Abstract
Background
Organisms are capable of developing different phenotypes by altering the genes they express. This phenotypic plasticity provides a means for species to respond effectively to environmental conditions. One of the most dramatic examples of phenotypic plasticity occurs in the highly social hymenopteran insects (ants, social bees, and social wasps), where distinct castes and sexes all arise from the same genes. To elucidate how variation in patterns of gene expression affects phenotypic variation, we conducted a study to simultaneously address the influence of developmental stage, sex, and caste on patterns of gene expression in Vespula wasps. Furthermore, we compared the patterns found in this species to those found in other taxa in order to investigate how variation in gene expression leads to phenotypic evolution.
Results
We constructed 11 different cDNA libraries derived from various developmental stages and castes of Vespula squamosa. Comparisons of overall expression patterns indicated that gene-expression differences distinguishing developmental stages were greater than expression differences differentiating sex or caste. Furthermore, we determined that certain sets of genes showed similar patterns of expression in the same phenotypic forms of different species. Specifically, larvae upregulated genes related to metabolism and genes possessing structural activity. Surprisingly, our data indicated that at least a few specific gene functions and at least one specific gene family are important components of caste differentiation across social insect taxa.
Conclusion
Despite research on various aspects of development originating from model systems, growth in understanding how development is related to phenotypic diversity relies on a growing literature of contrasting studies in non-model systems. In this study, we found that comparisons of patterns of gene expression with model systems highlighted areas of conserved and convergent developmental evolution across diverse taxa. Indeed, conserved biological functions across species implicated key functions related to how phenotypes are built. Finally, overall differences between social insect taxa suggest that the independent evolution of caste arose via distinct developmental trajectories.
Background
A fundamental goal of the burgeoning field of evolutionary developmental biology is to understand how differences in gene expression contribute to phenotypic diversity. Phenotypic plasticity, the ability of a single genotype to produce alternate forms of morphology, physiology or behavior in response to environmental conditions [1-3], provides a unique opportunity to investigate environmental influence on gene expression. Phenotypic plasticity is taxonomically widespread and usually results in continuous phenotypic variation [2,4]. However, some organisms exhibit phenotypic plasticity such that two or more discrete alternative phenotypes (without intermediate forms) are produced. This type of variation is called a polyphenism [5]. Because the phenotypic differences that exist among morphs can arise from an identical genome, polyphenisms provide an ideal means to explore how differential gene expression drives phenotypic diversity [6].
Highly social hymenopteran insects (ants, social bees, and social wasps) present one of the most striking examples of polyphenism. Hymenopteran queens, workers, and males all possess the same genes (although females are diploid and males are haploid), unlike many other animals, where sex chromosomes play a role in sex determination. Therefore, the phenotypic differences among hymenopteran social insect castes, as well as sexes, are derived from variation in gene expression.
In this study, we investigated the molecular underpinnings involved in the development of the social wasp Vespula squamosa. Vespula wasps are a particularly good taxon in which to study phenotypic evolution, for several reasons. First, Vespula wasps display distinct female castes; queens differ from workers in size, color, behavior, body proportions, and physiology [7,8] (Figure 1). Second, Vespula wasps display remarkable similarities to Apis bees, although the two taxa are only distantly related [9]. Moreover, the complex caste and social systems found in the two taxa arose via independent evolutionary events. This point is of fundamental importance, because comparative analysis of development in Vespula and Apis will reveal if analogous environmentally induced phenotypes are generated through similar patterns of gene expression. Finally, Vespula queens and workers are reared in distinct cells; this key feature allows the developmental fate of larvae to be known very early in ontogeny [7].
Figure 1.
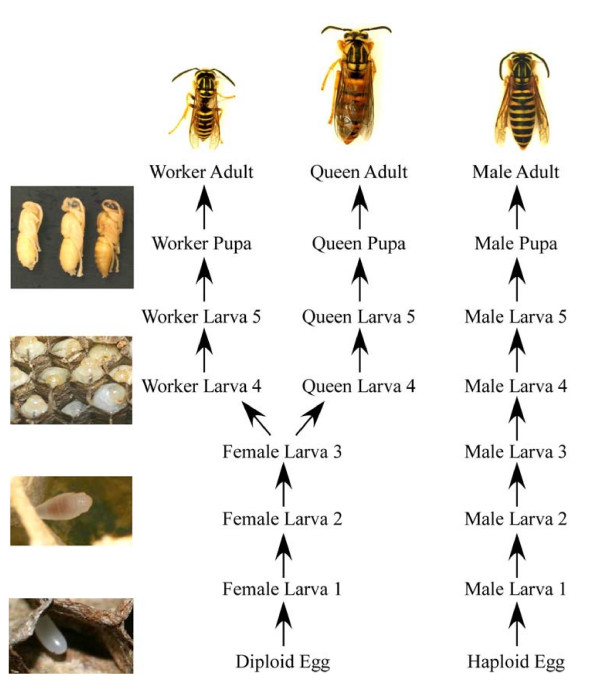
V. squamosa developmental pathways and stages. Haploid eggs develop into males. Diploid eggs give rise to workers and queens.
This study addressed the following three important questions related to the evolution and development of queens, workers and males in Vespula wasps:
(i) How do patterns of gene expression differ among developmentally distinct phenotypes?
(ii) Are sex-specific developmental patterns similar across insect species?
(iii) Do caste-specific developmental patterns display convergent evolution?
Results and discussion
Expression patterns and developmental stage
The most striking result from our data is that developmental stage (i.e., larva, pupa, and adult) plays a much larger role in establishing patterns of gene expression than either caste or sex (Figure 2). In fact, developmental time is the critical factor in grouping the libraries by overall expression pattern. Thus, individuals of the same developmental age express many genes in common regardless of their caste or sex.
Figure 2.
