Abstract
HOX genes encode transcription factors that control patterning and cell fates. Alterations in HOX expression have been clearly implicated in leukemia, but their role in most other malignant diseases remains unknown. By using degenerate reverse transcription–PCR and subsequent real-time quantitative assays, we examined HOX expression in lung cancer cell lines, direct tumor-control pairs, and bronchial epithelial cultures. As in leukemia, genes of the HOX9 paralogous group and HOXA10 were frequently overexpressed. For HOXB9, we confirmed that elevated RNA was associated with protein overexpression. In some cases, marked HOX overexpression was associated with elevated FGF10 and FGF17. During development, the WNT pathway affects cell fate, polarity, and proliferation, and WNT7a has been implicated in the maintenance of HOX expression. In contrast to normal lung and mortal short-term bronchial epithelial cultures, WNT7a was frequently reduced or absent in lung cancers. In immortalized bronchial epithelial cells, WNT7a was lost concomitantly with HOXA1, and a statistically significant correlation between the expression of both genes was observed in lung cancer cell lines. Furthermore, we identified a homozygous deletion of β-catenin in the mesothelioma, NCI-H28, associated with reduced WNT7a and the lowest overall cell line expression of HOXA1, HOXA7, HOXA9, and HOXA10, whereas HOXB9 levels were unaffected. Of note, both WNT7a and β-catenin are encoded on chromosome 3p, which undergoes frequent loss of heterozygosity in these tumors. Our results suggest that alterations in regulatory circuits involving HOX, WNT, and possibly fibroblast growth factor pathways occur frequently in lung cancer.
Keywords: HOXA1, HOXA7, HOXA9, HOXA10, HOXB9
Homeodomain-containing genes encode a set of master transcription factors which function during development to control pattern formation, differentiation, and proliferation. A common feature is the presence of a highly conserved 61-aa motif, the homeodomain (1). Mammals possess at least 39 class I HOX genes grouped into four major clusters (A–D). During development, HOX expression occurs temporally in accordance with both the position of a gene within its cluster and in a rostral-caudal manner (2). HOX genes in equivalent positions of different clusters (paralogs) are more closely related than adjacent genes in the same cluster. Recent evidence from chimeric gene experiments suggests that paralogous HOX loci may be functionally equivalent (3) and that it is the expression level of the paralogous group as a whole which is critical for correct development.
Deregulation of HOX genes has been observed in cancer, with the most convincing evidence coming from leukemia. For example, four different translocations in acute myeloid leukemia fuse the nucleoporin domain of NUP98 to the homeobox of a major or divergent HOX protein (4–6). Similarly, the T cell (10;14) chromosomal translocation in acute lymphoblastic leukemia results in overexpression of HOX11 (7) and the pre-B cell (1;19) translocation fuses the homeodomain of PBX1 with E2A proteins, leading to aberrant expression of WNT16 and fibroblast growth factor 15 (FGF15) (8, 9). Strikingly, HOXA9 was recently identified as the single gene whose expression was most correlated with treatment failure in acute myeloid leukemia (10). In solid tumors, rearrangements of HOX genes have not been reported. However, expression surveys have noted differences between normal and tumor samples in kidney and colon (11, 12). In melanomas, Caré et al. (13) reported that HOXB7 was constitutively expressed in melanomas and that antisense HOXB7 inhibited cellular proliferation and expression of basic fibroblast growth factor.
Few reports have dealt with HOX expression in lung cancer. Using Northern blots, Tiberio et al. (14) reported that few HOX genes were expressed in normal lung and these patterns were altered in small-cell lung cancer (SCLC) xenografts. Flagiello et al. (15) noted that retinoic acid treatment of SCLCs resulted in noncontiguous expression patterns in HOXB and HOXC genes. Last, Omatu (16) introduced HOXD3 into A549 cells and reported that this enhanced invasive properties. To further investigate the role of HOX genes in lung cancer, we used an initial degenerate reverse transcription–PCR (RT-PCR) approach to survey patterns of HOX expression in normal lung, lung cancer cell lines, and mesotheliomas which originate from the lung surface. These results identified differences in HOX expression both among tumors and compared with normal lung which were confirmed by quantitative real-time RT-PCR assays. We found that HOXB9, HOXA9, and HOXA10 were frequently up-regulated in lung cancer cell lines and direct tumors. At least for HOXB9, overexpression at the RNA level was correlated with protein overexpression. We observed that HOX overexpression may have functional consequences in terms of FGF expression. Finally, we identified a strong correlation between HOXA1 and WNT7a expression that may be linked to an autoregulatory loop in the WNT pathway (17).
Materials and Methods
Cell Lines and Tumors.
Twenty-five lung cancer cell lines (available from the American Type Culture Collection, Manassas, VA) and 25 matched tumor/nonmalignant lung pairs were obtained through the Colorado Lung Cancer Specialized Program of Research Excellence. The nonmalignant samples were obtained from a site equivalent, but removed from the tumor. Ten additional tumors were from pathology archives at the Centre Hospitalier Régional Universitaire (E.B.; Grenoble). The lung cancer cell lines have been described (18). GLC20 was provided by C. Buys (University of Groningen, Groningen, The Netherlands). Normal human bronchial epithelial (NHBE) cells were obtained from Clonetics (San Diego). In addition, three short-term bronchial epithelial cultures, 302–1, 604–2, and 2523, were derived from nonmalignant biopsies of heavy smokers. A fourth bronchial culture, TR5214–2, was immortalized with simian virus 40 large-T (tumor) antigen and examined at passages 50, 78, and 83. Human vascular endothelial cell (HUVEC) cells were obtained from BioWhittaker.
RNA Isolation and cDNA Preparation.
Total RNA was isolated by using Trizol (Life Technologies, Gaithersburg, MD) and further purified on RNeasy columns (Qiagen, Chatsworth, CA) with RNase-free DNase treatment. Absence of contaminating genomic DNA was verified by PCR using multiple primer pairs that amplify genomic DNA segments. Reverse transcription reactions used ≈2 μg of total RNA in 20 μl with 200 units of SuperScript II (Life Technologies) and random hexamers.
Homeobox Amplification by Using Degenerate Primers.
Two blocks of completely conserved amino acids, ELEKEF and KIWFQN, within the homeodomain were chosen for degenerate primers. These were ELEKEF [5′-GCT CTA GA(A/G) (C/T)T(A/C/G/T) GA(A/G) AA(A/G) GA(A/G) TT] and KIWFQN [5′-GGA ATT C(A/G)T T(C/T)T G(A/G)A ACC A(A/G/T)A T(C/T)T T]. PCR conditions consisted of 32–35 cycles of amplification (94°C, 1 min; 40°C, 1 min; and 72°C, 1 min) with 4.5 mM MgCl2 and 2 μM primers. A band of ≈120 bp was gel isolated and cloned into a T-vector. Individual clones were sequenced and analyzed by using blast.
Real-Time RT-PCR.
