22:199–202. doi: 10.1038/9722. [DOI] [PubMed] [Google Scholar]
- 18.Allikmets R, Seddon J M, Bernstein P S, Hutchinson A, Atkinson A, Sharma S, Gerrard B, Li W, Metzker M L, Wadelius C, et al. Hum Genet. 1999;104:449–453. doi: 10.1007/s004390050986. [DOI] [PubMed] [Google Scholar]
- 19.Lotery A J, Munier F L, Fishman G A, Weleber R G, Jacobson S G, Affatigato L M, Nichols B E, Schorderet D F, Sheffield V C, Stone E M. Invest Ophthalmol Visual Sci. 2000;41:1291–1296. [PubMed] [Google Scholar]
- 20.Marquardt A, Stohr H, Passmore L A, Kramer F, Rivera A, Weber B H. Hum Mol Genet. 1998;7:1517–1525. doi: 10.1093/hmg/7.9.1517. [DOI] [PubMed] [Google Scholar]
- 21.White K, Marquardt A, Weber B H. Hum Mutat. 2000;15:301–308. doi: 10.1002/(SICI)1098-1004(200004)15:4<301::AID-HUMU1>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 22.Nabi I R, Mathews A P, Cohen-Gould L, Gundersen D, Rodriguez-Boulan E. J Cell Sci. 1993;104:37–49. doi: 10.1242/jcs.104.1.37. [DOI] [PubMed] [Google Scholar]
- 23.Marmorstein A D, Bonilha V L, Chiflet S, Neill J M, Rodriguez-Boulan E. J Cell Sci. 1996;109:3025–3034. doi: 10.1242/jcs.109.13.3025. [DOI] [PubMed] [Google Scholar]
- 24.Marmorstein A D, Gan Y C, Bonilha V L, Finnemann S C, Csaky K G, Rodriguez-Boulan E. J Cell Biol. 1998;142:697–710. doi: 10.1083/jcb.142.3.697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marmorstein A D, Csaky K G, Baffi J, Lam L, Rahaal F, Rodriguez-Boulan E. Proc Natl Acad Sci USA. 2000;97:3248–3253. doi: 10.1073/pnas.070049497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dunn K C, Marmorstein A D, Bonilha V L, Rodriguez-Boulan E, Giordano F, Hjelmeland L M. Invest Ophthalmol Visual Sci. 1998;39:2744–2749. [PubMed] [Google Scholar]
- 27.Davis A A, Bernstein P S, Bok D, Turner J, Nachtigal M, Hunt R C. Invest Ophthalmol Visual Sci. 1995;36:955–964. [PubMed] [Google Scholar]
- 28.Hardy S, Kitamura M, Harris-Stansil T, Dai Y, Phipps M L. J Virol. 1997;71:1842–1849. doi: 10.1128/jvi.71.3.1842-1849.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Walter G, Scheidtmann K H, Carbone A, Laudano A P, Doolittle R F. Proc Natl Acad Sci USA. 1980;77:5197–5200. doi: 10.1073/pnas.77.9.5197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kohler G, Milstein C. Eur J Immunol. 1976;6:511–519. doi: 10.1002/eji.1830060713. [DOI] [PubMed] [Google Scholar]
- 31.Galfre G, Howe S C, Milstein C, Butcher G W, Howard J C. Nature (London) 1977;266:550–552. doi: 10.1038/266550a0. [DOI] [PubMed] [Google Scholar]
- 32.Marmorstein L Y, Ouchi T, Aaronson S A. Proc Natl Acad Sci USA. 1998;95:13869–13874. doi: 10.1073/pnas.95.23.13869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bordier C. J Biol Chem. 1981;256:1604–1607. [PubMed] [Google Scholar]
- 34.Marmorstein A D, Zurzolo C, Le Bivic A, Rodriguez-Boulan E. In: Cell Biology: A Laboratory Handbook. Celis J, editor. Vol. 4. New York: Academic; 1998. pp. 341–350. [Google Scholar]
- 35.Wing G L, Blanchard G C, Weiter J J. Invest Ophthalmol Visual Sci. 1978;17:601–607. [PubMed] [Google Scholar]
- 36.Feeney L. Invest Ophthalmol Visual Sci. 1978;17:583–600. [PubMed] [Google Scholar]
- 37.Bakall B, Marknell T, Ingvast S, Koisti M J, Sandgren O, Li W, Bergen A A, Andreasson S, Rosenberg T, Petrukhin K, et al. Hum Genet. 1999;104:383–389. doi: 10.1007/s004390050972. [DOI] [PubMed] [Google Scholar]
- 38.Liu S Y, Redmond T M. Arch Biochem Biophys. 1998;357:37–44. doi: 10.1006/abbi.1998.0817. [DOI] [PubMed] [Google Scholar]
- 39.Dunn K C, Aotaki-Keen A E, Putkey F R, Hjelmeland L M. Exp Eye Res. 1996;62:155–169. doi: 10.1006/exer.1996.0020. [DOI] [PubMed] [Google Scholar]