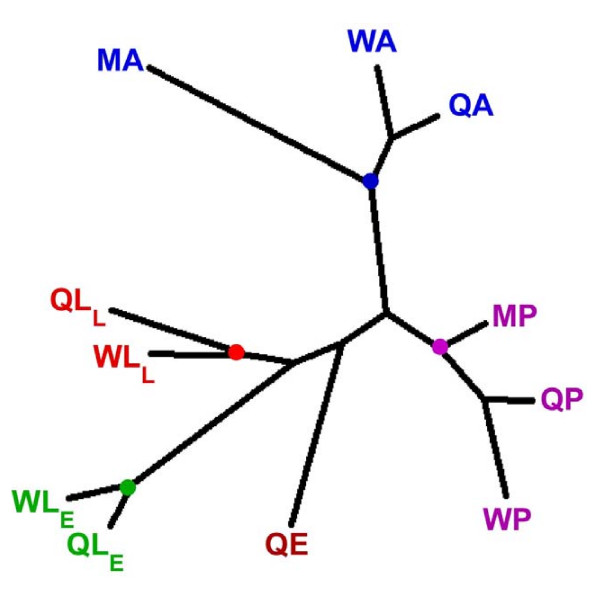
Relationships among V. squamosa life stages based upon EST frequencies. Colored dots indicate where libraries derived from developmentally similar stages cluster. Q, queen; W, worker; M, male; E, egg; LE, early larval instars; LL, late larval instars; P, pupa; A, adult.
Which genes contribute to the differences among V. squamosa libraries and thus provided insight into the molecular processes associated with development in this taxon? A general χ2 test (significance threshold at p < 0.01; [10]) identified 52 genes that were differentially expressed among libraries (Table 1). For example, hexamerin-like storage proteins (VSQ019, VSQ232, VSQ233, VSQ292; Table 1), which have been implicated in the development of other insect species (see below), also showed a distinctive pattern of upregulation during the late larval stages of V. squamosa. Additionally, the expressed sequence tags (ESTs) VSQ318 and VSQ031 are both members of the odorant-binding protein family (Table 1), which exhibit distinctive patterns of expression, falling along developmental lines. Interestingly, differentially expressed odorant-binding proteins have been implicated as key regulators of social behavior in other social insects [11].
Table 1.
Expression levels for 52 genes present at significantly different frequencies (p < 0.01) across cDNA libraries. Identity of putative homologous sequence and expectation (E) value of each match are presented. Raw number of sequences found in each library and the p value indicating significant differentiation of ESTs are also provided.
| Gene ID | GenBank ID | Homolog | e-value | QE | WL E | QL E | WL L | QL L | WP | QP | MP | WA | QA | MA | p |
| VSQ001 | EG326522 | Ctenocephalides felis serpin | 1e-021 | 0 | 0 | 0 | 3 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0.00239 |
| VSQ005 | EG326963 | Dermatobia hominis NADH dehydrogenase subunit 1 | 2e-034 | 0 | 0 | 0 | 3 | 3 | 0 | 0 | 0 | 0 | 1 | 0 | 0.00755 |
| VSQ018 | EG326808 | Blattella germanica similar to major allergen Bla g 1.02 | 5e-035 | 0 | 3 | 2 | 7 | 22 | 0 | 0 | 0 | 0 | 0 | 0 | <0.0001 |
| VSQ019 | EG326728 | Apis mellifera similar to arylphorin-like hexamerin | 1e-18 | 1 | 0 | 0 | 11 | 31 | 0 | 0 | 0 | 0 | 0 | 1 | <0.0001 |
| VSQ020 | EG326434 | Apis mellifera similar to glutamate receptor IB | 6e-039 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.00794 |
| VSQ031 | EG326500 | Polistes dominulus odorant-binding protein OBP-1 precursor | 8e-029 | 0 | 0 | 0 | 1 | 0 | 2 | 0 | 0 | 6 | 3 | 0 | 0.00226 |
| VSQ040 | EG325267 | Tetraodon nigroviridis unnamed protein product | 3e-08 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 5 | 0 | 0 | <0.0001 |
| VSQ048 | EG326323 | Human herpesvirus 6 U88 | 1e-28 | 0 | 1 | 5 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0.00123 |
| VSQ050 | EG326528 | Drosophila melanogaster TPA: HDC07203 | 8e-12 | 1 | 9 | 8 | 0 | 2 | 0 | 1 | 0 | 3 | 1 | 0 | <0.0001 |
| VSQ051 | EG325164 | No homology | 0 | 5 | 15 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | <0.0001 | |
| VSQ056 | EG326606 | No homology | 2 | 1 | 4 | 0 | 22 | 0 | 0 | 0 | 0 | 0 | 0 | <0.0001 | |
| VSQ058 | EG326432 | No homology | 0 | 16 | 4 | 3 | 7 | 0 | 0 | 1 | 0 | 0 | 2 | <0.0001 | |
| VSQ074 | EG327162 | Apis mellifera similar to ribosomal protein L35A | 3e-55 | 0 | 0 | 4 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0.00935 |
| VSQ089 | EG326517 | No homology | 0 | 3 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0.00662 | |
| VSQ096 | EG325485 | Drosophila melanogaster RNA polymerase II accessory factor rpb4 | 5e-47 | 0 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.00364 |
| VSQ122 | EG326342 | Apis mellifera similar to ENSANGP00000015316 | 2e-29 | 0 | 2 | 16 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | <0.0001 |
| VSQ156 | EG326487 | Anopheles gambiae ENSANGP00000014145 | 5e-13 | 0 | 4 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | <0.0001 |
| VSQ169 | EG326695 | Drosophila pseudoobscura GA11614-PA | 7e-70 | 0 | 0 | 1 | 1 | 5 | 0 | 0 | 0 | 1 | 0 | 0 | 0.00296 |
| VSQ173 | EG326465 | Apis mellifera similar to glucose dehydrogenase | 7e-67 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.00337 |
| VSQ176 | EG326537 | Apis mellifera similar to ribosomal protein S5 | 1e-90 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | <0.0001 |
| VSQ185 | EG326415 | Ixodes scapularis beta-adaptin | 1e-41 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.00337 |
| VSQ211 | EG326474 | No homology | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.00337 | |