An ABI5700 (PE Biosystems, Foster City, CA) with SYBR greenI fluorescence was used. Primers were designed to amplify segments of <200 bp to maximize efficiency. Primer sequences were as follows: HOXA1 forward 5′-ACC CCT CGG ACC ATA GGA TTA C-3′, reverse 5′-AAG GCG CAC TGA AGT TCT GTG-3′; HOXA7 forward 5′-ATC ACT CTA CCT CGT AAA ACC GAC AC-3′, reverse 5′-ACA TAA TAC GAA GAA CTC ATA ATT TTG ACC-3′; HOXA9 forward 5′-CAG AAC TGG TCG GTG ATT TAG GTA G-3′, reverse 5′-CAA CTG AAG TAA TGA AGG GCA GTG-3′; HOXA10 forward 5′-GAG AGC AGC AAA GCC TCG C-3′, reverse 5′-CCA GTG TCT GGT GCT TCG TG-3′; HOXB9 forward 5′-AAA AAG CGC TGT CCC TAC ACC-3′, reverse 5′-AGG AGT CTG GCC ACT TCG TG-3′; WNT7a forward 5′-TGC CCG GAC TCT CAT GAA C-3′, reverse 5′-GTG TGG TCC AGC ACG TCT TG-3′; and glyceraldehyde-3-phosphate dehydrogenase (G3PDH) forward 5′-TGC ACC ACC AAC TGC TTA GC-3′, reverse 5′-GGC ATG GAC TGT GGT CAT GAG-3′. Twenty-microliter reactions were used under conditions suggested by Applied Biosystems. Data were analyzed by using GeneAmp 5700 sds software (version 1.1) and converted into threshold cycle (Ct) values (see Results). Nonquantitative primers included FGF10 forward 5′-CAT TGT GCC TCA GCC TTT C-3′; reverse 5′-TCC ATT TTC CTC TAT CCT CTC C-3′; FGF17 forward 5′-TGC TGC CCA ACC TCA CTC-3′, and reverse 5′-TCT TTG CTC TTC CCG CTG-3′.
Immunofluorescence and Western Blots.
For immunofluorescence, cells were fixed as described (19). The anti-β-catenin mAb was C19220 (Transduction Laboratories, Lexington, KY) diluted 1:100. The secondary antibody was FITC-conjugated goat anti-mouse IgG (Jackson ImmunoResearch) diluted 1:100. Images were captured with a Vysis Smart Capture System on an Olympus BX60 microscope equipped with a Photometrix SenSys charge-coupled device camera. For HOXB9 Westerns, filters were incubated with anti-HOXB9 antibody (Berkeley Antibody, Richmond, CA) at 1:1000 dilution and processed as suggested by the manufacturer. To control for protein loading, parallel gels were incubated with anti-tubulin antibody (Ab-4, clone DM1A + DM1B, NeoMarkers, Freemont, CA) at 1:500 dilution. For detection, an enhanced chemiluminescence kit was used (Amersham International). To demonstrate specificity of the HOXB9 antibody, we prepared a glutathione S-transferase (GST)-HOXB9 fusion protein by using a bacterial expression construct (20) kindly provided by Laura Corbo (Centre Leon Berard, Lyon, France). GST-HOXB9 protein was affinity purified from 500 ml of BL21 cells as described by Harper and Speicher (21). For blocking, primary antibody was absorbed for 2 h with 50 μl of HOXB9 beads, or as a control, glutathione-agarose beads.
Results
RT-PCR Survey of HOX Expression by Using Degenerate Primers.
We initially surveyed HOX expression by using degenerate RT-PCR with primers corresponding to invariant amino acid sequences within the homeodomain (22). PCR products were subcloned and sequenced from 10 cell lines and normal lung. To assess PCR biases, we used genomic DNA as a template because the amplified segment is uninterrupted by an intron. These results, organized into paralogous groups, are shown in Table 1 as percentages. Only loci that gave products from either cDNA or genomic DNA are listed. This provided an estimate of the most abundantly expressed HOX genes and suggested differences among samples. For instance, whereas HOX9 paralogs and HOXA10 products were relatively abundant in several lung cancer cell lines, they were not detected in normal lung. This was interesting because HOXA9 is a known target of chromosomal alterations in leukemia (6) and HOXA9/HOXA10 coexpression has been similarly noted (23). Differences could occur in several ways such as overexpression in lung tumors, underexpression in nonmalignant lung, or alternatively, levels could be similar but masked by competing HOX genes.
Table 1.
HOX expression in lung cancers by degenerate PCR
| HOX | DNA control | Normal lung | GLC20 | H187 | H460 | H661 | H1648 | H322 | H226 | H290 | H513 | H28 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A1 | 11 | 5 | 14 | 24 | 25 | |||||||
| B1 | 12 | |||||||||||
| C1 | 3 | 2 | 5 | 4 | 11 | |||||||
| A2 | 2 | 3 | ||||||||||
| A3 | 3 | 13 | 4 | 16 | 2 | 34 | 2 | 33 | 2 | 2 | 8 | |
| B3 | 1 | 26 | 2 | 4 | 3 | 4 | 3 | 13 | 7 | 6 | ||
| A4 | 7 | 5 | 4 | 2 | 2 | 14 | 2 | 2 | 3 | 2 | ||
| B4 | 1 | 2 | 2 | |||||||||
| C4 | 2 | 24 | 10 | 3 | 2 | 6 | 13 | 9 | 6 | |||
| D4 | 1 | 2 | ||||||||||
| A5 | 3 | 8 | 2 | 6 | 8 | 2 | 4 | |||||
| B5 | 1 | |||||||||||
| A6 | 1 | 3 | ||||||||||
| B6 | 5 | 16 | 4 | 5 | 11 | 2 | ||||||
| C6/C8 | 2 | 2 | 4 | 20 | 10 | 33 | 3 | 7 | 13 | 2 | 6 | |
| A7 | 5 | 8 | 31 | 2 | 2 | 11 | ||||||
| B7 | 2 | 5 | 2 | 5 | 5 | 8 | ||||||
| B8 | 1 | 2 | 4 | 2 | 5 | 3 | ||||||
| D8 | 3 | 2 | 4 | 6 | 7 | 3 | ||||||
| A9 | 8 | 6 | 10 | 2 | 11 | 2 | 25 | |||||
| B9 | 5 | 19 | 20 | 18 | 17 | 10 | 3 | 2 | 53 | |||
| C9 | 6 | 2 | 14 | 4 | 5 | 7 | 5 | 3 | 10 | 2 | 3 | |
| D9 | 1 | 2 | 2 | 3 | ||||||||
| A10 | 5 | 13 | 2 | 5 | 29 | 3 | 25 | 25 | ||||
| D10 | 0 | 3 | 2 | |||||||||
| C12 | 2 | 4 | ||||||||||
| D12 | 0 | 2 | ||||||||||
| CDX1 | 1 | |||||||||||
| CDX2 | 0 | 2 | 8 | |||||||||
| GBX2 | 4 | 6 | 3 | 2 | 3 | 3 | ||||||
| Huh15 | 1 | 6 | 2 | 2 | 2 | 5 | ||||||
| Huh17 | 2 | |||||||||||
| Nkx-3.1 | 1 | 6 | ||||||||||
| Xlhbox8b | 1 | 2 |
The percentage of subclones corresponding to each identified HOX locus is listed. The sequence data did not discriminate between HOXC6 and C8, which are listed as C6/C8. The number of clones analyzed from each sample is as follows: DNA control (153), normal lung (62), CLC20-SCLC (48), H187-SCLC (51), H460-large cell (49), H661-large cell (42), H1648-adenocarcinoma (59), H322-bronchioalveolar (55), H226-squamous (36), H290-mesothelioma (61), H513-mesothelioma (44), H28-mesothelioma (36).
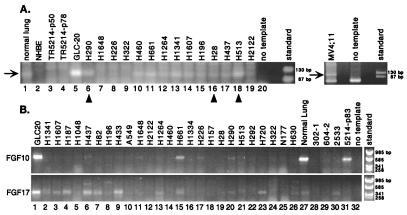
We also compared degenerate HOX RT-PCR band intensities as a crude measure of overall expression levels (Fig. 1A). Among lung cancers, there were substantial differences with GLC20 having marked overexpression, followed by NCI-H513. When quantitative real-time RT-PCR assays were applied (see below), these two cell lines had the highest overall levels of HOX expression. Shown for comparison are products from the acute leukemia cell line, MV4;11 (24), which contains a 4;11 chromosomal translocation affecting the MLL (Trithorax) gene. Whereas the degenerate HOX primers produced a band in normal lung, no products were detected in either mortal (NHBE) or immortalized (TR5214) bronchial epithelial cells. Because the lung is a highly vascular structure, other cell types such as endothelial or mesenchymal cells could contribute to the bulk of RT-PCR products. From quantitative assays (below), vascular endothelial cells (HUVEC) express several HOX genes at higher levels than the bronchial epithelium.
Figure 1.
RT-PCR amplification of HOX and FGF loci. (A) Degenerate RT-PCR products from the homeodomain (arrow) were amplified from nonmalignant (“normal”) lung, NHBE, two passages of T-antigen-immortalized bronchial epithelial cells (TR5214), and lung cancer cell lines. The leukemia line, MV4;11, is shown for comparison. (B) RT-PCR for FGF10 and FGF17 in lung tumor lines, a colorectal carcinoma, NCI-H630, and indicated controls.
In some developmental systems, as well as human cancers, HOX genes have been shown to affect FGF expression (13, 25, 26). Therefore, we screened a subset of lung cancer cell lines for expression of FGF1, FGF2, FGF5, FGF7, FGF9, FGF11 to FGF13, and FGF18. By nonquantitative RT-PCR, these were expressed in all lung cancers tested except for FGF2, which appeared to distinguish SCLCs with a classic phenotype from those with a variant phenotype (not shown). However, expression patterns for FGF10 and FGF17 were different (Fig. 1B). In the lung, FGF10 is produced by mesenchymal cells and induces branching of epithelial lung buds (27). Among 25 lung cancer cell lines, FGF10 was strongly expressed in only GLC20. Expression was also noted in the normal lung and the immortalized bronchial epithelial culture, 5214, but not in the short-term bronchial cultures. During development, FGF17 is preferentially expressed in the brain (28). In the cell lines, FGF17 was greater in SCLCs (lanes 1–9) and GLC20, again, had the highest expression. Thus, it appears that HOX overexpression may be associated with up-regulation of FGF genes.
Quantitative Real-Time RT-PCR and Western Blot Analysis.
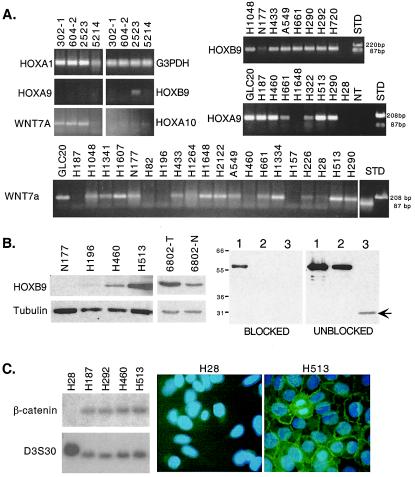
We developed quantitative RT-PCR assays for five HOX loci and WNT7a. In addition to cell lines and control cultures, we analyzed 35 non-SCLC resected tumors, of which 25 had adequate amounts of RNA from a matched nonmalignant portion of the lung. The results are expressed in terms of change in Ct values (ΔCt), which refer to the cycle number during exponential amplification at which the PCR product (measured in real-time by SYBR green fluorescence) crosses a set threshold. To adjust for variations in the amount of input RNA/cDNA, the average Ct values for each gene were normalized against average Ct values for the housekeeping gene G3PDH, i.e., ΔCt = average Ctspecific gene − average CtG3PDH. Because G3PDH is an abundant message, lower ΔCt values correspond to higher expression. These results are shown in Table 2, and selected examples of the PCRs are shown in Fig. 2A.
Table 2.