| VSQ212 | EG326484 | Anopheles gambiae ENSANGP00000023258 | 6e-07 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.00337 |
| VSQ232 | EG326810 | Apis mellifera hexamerin 70b | 6e-094 | 0 | 0 | 0 | 3 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0.0035 |
| VSQ233 | EG326613 | Camponotus festinates hexamerin 2 | 4e-054 | 0 | 0 | 0 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0.00314 |
| VSQ249 | EG326612 | No homology | 0 | 0 | 0 | 2 | 4 | 0 | 0 | 1 | 0 | 0 | 0 | 0.00709 | |
| VSQ254 | EG325941 | Apis mellifera short-chain dehydrogenase/reductase | 3e-41 | 0 | 0 | 0 | 0 | 2 | 1 | 5 | 0 | 0 | 0 | 0 | <0.0001 |
| VSQ280 | EG326741 | Apis mellifera similar to apontic CG5393-PB, isoform B | 6e-21 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 3 | 0 | 0.00481 |
| VSQ292 | EG326813 | Apis mellifera hexamerin 70b | 9e-32 | 0 | 0 | 0 | 3 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0.0021 |
| VSQ296 | EG326805 | Vespula vulgaris cytochrome b | 1e-067 | 0 | 0 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0.00163 |
| VSQ303 | EG326821 | Lysiphlebus testaceipes ribosomal protein L3 variant 1 | 3e-68 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.00794 |
| VSQ307 | EG326924 | Apis mellifera similar to eukaryotic initiation factor 4A | 6e-74 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.00794 |
| VSQ318 | EG326264 | Drosophila yakuba odorant-binding protein 56e | 8e-15 | 2 | 0 | 0 | 1 | 0 | 7 | 4 | 6 | 1 | 1 | 2 | 0.00158 |
| VSQ338 | EG325431 | Apis mellifera similar to bellwether CG3612-PA isoform 1 | e-105 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 4 | 0 | 0 | 0.00648 |
| VSQ349 | EG325944 | Tenebrio molitor 86 kDa early-staged encapsulation inducing protein | 5e-14 | 0 | 0 | 0 | 1 | 0 | 0 | 3 | 0 | 0 | 0 | 0 | 0.00326 |
| VSQ352 | EG326946 | No homology | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.00794 | |
| VSQ363 | EG326968 | No homology | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 6 | <0.0001 | |
| VSQ378 | EG326993 | No homology | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 0.00014 | |
| VSQ389 | EG327083 | No homology | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 9 | <0.0001 | |
| VSQ437 | EG327159 | Vespa crabro chemosensory protein | 4e-61 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 1 | 0.00376 |
| VSQ445 | EG325384 | Periplaneta americana Cr-PII allergen | 8e-16 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 5 | 1 | 1 | 0.00311 |
| VSQ463 | EG326174 | No homology | 0 | 0 | 0 | 0 | 0 | 4 | 4 | 0 | 0 | 1 | 0 | <0.0001 | |
| VSQ501 | EG325247 | Apis mellifera similar to CG14934-PA | 2e-77 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 5 | 1 | 0 | 0.00059 |
| VSQ565 | EG325238 | No homology | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 4 | 0 | 0 | 0.00101 | |
| VSQ581 | EG325293 | Apis mellifera similar to kazal-type proteinase inhibitor | 9e-026 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 4 | 0 | 0 | 0.00847 |
| VSQ682 | EG325660 | Lysiphlebus testaceipes ribosomal protein L3 variant 1 | 2e-76 | 7 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| VSQ709 | EG325659 | Pisum sativum putative senescence-associated protein | 4e-15 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.00066 |
| VSQ803 | EG325675 | Anopheles gambiae ENSANGP00000011747 | 9e-33 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 0 | 0 | 0 | 0.00677 |
| VSQ850 | EG326242 | No homology | 0 | 0 | 0 | 0 | 0 | 6 | 0 | 2 | 0 | 0 | 0 | 0 | |
| VSQ943 | EG326146 | Human coxsackievirus polyprotein | 4e-05 | 0 | 0 | 0 | 0 | 0 | 17 | 29 | 0 | 0 | 0 | 0 | 0 |
| VSQ954 | EG326048 | No homology | 0 | 0 | 0 | 0 | 0 | 2 | 4 | 0 | 0 | 0 | 0 | 0.00016 | |
| VSQ955 | EG325987 | Apis mellifera hypothetical protein XP_397583 | 3e-12 | 0 | 0 | 0 | 0 | 0 | 1 | 3 | 0 | 0 | 0 | 0 | 0.00346 |
EST, expressed sequence tag.
Q, queen; W, worker; M, male; E, egg; LE, early larval instars; LL, late larval instars; P, pupa; A, adult.
Two other genes show patterns of expression similar to those observed in other species. First, VSQ445, which is homologous to the German cockroach major allergen Bla g 1, is upregulated in adult females, as is the case in cockroaches [12]. Second, VSQ709, which is upregulated in queen eggs, most closely matches the Pisum sativum putative senescence-associated protein. Surprisingly, Sharf et al [13] found that the same gene homolog was upregulated in immature reproductives of the termite Reticulitermes flavipes. They suggest that this gene may play a role in ribosomal filtering [14], owing to the similarity of R. flavipes (and VSQ709) transcript to 28s rRNA-like sequences.
Finally, several genes of unknown function or those with low or no known homology also showed characteristic expression patterns at particular developmental stages. For example, both VSQ056 and VSQ058 were upregulated during the early and late larval developmental stages, whereas VSQ943 exhibits a striking pattern of upregulation during the female pupal stages (Table 1). The role of these genes is currently unknown, but their expression patterns indicate that further research into their functions is warranted.
The general expression patterns found in V. squamosa support previous studies that have investigated gene-expression patterns among developmental stages. For example, Mathavan et al [15] found clearly demarcated transcript clusters in five different developmental stages during embryogenesis of the zebrafish. Wagner et al [16] also found an orderly progression in transcription through time during embryogenesis of the mouse. Working in a taxon more closely related to wasps, Arbeitman et al [17] discovered that the major breaks in gene-expression clustering of Drosophila melanogaster occurred between life stages. Similar to our results, Arbeitman et al found that expression patterns for both male and female adults grouped closely together despite the apparent morphological, physiological and behavioral differences that exist between sexes. In contrast to our results, they found that larval expression was more similar to that of adults, whereas expression of embryos was more similar to that of pupae. The V. squamosa data suggest a more temporal pattern, with EST frequencies more similar between adults and pupae than between adults and larvae.
To investigate patterns of gene expression in V. squamosa further, we used information from the gene ontology (GO) classifications [18]. Use of GO classifications enabled us to determine whether there were conserved biological functions across species that demonstrated how phenotypes were built. In general, gene function may be remarkably conserved, with broad temporal patterns in gene class utilization showing similar patterns through mouse embryogenesis and development of D. melanogaster [19]. To determine the extent to which conserved biological functions persist across species, we searched for GO similarities to other insects.
Goodisman et al [20] compared gene function similarities between Camponotus festinatus ants and D. melanogaster, and found that some patterns persisted between these species. In particular, the larvae of both species upregulated genes that were involved in protein production and possessed structural activity relative to those in adults. For both of these GO functions, the patterns hold true in V. squamosa when ESTs that show significant similarity to D. melanogaster genes are considered. The mean ± SE number of transcripts associated with protein metabolism is 51.5 ± 15.5 in larvae and 25.3 ± 4.8 in adults, and the mean ± SE number of transcripts possessing structural activity is 16.5 ± 4.5 in larvae and 5.3 ± 0.7 in adults (Figure 3). As with other studies, our results suggest that genes expressed throughout immature stages of holometabolous insects are associated with growth.
Figure 3.
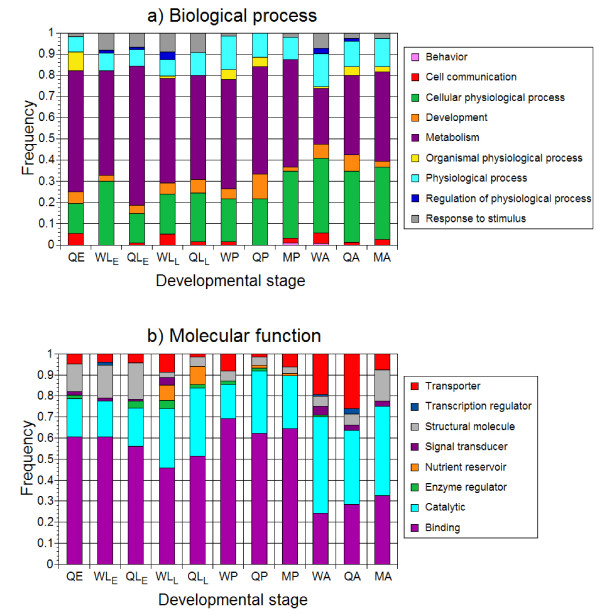
Proportion of transcripts from 11 cDNA libraries falling into different categories for (A) biological processes and (B) molecular function gene ontology. Q, queen; W, worker; M, male; E, egg; LE, early larval instars; LL, late larval instars; P, pupa; A, adult.
Expression patterns and sex differences
Sex influences patterns of gene expression in V. squamosa. Within both branches of the neighbor-joining tree that contain both sexes (pupal and adult), the male patterns of gene expression are more different and hence diverge before the two female castes (Figure 2). Thus, despite the dramatic phenotypic differences between queens and workers, they are still more similar to each other in terms of gene expression than either is to males. This result is consistent with studies in Caenorhabditis elegans, Anopheles gambiae, and D. melanogaster, which have demonstrated that the sexes differ substantially in the genes they express [21-23].
Our analysis of sequenced ESTs uncovered another interesting pattern regarding sex in V. squamosa. We found that the proportion of ESTs matching known sequences in GenBank varied significantly among libraries (G10 = 102.60; p < 0.0001). Specifically, the differences in the proportion of genes displaying homology in the adult male and adult female libraries is striking (Figure 4), with adult females exhibiting significantly higher (G1 = 25.51, p < 0.001) numbers of homologs (adult workers 69%, adult queens 53%) compared with adult males (26%). One possible explanation for these differences is that genes expressed in adult males evolve particularly rapidly relative to those expressed in females and at other developmental stages. This result is consistent with studies in other taxa that have shown that male-specific genes evolve rapidly [24-26]. Indeed, Singh and Kulathinal [27] deduced from comparative analyses of genome evolution that much de novo gene evolution occurs among male-biased genes. Our data indicate that similar processes may operate in social insects. Whether such putatively rapidly evolving genes are exclusively or primarily expressed in sex-specific tissues, as has been found to be the case in other taxa [24], represents an area of future research. Regardless, our suggestion that genes expressed in males may evolve differently from those expressed in females is notable because it points to the importance of males in the evolution of social-insect populations, a subject that until recently has been largely ignored [28].
Figure 4.