Quantitative RT-PCR analysis of HOX expression
| Controls | ΔCt
|
||||||
|---|---|---|---|---|---|---|---|
| A1 | A7 | A9 | B9 | A10 | WNT7A | ||
| 302-1 | NHBE | 8.9 | Ab. | Ab. | 16.1 | 15.4 | 4.5 |
| 604-2 | NHBE | 9.8 | Ab. | Ab. | 18.0 | 15.6 | 5.0 |
| 2523 | NHBE | 8.9 | Ab. | Ab. | 15.0 | 15.8 | 4.8 |
| 5214 | T-Ag, p83 | 17.3 | Ab. | Ab. | 15.7 | 15.4 | 16.6 |
| Nonmalignant Lung | 10.8 | 7.7 | 15.3 | 12.9 | 11.6 | 6.1 | |
| HUVEC | Endothel. | 12.8 | 8.7 | 10.1 | 10.0 | 9.1 | Ab. |
| Cell lines | |||||||
| GLC20 | SCLC | 11.1 | 6.8 | 11.1 | 6.8 | 6.9 | 8.2 |
| H187 | SCLC | 11.5 | 7.1 | 9.7 | 9.8 | 7.6 | 19.5 |
| H1048 | SCLC | 10.8 | 7.5 | 9.2 | 10.1 | 7.8 | 15.3 |
| H1341 | SCLC | 12.4 | 15.2 | 8.1 | 8.3 | 10.6 | 15.4 |
| H1607 | SCLC | 9.3 | 13.3 | 18.1 | 6.7 | 15.5 | 5.9 |
| N177 | SCLC | 17.8 | 10.3 | 11.3 | 19.0 | 9.2 | 15.4 |
| H82 | SCLC | 11.7 | 11.1 | 17.6 | 8.4 | 9.9 | 19.9 |
| H196 | SCLC | 8.9 | 12.9 | 19.7 | 11.5 | 6.5 | 19.3 |
| H433 | SCLC | 12.0 | 12.0 | 10.6 | 11.9 | 7.4 | 15.8 |
| H437 | SCLC | 9.1 | 9.9 | 15.5 | 9.2 | 8.4 | 18.8 |
| H1264 | Adeno. | 14.8 | 9.5 | 9.7 | 8.7 | 8.6 | 19.2 |
| H1648 | Adeno. | 8.5 | 12.8 | 20.6 | 11.4 | 15.5 | 10.3 |
| H2122 | Adeno. | 11.4 | 12.4 | 19.6 | 11.0 | 15.0 | 11.5 |
| A549 | Adeno. | 12.4 | 9.7 | 14.3 | 8.4 | 9.4 | 14.6 |
| H460 | Lg. Cell | 14.1 | 10.1 | 9.2 | 6.4 | 9.0 | 20.3 |
| H661 | Lg. Cell | 10.1 | 5.6 | 16.2 | 6.6 | 7.7 | 14.7 |
| H1334 | Lg. Cell | 10.1 | 13.7 | 16.4 | 10.4 | 8.2 | 9.3 |
| H157 | Squam. | 11.5 | 15.5 | 18.8 | 9.5 | 13.1 | 19.9 |
| H226 | Squam. | 11.5 | 15.3 | 14.3 | 9.4 | 6.7 | 18.8 |
| H28 | Meso. | 13.4 | 14.9 | 20.6 | 6.8 | 14.9 | 18.7 |
| H513 | Meso. | 7.8 | 7.2 | 14.6 | 6.7 | 5.9 | 6.2 |
| H290 | Meso. | 8.8 | 12.5 | 12.5 | 8.3 | 6.8 | 9.8 |
| H322 | BronchAlv. | 9.8 | 14.5 | 16.7 | 13.4 | 9.0 | 7.6 |
| H292 | Carcinoid | 9.6 | 12.2 | 9.9 | 11.9 | 7.3 | 6.8 |
| H720 | Carcinoid | 12.0 | 7.4 | 14.9 | 6.4 | 6.9 | 19.4 |
| Direct tumors | |||||||
| tumor03 | Adeno. | 8.6 | 8.8 | 9.1 | 6.8 | 6.1 | 14.5 |
| tumor07 | Adeno. | 14.3 | 13.9 | 12.8 | 16.9 | 16.2 | 15.7 |
| tumor08 | Adeno. | 13.5 | 11.2 | 13.0 | 15.8 | 5.9 | 11.1 |
| 3279-T | Adeno. | 11.0 | 10.0 | 17.7 | 9.5 | 10.1 | 7.4 |
| 3483-T | Adeno. | 12.1 | 10.0 | 16.8 | 9.4 | 8.7 | 8.8 |
| 3662-T | Adeno. | 11.6 | 12.2 | 17.6 | 14.4 | 14.0 | 14.0 |
| 3663-T | Adeno. | 7.9 | 10.6 | 13.4 | 12.2 | 8.3 | 9.0 |
| 3740-T | Adeno. | 10.6 | 9.1 | 16.6 | 13.2 | 8.6 | 9.8 |
| 6160-T | Adeno. | 13.1 | 15.4 | 18.4 | 15.5 | 14.4 | 11.2 |
| 6802-T | Adeno. | 9.5 | 15.5 | 14.6 | 5.7 | 9.0 | 8.1 |
| 7075-T | Adeno. | 13.8 | 16.5 | 18.0 | 15.5 | 14.3 | 13.4 |
| 7257-T | Adeno. | 12.2 | 13.5 | 15.6 | 11.7 | 10.6 | 10.4 |
| 7332-T | Adeno. | 10.4 | 13.7 | 14.3 | 11.8 | 11.8 | 10.6 |
| 7343-T | Adeno. | 14.9 | 16.2 | 18.2 | 12.8 | 13.9 | 11.3 |
| 7345-T | Adeno. | 10.9 | 13.3 | 15.9 | 15.1 | 8.4 | 13.0 |
| 7566-T | Adeno. | 12.5 | 15.0 | 16.4 | 5.4 | 9.2 | 17.0 |
| 2520-T | BronchAlv. | 12.4 | 12.9 | 16.1 | 7.5 | 11.6 | 9.9 |
| 6157-T | Carcinoid | 15.1 | 16.2 | 18.3 | 16.1 | 14.7 | 12.3 |
| 6374-T | Carcinoid | 15.4 | 16.8 | 17.8 | 15.3 | 15.9 | 12.0 |
| 2665-T | Carcinoid | 14.6 | 16.8 | 18.2 | 16.8 | 14.0 | 18.0 |
| 6206-T | Lg. Cell | 8.4 | 12.1 | 16.2 | 8.8 | 7.3 | 15.4 |
| 7362-T | Meso. | 9.4 | 14.3 | 12.7 | 12.3 | 7.2 | 18.0 |
| tumor02 | Squam. | 9.5 | 10.4 | 12.7 | 13.4 | 8.5 | 7.3 |
| tumor11 | Squam. | 12.1 | 10.4 | 12.9 | 12.6 | 8.5 | 12.2 |
| tumor12 | Squam. | 10.3 | 13.2 | 12.9 | 13.5 | 8.6 | 15.8 |
| tumor13 | Squam. | 10.6 | 9.0 | 13.9 | 13.6 | 8.9 | 17.0 |
| tumor14 | Squam. | 11.8 | 10.2 | 17.0 | 15.0 | 8.9 | 10.1 |
| tumor15 | Squam. | 10.3 | 10.0 | 10.5 | 16.1 | 9.4 | 8.2 |
| tumor17 | Squam. | 13.0 | 9.3 | 10.7 | 13.4 | 8.7 | 8.1 |
| 3041-T | Squam. | 11.0 | 8.2 | 16.8 | 13.2 | 12.0 | 9.4 |
| 6406-T | Squam. | 9.6 | 15.8 | 17.0 | 11.0 | 8.8 | 12.5 |
| 6792-T | Squam. | 11.7 | 16.1 | 17.2 | 16.4 | 8.6 | 13.1 |
| 7259-T | Squam. | 10.0 | 12.6 | 12.8 | 9.7 | 9.7 | 12.8 |
| 7466-T | Squam. | 10.6 | 15.2 | 15.4 | 13.7 | 12.4 | 8.0 |
| 7565-T | Squam. | 9.4 | 12.6 | 13.9 | 9.1 | 8.3 | 9.5 |
Values represent ΔCts obtained from 6 controls, 25 lung tumor cell lines, and 35 resected tumors, as indicated. Nonmalignant lung values represent the average of 25 independent resections, which were tumor free. The All HOX column contains the sum of ΔCt values for the 5 HOX loci analyzed. The average standard deviation was 0.12 cycles.
Figure 2.
(A) (Upper Left) Quantitative RT-PCR products for G3PDH, HOXA1, HOXA9, HOXA10, HOXB9, and WNT7a from NHBE cells and TR5214, passage 83. (Upper Right) Examples of quantitative RT-PCR products from tumor lines for HOXB9 and HOXA9. NT, no template. (Lower) Similar products shown for WNT7a. (B) Western blot for HOXB9. (Left) The indicated cell lines and direct tumor pair, as described in the text, were analyzed for the 30-kDa HOXB9 protein. Tubulin (55 kDa) served as a loading control. (Right) Identical Western blots contained (lane 1) 3-fold and (lane 2) 1-fold amounts of GST-HOXB9 fusion protein and (lane 3) tumor NCI-H513. No signal was observed for the endogenous HOXB9 (arrow) when the primary antibody was preincubated with GST-HOXB9-bound beads (blocked). (C) Southern blot demonstrating the specific absence of the β-catenin gene in EcoRI-digested genomic DNA from NCI-H28. A control probe (D3S30) is shown for comparison. Immunofluorescence confirmed loss of β-catenin protein in NCI-H28. Nuclei were visualized with 4′,6-diamidino-2-phenylindole (DAPI).
Neither HOXA9 nor HOXA7 was expressed in control bronchial epithelial cultures, mortal or immortal. While HOXA10 and HOXB9 could be detected, over 30 PCR cycles were required (e.g., CtG3PDH + CtHOXB9). In contrast, HOXA1 and WNT7a were relatively abundant (ΔCt range = 4–9). Interestingly, in the T-antigen-immortalized culture, expression of both HOXA1 and WNT7a was reduced by 8–9 cycles. Control ΔCt values for the nonmalignant lung were obtained from an average of 25 resected samples. However, there was little variation among these (not shown), suggesting that altered HOX expression is not a common feature of normal-appearing airways. Because the lung is an extremely vascular structure, we included HUVEC as a control. Each HOX gene tested was expressed in HUVEC cells, but WNT7a was absent. At least for HOXA1, HOXA9, HOXB9, and HOXA10, the ΔCt values from nonmalignant lung were intermediate with those obtained in the mortal bronchial epithelial and HUVEC cultures.
In agreement with our initial degenerate RT-PCR survey, HOXA9 and HOXB9 were overexpressed in many lung cancer cell lines compared with nonmalignant lung. When at least two PCR cycle differences were used, HOXB9 overexpression occurred in 18/25 (72%) of the cell lines and HOXA9 overexpression was identified in 10/25 (40%). Nine cell lines (GLC20, NCI-H187, H1048, H1341, H433, H1264, H460, H290, and H292) overexpressed both genes. Similarly, HOXA10 was overexpressed in 18/25. Of the cell lines that overexpressed both HOXA9 and HOXB9, all but one (H1341) had elevated HOXA10. In the direct tumors, overexpression of these same genes was observed, but less frequently. For instance, HOXB9 was overexpressed in 9/35 (26%), HOXA9 in 11/35 (31%), and HOXA10 in 20/35 (57%). Thus, overexpression of HOX9 paralogous genes and HOXA10 occurs frequently in lung cancer. It is also possible that overexpression of these genes is selected for in cell lines, often associated with more aggressive tumors.
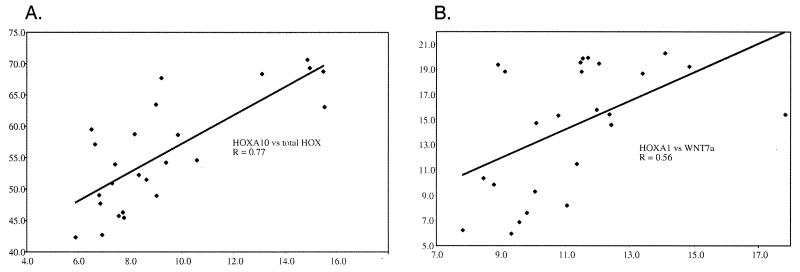
These data were analyzed by using both parametric (Pearson) and nonparametric (Spearman) correlation coefficients in conjunction with the University of Colorado Cancer Center Biostatistics Core. In the lung cancer cell lines, which are free of contaminating nonmalignant components, HOXA9 expression was significantly correlated with both HOXA10 (Pearson r = 0.559; P = 0.0036) and HOXA7 (Spearman r = 0.404; P = 0.0453). Likewise, HOXA7 expression was correlated with HOXA10 (Pearson r = 0.472, P = 0.0166). In contrast, neither HOXA1 nor HOXB9 expression was correlated with any other HOX locus (see below). However, there was a significant correlation (Spearman r = 0.43; P = 0.0319) when HOXB9 was compared with overall HOX expression measured as the sum of the individual ΔCt values for HOXA1, HOXA7, HOXA9, HOXA10, and HOXB9. Nevertheless, this relationship was weaker than the observed correlations between HOXA7, HOXA9, or HOXA10 (Fig. 3A) with the total HOX score (Spearman r = 0.768, P = 0.0001; r = 0.676, P = 0.0002; and r = 0.684, P = 0.0002, respectively).
Figure 3.
Correlation plots for HOX gene expression in cell lines. (A) ΔCt values for HOXA10 (x axis) are plotted against the sum of ΔCt values for all HOX loci analyzed (y axis). The trend line is shown along with the correlation coefficient (Pearson r = 0.77; P = 0.0001). (B) Similar analysis for HOXA1 (x axis) vs. WNT7a (y axis). In this case, the Spearman correlation coefficient is r = 0.56 with P = 0.0038. Note that the trend line is skewed by the presence of one outlying data point.
For HOXB9, we confirmed that elevated RNA levels were associated with elevated protein expression (Fig. 2B Left). Two cell lines (NCI-H513 and H460) predicted to express HOXB9 protein on the basis of their low ΔCt values were compared with cell lines (N177 and NCI-H196) with much higher ΔCt values. As anticipated, HOXB9 protein was identifiable by Western blot in extracts from NCI-H513 and H460, but not from H196 and N177. Anti-tubulin antibody was used to assess protein loading. We also confirmed that protein overexpression occurs in direct tumors, as can be seen in the matched pair, 6802. To demonstrate that the antibody was specific, we produced GST-HOXB9 protein. The antibody detected recombinant protein (Fig. 2B Right), and in addition, excess GST-HOXB9 attached to glutathione agarose beads was capable of blocking the endogenous HOXB9 band in H513 extracts (arrow). In summary, elevated HOXB9 RNA correlates with elevated HOXB9 protein in cell lines and direct tumors. However, the degree of protein overexpression was less than what would have been predicted from the RNA measurements alone. How factors such as alternative splicing, translational regulation, or protein stability, either alone or in combination, affect HOXB9 levels detectable by using this antibody is unknown.
A Relationship Between HOXA1 and WNT7a Expression in Lung Tumors.
In contrast to the above results, there were no cell lines with elevated HOXA1 levels that differed from mortal bronchial epithelial cells by more than two PCR cycles and only one, H513, had expression that was greater by one PCR cycle. If the nonmalignant lung value is taken as the reference, then three cells lines (NCI-H513, H1648, and H290) had elevated HOXA1 expression by two PCR cycles. Interestingly, NCI-H513 and H290 represent two of the three mesotheliomas. This relationship becomes more obvious when WNT7a expression is considered (see below). Among direct tumors, there were no examples of significantly elevated HOXA1 expression. There was only one mesothelioma (7362) in the set of direct tumors and it was among the highest 10% in terms of HOXA1 expression.