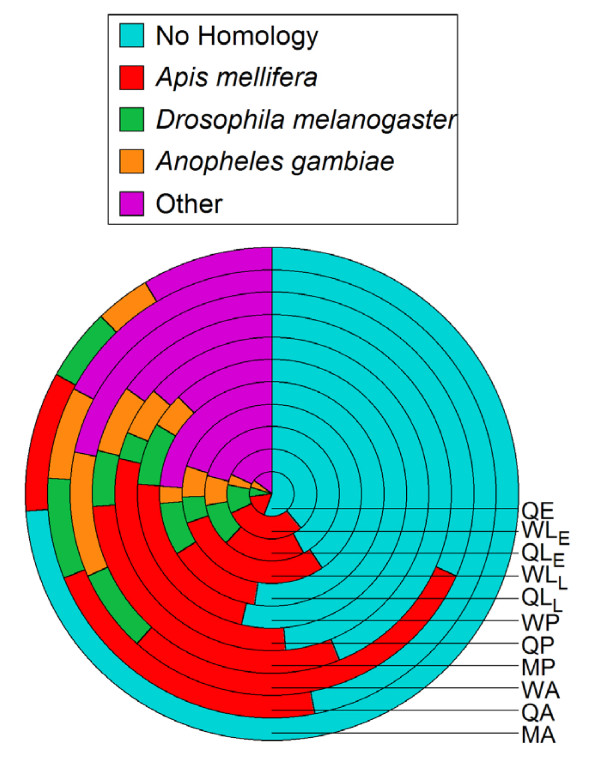
Distribution of GenBank Blast matches (expectation (e) values < 10-5) within each V. squamosa cDNA library. Q, queen; W, worker; M, male; E, egg; LE, early larval instars; LL, late larval instars; P, pupa; A, adult.
Expression patterns and development of caste
The defining feature of social insects is the division of individuals into reproductive and sterile castes [29]. Therefore, considerable research has focused on identifying genes that are differentially expressed between castes. For example, the molecular basis of caste differences has been investigated in bees [30-35], ants [36,37], wasps [38], and termites [13,39-41]. Overall, these studies represent at least five independent evolutionary events leading to sociality (summarized by Sumner et al [38]).
How does caste development in V. squamosa compare with these other taxa? Four major trends arise from our analyses. First, as might be expected, EST chord distances among castes at earlier life stages are more similar (WLE-QLE = 1.08) than at later life stages (WA-QA distance = 1.23), with intermediate stages at intermediate distances (see Table 1 for library definitions). This result indicates that as castes diverge phenotypically, physiologically, and behaviorally, patterns of gene expression also become increasingly divergent.
Second, our data contrast with a pattern of development identified in honeybees. Evans and Wheeler [31] suggested that patterns of expression of worker-destined larvae and younger bipotent larvae were more similar than between queen-destined larvae and bipotent larvae in A. mellifera. However, in V. squamosa, we found the opposite pattern when either early worker larvae (chord distances: WLE-WLL = 1.28, WLE-QLL = 1.21) or early queen larvae (chord distances: QLE-WLL = 1.29, QLE-QLL = 1.16) were used in the comparison. Thus, the trends in our data indicate that young larvae are more similar to queen-destined larvae than to worker-destined larvae in V. squamosa. Additionally, the observed differences in chord distances between worker or queen early larvae and worker or queen late larvae suggest that even though young larvae (i.e., WLE and QLE) are potentially bipotent, they may express different genes.
Third, despite the differences in overall patterns of caste differentiation mentioned above, some patterns of gene function are conserved. Specifically, early queen larvae of V. squamosa express more genes related to metabolism (G1 = 4.70, p < 0.05) than do similarly aged worker larvae (Figure 3). A similar pattern has been found in both the wasp P. canadensis [38] and the bee A. mellifera [31]. It is unclear why genes associated with metabolism show increased expression only in certain stages of queen development. It is possible that overexpression of metabolic genes early in ontogeny is sufficient to spur rapid growth in Apis queens, which develop faster than Apis workers. Similarly, Vespula queens are fed more than Vespula workers in the early larval instars [42], which may be a consequence of higher metabolic rates at these early stages and may ultimately lead to the large size differences observed between the castes. Regardless, queen production seems to be associated with increased energy production in hymenopteran social insects. This implies that gene function related to caste development may be conserved.
Fourth, the hexamerin gene family, which plays a significant role in caste differentiation in A. mellifera [30], B. terrestris [33] and R. flavipes [13,40,41], also shows significant differential expression in V. squamosa (VSQ019, VSQ232, VSQ233, VSQ292; Table 1). The arylphorin-like hexamerin most highly expressed in V. squamosa (VSQ019) is a methionine-rich member of the hexamerin family that participates in the storage of amino acids accumulated during larval development [43]. Moreover, as is the case in other social insect taxa, the different hexamerin ESTs in V. squamosa exhibit different expression patterns between queen and worker castes. VSQ019 and VSQ233 are upregulated in queen-destined larvae of V. squamosa, whereas VSQ232 and VSQ292 (both hexamerin 70b-like ESTs) are more highly expressed in the late larvae of workers relative to queens. The similarity of this gene-expression pattern among these species suggests that some specific pathways are conserved during social-insect evolution.
Conclusion
We conducted the first study to simultaneously address the influence of developmental stage, sex, and caste on patterns of gene expression. We found that patterns of expression are more similar across castes for a specific developmental stage than within castes at different stages. Similar to other insect taxa, larvae of our study taxon V. squamosa upregulate genes related to metabolism and possessing structural activity. Furthermore, our data provide a provocative example of divergent selection pressures for genes expressed differentially between the sexes. We also discovered that V. squamosa and A. mellifera castes, which arose via independent evolutionary events, may develop through different trajectories. Nevertheless, at least a few specific gene functions and at least one specific gene family appear to be conserved components of caste differentiation. Overall, our results illustrate how the study of phenotypic diversity arising from patterns of gene expression can illuminate evolutionary effects of development in animal taxa.