We wondered if other pathways known to affect HOX expression during normal development might be operative in lung tumors. The WNT pathway (29–31) was an obvious candidate because there are several well-described interactions between these pathways in model organisms plus other reports of interactions involving human cancers (see Discussion). We carried out a survey of WNT pathway components in this same collection of lung cancer cell lines (to be described elsewhere). Interestingly, compared with either NHBE cells or nonmalignant lung, WNT7a expression was almost uniformly reduced in the tumors. In the cell lines, WNT7a was undetectable in 10 of 26 (38%) (Fig. 2A Bottom). Moreover, WNT7a was lost concomitantly with HOXA1 in T-antigen-immortalized bronchial epithelial cells (Fig. 2A Left).
Chromosome 3p21.3 is recognized as a somatic deletion target in lung cancer (32) and includes the gene for β-catenin. In a PCR survey for homozygous deletions affecting 3p, we discovered that β-catenin was missing in the mesothelioma, NCI-H28. This was confirmed by Southern blot analysis and at the protein level by immunofluorescence (Fig. 2C). The absence of β-catenin should result in down-regulation or loss of genes dependent on an activated WNT pathway. In Drosophila, a positive autoregulatory WNT feedback loop has been postulated (17), and in the mouse uterus, WNT7a has been associated with maintenance of some HOXA loci (33). Strikingly, loss of β-catenin in NCI-H28 was associated with loss of WNT7a and the lowest overall expression of HOX loci among all cell lines despite the fact that HOXB9 appeared relatively unaffected. Whereas, as mentioned above, HOXA1 expression did not correlate with any other HOX genes, or with the total HOX score, it was strongly associated in the cell lines with WNT7a expression when either Pearson (r = 0.506, P = 0.0099) or Spearman (r = 0.558, P = 0.0038) correlation coefficients were used (Fig. 3B).
Discussion
Our results indicate that lung cancers are associated with frequent and substantial changes in HOX gene expression. Moreover, this expression is not haphazard but rather regulated and appears integrated with the WNT and FGF pathways. The degenerate PCR approach provided an initial estimate of the most abundantly expressed HOX genes in lung cancer cell lines and normal lung. However, not all HOX loci could be amplified, and clearly, there are more genes of interest not yet investigated by quantitative analysis. For example, HOXA5 was recently shown to positively regulate p53 expression (34). From the degenerate analysis (Table 1), HOXA5 levels varied from 0% to 8%. Considerably more variation is suggested for other genes such as the HOX3, HOX4, HOX6, and HOX7 paralogs (Table 1). We were initially surprised that the degenerate PCR results did not detect more differences among cell types, especially SCLC vs. non-SCLC. However, Anbazhagan et al. (35) recently described similar findings by using cDNA arrays in which the patterns of gene expression for SCLC were more similar to epithelial cells than neuroendocrine tumors.
Interestingly, as in leukemia (23, 36), genes of the HOX9 paralogous group as well as HOXA10 were frequently overexpressed in lung cancers. Chen and Capecchi (37) demonstrated that HOX9 paralogous genes influenced mammary gland proliferation and suggested they should be examined in mammary carcinomas. Increased expression of HOX9 paralogous genes was not simply related to proliferation because they were transcriptionally silent in the simian virus 40-immortalized epithelial culture, TR5214. In leukemia, HOXA9 expression has been associated with a poor outcome (10). Whereas we do not have clinical outcomes for these patients, the finding that HOX9 genes were more frequently overexpressed in lung cancer cell lines than in resectable primary tumors suggests that a similar relationship might exist. In several tumors, such as neuroblastoma and melanoma, cell lines are more readily established from biologically aggressive tumors and metastatic sites. From our quantitative results, there are subsets of cell lines and direct tumors that overexpress most of the genes tested and vice versa. Thus, HOX genes may provide important biomarkers and possible therapeutic targets.
To search for consequences of altered HOX expression, we looked at FGFs, which are known target genes in some instances (38, 39). In general, most FGFs tested were widely expressed, although FGF10 and FGF17 were more restricted. Interestingly, GLC20, which had the highest overall HOX expression by both degenerate RT-PCR and quantitative analyses, also had the highest expression for both FGF10 and FGF17. In the breast cancer cell line, SkBr3, HOXB7 was shown to induce basic FGF (39). While we have not yet examined HOXB7, the paralogous HOXA7 was most highly expressed in GLC20 and H661 (ΔCt = 6.8 and 5.6, respectively), both of which expressed FGF10. During lung development, FGF10 is produced by mesenchymal cells and influences epithelial branching (40) and FGFs can influence proliferation, transformation, and angiogenesis (36, 41, 42). Thus, in lung cancer, HOX overexpression could function in part by up-regulating FGFs.
The apparent alteration of HOX genes in lung cancers suggested that pathways affecting these loci might be altered. In Caenorhabditis elegans, the WNT pathway controls expression of HOX genes including mab-5 during development of the migratory QL neuroblast (43) and lin-39 during vulval development (30). HOX loci can also regulate Wnt expression in both Drosophila development and human leukemias (31, 38, 44). We found that WNT7a was significantly correlated with HOXA1 in several instances. In lung cancer cell lines, the only significant HOXA1 correlation was with WNT7a. In the bronchial epithelial cultures, WNT7a and HOXA1 expression were lost concomitantly after immortalization with large-T antigen. Finally, a homozygous deletion of β-catenin in the mesothelioma H28 was associated with loss of both genes which were otherwise strongly expressed in the two other mesotheliomas, H513 and H290. In the mouse, a knockout of WNT7a was associated with uterine alterations and loss of HoxA10 and HoxA11 (45). HoxA1 expression was not mentioned.
Similar relationships were observed in the direct tumors. However, the expression of all of the HOX genes examined in HUVEC cells suggests that contamination by nonmalignant stroma may complicate the analysis. WNT7a was not expressed by HUVEC cells, and interestingly, was substantially down-regulated in most tumors. WNT7a is located at 3p25, a common site for genetic loss in lung cancer (46). Thus, it is possible that 3p alterations, or epigenetic changes, could lead to reduced WNT7a expression and loss of target genes including HOX loci. Likewise, imbalances have been noted for the chromosome arms containing the HOXA and HOXB clusters (7p and 17q, respectively), but these changes have not been sufficiently delineated to suggest the HOX clusters as targets. The WNT pathway has been implicated in differentiation processes in various settings including avian mesoderm (47), neural crest (48), hair follicle development (49), and axonal remodeling (50). However, WNT7a is also weakly transforming when expressed in C57MG cells (51). However, based on its frequent loss, we hypothesize that WNT7a signaling in lung epithelial cells affects differentiation, possibly through corresponding changes in HOX expression. HOX genes can also be influenced by other signaling pathways, including transforming growth factor β (TGF-β) (52). Interestingly, TGF-β signaling can directly interact with WNT signaling by a complex between β-catenin/T-cell factor/Leukemia enhancer factor and Smad4 on the Xtwn promoter (53). Because TGF-β signaling is commonly disrupted in epithelial tumors (54), it is plausible that both the WNT and TGF-β pathways may synergistically interact to affect specific HOX gene expression.