Methods
cDNA library construction, processing and assembling
We constructed 11 directional cDNA libraries from several developmental stages of the wasp V. squamosa. The 11 libraries contained copies of transcripts obtained from: (i) eggs collected from queen cells; pooled female larvae from the first three early larval instars sampled from (ii) queen cells and (iii) worker cells; pooled female larvae of the fourth and fifth late larval instars sampled from (iv) queen cells and (v) worker cells; (vi) queen pupae; (vii) worker pupae; (viii) male pupae; (ix) queen adults; (x) worker adults; and (xi) male adults. The cDNA libraries were synthesized using a commercial construction kit (pBluescript® II XR cDNA Library Construction Kit; Stratagene, La Jolla, CA, USA). In total, 4224 independent clones were isolated from these libraries, and 3388 single-pass sequences were obtained using the SK primer. After cloned sequences were filtered for vector contamination and quality, we obtained 2144 expressed sequence tags (ESTs; GenBank accession numbers: EG325041–EG327184).
EST processing and assembling
These ESTs were grouped into clusters using the BLASTN algorithm [44]. When sequences from all 11 libraries were analyzed in parallel, 760 sequences were unique, and the remaining sequences formed 294 clusters of two or more sequences, giving a total of 1054 unigenes (Table 2). Within each library, the mean ± SE number of ESTs was 194 ± 9.2 (range 147–233), and the frequency of private ESTs was 0.38 ± 0.049 (Table 2) (range 16.8–66.3). Furthermore, the gene diversity [45] for each library, which represents the probability of drawing two distinct sequences from a library by chance, ranged from 0.970 to 0.994, indicating that the libraries contained many unique sequences (Table 2). BLASTX similarity searches [44] indicated that 52% of all the ESTs showed similarity to known sequences (e<10-5; Figure 4), a frequency not substantially different from previous studies in other Hymenoptera [20,46].
Table 2.
Numbers and clustering of ESTs among different V. squamosa libraries. n corresponds to the total number of ESTs sequenced per library. Contigs are ESTs represented multiple times while Singletons are ESTs present only once per library. Unigenes equal the total number of different ESTs in a library. Private EST's are sequences that occur only in that specific library. Gene diversity is the probability of drawing two distinct sequences from a library by chance.
| Library | QE | WL E | QL E | WL L | QL L | WP | QP | MP | WA | QA | MA | Total |
| n | 196 | 147 | 208 | 145 | 231 | 172 | 177 | 233 | 222 | 196 | 206 | 2133 |
| Contigs | 15 | 26 | 37 | 24 | 35 | 19 | 25 | 26 | 34 | 27 | 18 | 294 |
| Singletons | 151 | 68 | 94 | 94 | 61 | 108 | 83 | 170 | 158 | 116 | 112 | 760 |
| Unigenes | 166 | 94 | 131 | 118 | 96 | 127 | 108 | 196 | 192 | 143 | 130 | 1054 |
| Private ESTs | 114 | 37 | 50 | 53 | 33 | 65 | 46 | 135 | 120 | 82 | 88 | 823 |
| Gene diversity | 0.998 | 0.982 | 0.986 | 0.991 | 0.957 | 0.987 | 0.970 | 0.998 | 0.997 | 0.986 | 0.994 |
EST, expressed sequence tag.
Q, queen; W, worker; M, male; E, egg; LE, early larval instars; LL, late larval instars; P, pupa; A, adult
Digital gene-expression analysis
We clustered the 11 libraries using the neighbor-joining method based on chord distances derived from library EST frequencies in order to gain an understanding of how patterns of gene expression were associated with development [47]. Furthermore, we explored variation in the genes expressed among libraries using digital methods [48]. This approach uses large-scale non-normalized random 3' -end cDNA library sequencing [49], but is extensible to any methodical sequencing strategy. The level of expression within each tissue is estimated from the number of cognate ESTs found in each library, under the assumption that it is proportional to the transcript frequencies [50,51]. These tests were conducted with the software program IDEG6 [52]. Overall, these methods may not provide accurate estimates of the absolute frequencies of particular genes, if certain gene sequences are subject to cloning biases. In addition, these techniques are unlikely to detect genes expressed at low levels, such as those with regulatory functions. Nevertheless, this approach can be reliably used to detect genes differentially expressed among libraries.
Authors' contributions
EH and MG conceived the study and participated in its design and coordination. EH carried out the library construction, DNA sequencing, and EST analysis. EH and MG outlined the manuscript together and EH drafted the manuscript. Both authors have read and approved the final manuscript
Acknowledgments
Acknowledgements
We thank the Georgia Institute of Technology, University of Central Florida and the U.S. National Science Foundation (DEB-0640690) for providing the funds necessary for conducting this research, N. Ueda and D. Williams for help with analyses, and J. Evans for comments on an earlier version of this manuscript.
Contributor Information
Eric A Hoffman, Email: eahoffma@mail.ucf.edu.
Michael AD Goodisman, Email: michael.goodisman@biology.gatech.edu.
References
- West-Eberhard MJ. Developmental plasticity and evolution. Oxford: Oxford University Press; 2003. [Google Scholar]
- Dewitt TJ, Scheiner SM, (eds) Phenotypic plasticity: functional and conceptual approaches. Oxford: Oxford University Press; 2004. [Google Scholar]
- Schlichting CD, Smith H. Phenotypic plasticity: linking molecular mechanisms with evolutionary outcomes. Evol Ecol. 2002;16:189–211. [Google Scholar]
- Pigliucci M. Phenotypic plasticity: beyond nature and nurture. Baltimore, MD: Johns Hopkins University Press; 2001. [Google Scholar]
- Nijhout HF. Development and evolution of adaptive polyphenisms. Evol Dev. 2003;5:9–18. doi: 10.1046/j.1525-142x.2003.03003.x. [DOI] [PubMed] [Google Scholar]
- Evans JD, Wheeler DE. Gene expression and the evolution of insect polyphenisms. Bioessays. 2001;23:62–68. doi: 10.1002/1521-1878(200101)23:1<62::AID-BIES1008>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- Spradbery JP. Wasps: An account of the biology and natural history of solitary and social wasps. London: Sidgwick & Jackson; 1973. [Google Scholar]
- Greene A. Dolichovespula and Vespula. In: Ross KG, Matthews RW, editor. The social biology of wasps. Ithaca: Comstock Publishing Associates; 1991. pp. 263–305. [Google Scholar]
- Brothers DJ. Phylogeny and evolution of wasps, ants and bees (Hymenoptera, Chrysidoidea, Vespoidea and Apoidea) Zool Scripta. 1999;21:233–249. [Google Scholar]
- Romualdi C, Bortoluzzi S, D'Alessi F, Danieli GA. IDEG6: a web tool for detection of differentially expressed genes in multiple tag sampling experiments. Phys Genomics. 2003;12:159–162. doi: 10.1152/physiolgenomics.00096.2002. [DOI] [PubMed] [Google Scholar]
- Krieger MJB, Ross KG. Identification of a major gene regulating complex social behavior. Science. 2002;295:328–332. doi: 10.1126/science.1065247. [DOI] [PubMed] [Google Scholar]
- Gore JC, Schal C. Gene expression and tissue distribution of the major human allergen Bla g 1 in the German cockroach, Blattella germanica L. (Dictyoptera : Blattellidae) J Med Entomol. 2004;41:953–960. doi: 10.1603/0022-2585-41.5.953. [DOI] [PubMed] [Google Scholar]
- Scharf ME, Wu-Scharf D, Zhou X, Pittendrigh BR, Bennett GW. Gene expression profiles among immature and adult reproductive castes of the termite Reticulitermes flavipes. Ins Mol Biol. 2005;14:31–44. doi: 10.1111/j.1365-2583.2004.00527.x. [DOI] [PubMed] [Google Scholar]
- Mauro VP, Edelman GM. The ribosome filter hypothesis. Proc Natl Acad Sci USA. 2002;99:12031–12036. doi: 10.1073/pnas.192442499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathavan S, Lee SGP, Mak A, Miller LD, Murthy KRK, Govindarajan KR, Tong Y, Wu YL, Lam SH, Yang H, et al. Transcriptome analysis of zebrafish embryogenesis using microarrays. PLOS Genet. 2005;1:260–276. doi: 10.1371/journal.pgen.0010029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner A. Energy constraints on the evolution of gene expression. Mol Biol Evol. 2005;22:1365–1374. doi: 10.1093/molbev/msi126. [DOI] [PubMed] [Google Scholar]
- Arbeitman MN, Furlong EEM, Imam F, Johnson E, Null BH, Baker BS, Krasnow MA, Scott MP, Davis RW, White KP. Gene expression during the life cycle of Drosophila melanogaster. Science. 2002;297:2270–2275. doi: 10.1126/science.1072152. [DOI] [PubMed] [Google Scholar]
- The Gene Ontology Consortium Gene Ontology: tool for the unification of biology. Nature Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner RA, Tabibiazar R, Liao A, Quertermous T. Genome-wide expression dynamics during mouse embryonic development reveal similarities to Drosophila development. Dev Biol. 2005;288:595–611. doi: 10.1016/j.ydbio.2005.09.036. [DOI] [PubMed] [Google Scholar]
- Goodisman MAD, Isoe J, Wheeler DE, Wells MA. Evolution of insect metamorphosis: A microarray-based study of larval and adult gene expression in the ant Camponotus festinatus. Evolution. 2005;59:858–870. [PubMed] [Google Scholar]
- Jiang M, Ryu J, Kiraly M, Duke K, Reinke V, Kim SK. Genome-wide analysis of developmental and sex-regulated gene expression profiles in Caenorhabditis elegans. Proc Natl Acad Sci USA. 2001;98:218–223. doi: 10.1073/pnas.011520898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marinotti O, Calvo E, Nguyen QK, Dissanayake S, Ribeiro JMC, James AA. Genome-wide analysis of gene expression in adult Anopheles gambiae. Ins Mol Biol. 2006;15:1–12. doi: 10.1111/j.1365-2583.2006.00610.x. [DOI] [PubMed] [Google Scholar]
- Arbeitman MN, Fleming AA, Siegal ML, Null BH, Baker BS. A genomic analysis of Drosophila somatic sexual differentiation and its regulation. Development. 2004;131:2007–2021. doi: 10.1242/dev.01077. [DOI] [PubMed] [Google Scholar]
- Swanson WJ, Vacquier VD. Reproductive protein evolution. Annu Rev Ecol Syst. 2002;33:161–179. [Google Scholar]
- Cutter AD, Ward S. Sexual and temporal dynamics of molecular evolution in C. elegans development. Mol Biol Evol. 2005;22:178–188. doi: 10.1093/molbev/msh267. [DOI] [PubMed] [Google Scholar]
- Meiklejohn CD, Parsch J, Ranz JM, Hartl DL. Rapid evolution of male-biased gene expression in Drosophila. Proc Natl Acad Sci USA. 2003;100:9894–9899. doi: 10.1073/pnas.1630690100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh RS, Kulathinal RJ. Male sex drive and the masculinization of the genome. Bioessays. 2005;27:518–525. doi: 10.1002/bies.20212. [DOI] [PubMed] [Google Scholar]