Acknowledgments
We thank Dr. Gary Miller for his help with implementing the HOX degenerate PCR, and Dr. James Murphy and Chan Zeng in the Biostatistics Core for their assistance; and we acknowledge the generous equipment support from the Lulu Frankel Foundation (H.D.) and the La Caixa Fellowship award to R.C. This work was supported by the University of Colorado Lung Cancer Specialized Program of Research Excellence Grant CA5187–07.
Abbreviations
- RT-PCR
reverse transcription–PCR
- FGF
fibroblast growth factor
- SCLC
small-cell lung cancer
- NHBE
normal human bronchial epithelial
- GST
glutathione S-transferase
- Ct
threshold cycle
- HUVEC
human vascular endothelial cell
- G3PDH
glyceraldehyde-3-phosphate dehydrogenase
References
- 1.Scott M P, Tamkun J W, Hartzell G W., III Biochim Biophys Acta. 1989;989:25–48. doi: 10.1016/0304-419x(89)90033-4. [DOI] [PubMed] [Google Scholar]
- 2.Graham A, Papalopulu N, Krumlauf R. Cell. 1989;57:367–378. doi: 10.1016/0092-8674(89)90912-4. [DOI] [PubMed] [Google Scholar]
- 3.Greer J M, Puetz J, Thomas K R, Capecchi M R. Nature (London) 2000;403:661–665. doi: 10.1038/35001077. [DOI] [PubMed] [Google Scholar]
- 4.Raza-Egilmez S Z, Jani-Sait S N, Grossi M, Higgins M J, Shows T B, Aplan P D. Cancer Res. 1998;58:4269–4273. [PubMed] [Google Scholar]
- 5.Nakamura T, Yamazaki Y, Hatano Y, Miura I. Blood. 1999;94:741–747. [PubMed] [Google Scholar]
- 6.Borrow J, Shearman A M, Stanton V P, Jr, Becher R, Collins T, Williams A J, Dube I, Katz F, Kwong Y L, Morris C, et al. Nat Genet. 1996;12:159–167. doi: 10.1038/ng0296-159. [DOI] [PubMed] [Google Scholar]
- 7.Hatano M, Roberts C W, Minden M, Crist W M, Korsmeyer S J. Science. 1991;253:79–82. doi: 10.1126/science.1676542. [DOI] [PubMed] [Google Scholar]
- 8.Nourse J, Mellentin J D, Galili N, Wilkinson J, Stanbridge E, Smith S D, Cleary M L. Cell. 1990;60:535–545. doi: 10.1016/0092-8674(90)90657-z. [DOI] [PubMed] [Google Scholar]
- 9.Kamps M P, Murre C, Sun X H, Baltimore D. Cell. 1990;60:547–555. doi: 10.1016/0092-8674(90)90658-2. [DOI] [PubMed] [Google Scholar]
- 10.Golub T R, Slonim D K, Tamayo P, Huard C, Gaasenbeek M, Mesirov J P, Coller H, Loh M L, Downing J R, Caligiuri M A, et al. Science. 1999;286:531–537. doi: 10.1126/science.286.5439.531. [DOI] [PubMed] [Google Scholar]
- 11.Cillo C, Barba P, Freschi G, Bucciarelli G, Magli M C, Boncinelli E. Int J Cancer. 1992;51:892–897. doi: 10.1002/ijc.2910510610. [DOI] [PubMed] [Google Scholar]
- 12.De Vita G, Barba P, Odartchenko N, Givel J C, Freschi G, Bucciarelli G, Magli M C, Boncinelli E, Cillo C. Eur J Cancer. 1993;29A:887–893. doi: 10.1016/s0959-8049(05)80432-0. [DOI] [PubMed] [Google Scholar]
- 13.Care A, Silvani A, Meccia E, Mattia G, Stoppacciaro A, Parmiani G, Peschle C, Colombo M P. Mol Cell Biol. 1996;16:4842–4851. doi: 10.1128/mcb.16.9.4842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tiberio C, Barba P, Magli M C, Arvelo F, Le Chevalier T, Poupon M F, Cillo C. Int J Cancer. 1994;58:608–615. doi: 10.1002/ijc.2910580426. [DOI] [PubMed] [Google Scholar]
- 15.Flagiello D, Gibaud A, Dutrillaux B, Poupon M F, Malfoy B. FEBS Lett. 1997;415:263–267. doi: 10.1016/s0014-5793(97)01118-6. [DOI] [PubMed] [Google Scholar]
- 16.Omatu T. Hokkaido Igaku Zasshi. 1999;74:367–376. [PubMed] [Google Scholar]
- 17.Hooper J E. Nature (London) 1994;372:461–464. doi: 10.1038/372461a0. [DOI] [PubMed] [Google Scholar]
- 18.Phelps R M, Johnson B E, Ihde D C, Gazdar A F, Carbone D P, McClintock P R, Linnoila R I, Matthews M J, Bunn P A, Jr, Carney D, et al. J Cell Biochem Suppl. 1996;24:32–91. doi: 10.1002/jcb.240630505. [DOI] [PubMed] [Google Scholar]
- 19.Hirsch E, Hu L J, Prigent A, Constantin B, Agid Y, Drabkin H, Roche J. Brain Res. 1999;823:67–79. doi: 10.1016/s0006-8993(99)01103-8. [DOI] [PubMed] [Google Scholar]
- 20.Prevot D, Voeltzel T, Birot A M, Morel A P, Rostan M C, Magaud J P, Corbo L. J Biol Chem. 2000;275:147–153. doi: 10.1074/jbc.275.1.147. [DOI] [PubMed] [Google Scholar]
- 21.Harper S, Speicher D W. In: Current Protocols in Protein Science. Coligan J E, Dunn B M, Ploegh H L, Speicher D W, Wingfield P T, editors. New York: Wiley; 1995. pp. 6.6.1–6.6.19. [Google Scholar]
- 22.Scott M P, Tamkun J W, Hartzell G W., III Biochim Biophys Acta. 1989;989:25–48. doi: 10.1016/0304-419x(89)90033-4. [DOI] [PubMed] [Google Scholar]
- 23.Celetti A, Barba P, Cillo C, Rotoli B, Boncinelli E, Magli M C. Int J Cancer. 1993;53:237–244. doi: 10.1002/ijc.2910530211. [DOI] [PubMed] [Google Scholar]
- 24.Lange B, Valtieri M, Santoli D, Caracciolo D, Mavilio F, Gemperlein I, Griffin C, Emanuel B, Finan J, Nowell P, et al. Blood. 1987;70:192–199. [PubMed] [Google Scholar]
- 25.McWhirter J R, Goulding M, Weiner J A, Chun J, Murre C. Development (Cambridge, UK) 1997;124:3221–3232. doi: 10.1242/dev.124.17.3221. [DOI] [PubMed] [Google Scholar]