- Boomsma JJ, Baer B, Heinze J. The evolution of male traits in social insects. Annu Rev Entomol. 2005;50:395–420. doi: 10.1146/annurev.ento.50.071803.130416. [DOI] [PubMed] [Google Scholar]
- Wilson EO. The insect societies. Cambridge: Harvard University Press; 1971. [Google Scholar]
- Evans JD, Wheeler DE. Differential gene expression between developing queens and workers in the honey bee, Apis mellifera. Proc Natl Acad Sci USA. 1999;96:5575–5580. doi: 10.1073/pnas.96.10.5575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans JD, Wheeler DE. Expression profiles during honeybee caste determination. Genome Biol. 2000;2:1–6. doi: 10.1186/gb-2000-2-1-research0001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hepperle C, Hartfelder K. Differentially expressed regulatory genes in honey bee caste development. Naturwissenschaften. 2001;88:113–116. doi: 10.1007/s001140000196. [DOI] [PubMed] [Google Scholar]
- Pereboom JJM, Jordan WC, Sumner S, Hammond RL, Bourke AFG. Differential gene expression in queen-worker caste determination in bumble-bees. Proc Roy Soc Lond B. 2005;272:1145–1152. doi: 10.1098/rspb.2005.3060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartfelder K, Makert GR, Judice CC, Pereira GAG, Santana WC, Dallacqua R, Bitondi MMG. Physiological and genetic mechanisms underlying caste development, reproduction and division of labor in stingless bees. Apidologie. 2006;37:144–163. [Google Scholar]
- Judice CC, Carazzole MF, Festa F, Sogayar MC, Hartfelder K, Pereira GAG. Gene expression profiles underlying alternative caste phenotypes in a highly eusocial bee, Melipona quadrifasciata. Ins Mol Biol. 2006;15:33–44. doi: 10.1111/j.1365-2583.2005.00605.x. [DOI] [PubMed] [Google Scholar]
- Sameshima S, Miura T, Matsumoto T. Wing disc development during caste differentiation in the ant Pheidole megacephala (Hymenoptera: Formicidae) Evol Dev. 2004;6:336–341. doi: 10.1111/j.1525-142X.2004.04041.x. [DOI] [PubMed] [Google Scholar]
- Tian H, Vinson SB, Coates CJ. Differential gene expression between alate and dealate queens in the red imported fire ant, Solenopsis invicta Buren (Hymenoptera: Formicidae) Ins Biochem Mol Biol. 2004;34:937–949. doi: 10.1016/j.ibmb.2004.06.004. [DOI] [PubMed] [Google Scholar]
- Sumner S, Pereboom JJM, Jordan WC. Differential gene expression and phenotypic plasticity in behavioural castes of the primitively eusocial wasp, Polistes canadensis. Proc Roy Soc Lond B. 2006;273:19–26. doi: 10.1098/rspb.2005.3291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scharf ME, Wu-Scharf D, Pittendrigh BR, Bennett GW. Caste- and development-associated gene expression in a lower termite. Genome Biol. 2003;4:R62.61–R62.11. doi: 10.1186/gb-2003-4-10-r62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou XG, Oi FM, Scharf ME. Social exploitation of hexamerin: RNAi reveals a major caste-regulatory factor in termites. Proc Natl Acad Sci USA. 2006;103:4499–4504. doi: 10.1073/pnas.0508866103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Tarver MR, Bennett GW, Oi FM, Scharf ME. Two hexamerin genes from the termite Reticulitermes flavipes : sequence, expression, and proposed functions in caste regulation. Gene. 2006;376:47–58. doi: 10.1016/j.gene.2006.02.002. [DOI] [PubMed] [Google Scholar]
- Edwards RE. Social wasps: their biology and control. East Grinstead: Rentokil; 1980. [Google Scholar]
- Telfer WH, Kunkel JG. The function and evolution of insect storage hexamers. Annu Rev Entomol. 1991;36:205–228. doi: 10.1146/annurev.en.36.010191.001225. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Nei M. Molecular evolutionary genetics. New York: Columbia University Press; 1987. [Google Scholar]
- Whitfield CW, Band MR, Bonaldo MF, Kumar CG, Liu L, Pardinas JR, Robertson HM, Soares MB, Robinson GE. Annotated expressed sequence tags and cDNA microarrays for studies of brain and behavior in the honey bee. Genome Res. 2002;12:555–566. doi: 10.1101/gr.5302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page RDM, Holmes EC. Molecular evolution: a phylogenetic approach. Oxford: Blackwell Science; 1998. [Google Scholar]
- Audic S, Claverie J-M. The significance of digital gene expression profiles. Genome Res. 1997;7:986–995. doi: 10.1101/gr.7.10.986. [DOI] [PubMed] [Google Scholar]
- Okubo K, Hori N, Matoba R, Niiyama T, Fukushima A, Kojima Y, Matsubara K. Large-scale cDNA sequencing for analysis of quantitative and qualitative aspects of gene-expression. Nature Genet. 1992;2:173–179. doi: 10.1038/ng1192-173. [DOI] [PubMed] [Google Scholar]
- Megy K, Audic S, Claverie J-M. Heart-specific genes revealed by expressed sequence tag (EST) sampling. Genome Biol. 2002;3:research0074.1–0074.11. doi: 10.1186/gb-2002-3-12-research0074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romualdi C, Bortoluzzi S, Danieli GA. Detecting differentially expressed genes in multiple tag sampling experiments: comparative evaluation of statistical tests. Human Mol Genet. 2001;10:2133–2141. doi: 10.1093/hmg/10.19.2133. [DOI] [PubMed] [Google Scholar]
- IDEG6: (web tool for the identification of differentially expressed genes in multiple tag sampling experiments) http://telethon.bio.unipd.it/bioinfo/IDEG6_form/index.html [DOI] [PubMed]