- 26.Care A, Silvani A, Meccia E, Mattia G, Peschle C, Colombo M P. Oncogene. 1998;16:3285–3289. doi: 10.1038/sj.onc.1201875. [DOI] [PubMed] [Google Scholar]
- 27.Bellusci S, Grindley J, Emoto H, Itoh N, Hogan B L. Development (Cambridge, UK) 1997;124:4867–4878. doi: 10.1242/dev.124.23.4867. [DOI] [PubMed] [Google Scholar]
- 28.Hoshikawa M, Ohbayashi N, Yonamine A, Konishi M, Ozaki K, Fukui S, Itoh N. Biochem Biophys Res Commun. 1998;244:187–191. doi: 10.1006/bbrc.1998.8239. [DOI] [PubMed] [Google Scholar]
- 29.Maloof J N, Whangbo J, Harris J M, Jongeward G D, Kenyon C. Development (Cambridge, UK) 1999;126:37–49. doi: 10.1242/dev.126.1.37. [DOI] [PubMed] [Google Scholar]
- 30.Eisenmann D M, Maloof J N, Simske J S, Kenyon C, Kim S K. Development (Cambridge, UK) 1998;125:3667–3680. doi: 10.1242/dev.125.18.3667. [DOI] [PubMed] [Google Scholar]
- 31.Rauskolb C, Wieschaus E. EMBO J. 1994;13:3561–3569. doi: 10.1002/j.1460-2075.1994.tb06663.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kok K, Naylor S L, Buys C H. Adv Cancer Res. 1997;71:27–92. doi: 10.1016/s0065-230x(08)60096-2. [DOI] [PubMed] [Google Scholar]
- 33.Parr B A, McMahon A P. Nature (London) 1998;395:707–710. doi: 10.1038/27221. [DOI] [PubMed] [Google Scholar]
- 34.Raman V, Martensen S A, Reisman D, Evron E, Odenwald W F, Jaffee E, Marks J, Sukumar S. Nature (London) 2000;405:974–978. doi: 10.1038/35016125. [DOI] [PubMed] [Google Scholar]
- 35.Anbazhagan R, Tihan T, Bornman D M, Johnston J C, Saltz J H, Weigering A, Piantadosi S, Gabrielson E. Cancer Res. 1999;59:5119–5122. [PubMed] [Google Scholar]
- 36.Thorsteinsdottir U, Sauvageau G, Hough M R, Dragowska W, Lansdorp P M, Lawrence H J, Largman C, Humphries R K. Mol Cell Biol. 1997;17:495–505. doi: 10.1128/mcb.17.1.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen F, Capecchi M R. Proc Natl Acad Sci USA. 1999;96:541–546. doi: 10.1073/pnas.96.2.541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McWhirter J R, Goulding M, Weiner J A, Chun J, Murre C. Development (Cambridge, UK) 1997;124:3221–3232. doi: 10.1242/dev.124.17.3221. [DOI] [PubMed] [Google Scholar]
- 39.Care A, Silvani A, Meccia E, Mattia G, Peschle C, Colombo M P. Oncogene. 1998;16:3285–3289. doi: 10.1038/sj.onc.1201875. [DOI] [PubMed] [Google Scholar]
- 40.Bellusci S, Grindley J, Emoto H, Itoh N, Hogan B L. Development (Cambridge, UK) 1997;124:4867–4878. doi: 10.1242/dev.124.23.4867. [DOI] [PubMed] [Google Scholar]
- 41.Coulier F, Pontarotti P, Roubin R, Hartung H, Goldfarb M, Birnbaum D. J Mol Evol. 1997;44:43–56. doi: 10.1007/pl00006120. [DOI] [PubMed] [Google Scholar]
- 42.Hajitou A, Baramova E N, Bajou K, Noe V, Bruyneel E, Mareel M, Collette J, Foidart J M, Calberg-Bacq C M. Oncogene. 1998;17:2059–2071. doi: 10.1038/sj.onc.1202126. [DOI] [PubMed] [Google Scholar]
- 43.Maloof J N, Whangbo J, Harris J M, Jongeward G D, Kenyon C. Development (Cambridge, UK) 1999;126:37–49. doi: 10.1242/dev.126.1.37. [DOI] [PubMed] [Google Scholar]
- 44.McWhirter J R, Neuteboom S T, Wancewicz E V, Monia B P, Downing J R, Murre C. Proc Natl Acad Sci USA. 1999;96:11464–11469. doi: 10.1073/pnas.96.20.11464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Miller C, Sassoon D A. Development (Cambridge, UK) 1998;125:3201–3211. doi: 10.1242/dev.125.16.3201. [DOI] [PubMed] [Google Scholar]
- 46.Hibi K, Takahashi T, Yamakawa K, Ueda R, Sekido Y, Ariyoshi Y, Suyama M, Takagi H, Nakamura Y, Takahashi T. Oncogene. 1992;7:445–449. [PubMed] [Google Scholar]
- 47.Eisenberg C A, Gourdie R G, Eisenberg L M. Development (Cambridge, UK) 1997;124:525–536. doi: 10.1242/dev.124.2.525. [DOI] [PubMed] [Google Scholar]
- 48.Saint-Jeannet J P, He X, Varmus H E, Dawid I B. Proc Natl Acad Sci USA. 1997;94:13713–13718. doi: 10.1073/pnas.94.25.13713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.DasGupta R, Fuchs E. Development (Cambridge, UK) 1999;126:4557–4568. doi: 10.1242/dev.126.20.4557. [DOI] [PubMed] [Google Scholar]
- 50.Hall A C, Lucas F R, Salinas P C. Cell. 2000;100:525–535. doi: 10.1016/s0092-8674(00)80689-3. [DOI] [PubMed] [Google Scholar]
- 51.Shimizu H, Julius M A, Giarre M, Zheng Z, Brown A M, Kitajewski J. Cell Growth Differ. 1997;8:1349–1358. [PubMed] [Google Scholar]
- 52.Kloen P, Visker M H, Olijve W, van Zoelen E J, Boersma C J. Biochem Biophys Res Commun. 1997;233:365–369. doi: 10.1006/bbrc.1997.6458. [DOI] [PubMed] [Google Scholar]
- 53.Nishita M, Hashimoto M K, Ogata S, Laurent M N, Ueno N, Shibuya H, Cho K W. Nature (London) 2000;403:781–785. doi: 10.1038/35001602. [DOI] [PubMed] [Google Scholar]
- 54.Markowitz S, Wang J, Myeroff L, Parsons R, Sun L, Lutterbaugh J, Fan R S, Zborowska E, Kinzler K W, Vogelstein B, et al. Science. 1995;268:1336–1338. doi: 10.1126/science.7761852. [DOI] [PubMed] [Google Scholar]