Abstract
Aims
To identify the molecular basis for a low CYP1A2 metabolic status, as determined by a caffeine phenotyping test, in a 71-year-old, nonsmoking, Caucasian woman who presented with very high clozapine concentrations despite being administered a standard dose of the drug.
Methods
The nucleotide sequence of the 7 exons, exon-intron boundaries and 5′-flanking region of the CYP1A2 gene was analysed by direct sequencing.
Results
Only one heterozygous point mutation was identified in the donor splice site of intron 6 (3534G > A) of CYP1A2. This mutation could cause abnormal RNA splicing and therefore lead to a truncated nonfunctional enzyme. No other carrier of this mutation was identified in a population of 100 unrelated healthy Caucasians.
Conclusions
This is the first report of a splice-site mutation affecting the CYP1A2 gene. This polymorphism is a likely explanation for the low CYP1A2 activity associated with high clozapine concentrations in this patient.
Keywords: CYP1A2, cytochrome P450, pharmacogenetics, polymorphism, splicing mutation
Introduction
Cytochrome P450 1A2 (CYP1A2) is an enzyme implicated in the metabolic activation of environmental carcinogens, as well as in the metabolism of several drugs [1]. There are pronounced interindividual differences in CYP1A2 activity among humans, although the existence of a true phenotypic polymorphism in enzyme activity has not been unequivocally demonstrated to date [1–3]. Overall, it is recognized that CYP1A2-mediated slow and intermediate metabolizers represent about 50% of Caucasians [1]. Variability in CYP1A2 activity may influence individual susceptibility to cancer risk and therapeutic efficacy of some drugs. Different factors such as gender, race and environmental exposure to inducers or inhibitors are responsible for interindividual differences in the CYP1A2 phenotype [1]. Induction of CYP1A2 expression by smoking and inhibition of activity by oral contraceptives, for example, partly explain the variation in in vivo enzyme activity [1, 4]. Several genetic polymorphisms have been identified, in particular in the 5′-flanking region and in intron 1 of CYP1A2 [5–7, see also http://www.imm.ki.se/CYPalleles/cyp1a2.htm]. Some of these polymorphisms may be associated with altered inducibility of gene expression in smokers [6–8]. However, no genetic polymorphism has been reported to date that results in the expression of a CYP1A2 protein lacking catalytic activity.
Methods
A 71-year-old, nonsmoking, Caucasian woman, hospitalized with a schizoaffective disorder, mania type diagnosis (ICD 10 F25.0), was suspected of presenting with an overdose of clozapine, an antipsychotic drug. During the period of the study, the patient exhibited slight hypoalbuminaemia (35 g l−1, normal range: 37–51 g l−1), but with no known somatic disease or organ dysfunction, and with normal hepatic and renal function as assessed by standard clinical chemistry tests performed on several occasions (total bilirubin 17 µmol l−1; aspartate aminotransferase 14 U l−1; alanine aminotransferase 9 U l−1; alkaline phosphatase 25 U l−1; γ-glutamyltransferase 13 U l−1; urea 5.6 mmol l−1; creatinine 82 µmol l−1; Na+ 1.42 mmol l−1; K+ 4.0 mmol l−1). Clozapine and N-desmethylclozapine (norclozapine) were measured by gas chromatography with the use of a nitrogen phosphorus detector, according to the method described by Bondesson & Lindström [9] with slight modifications. Intraday and interday coefficients of variation for the determination of clozapine and norclozapine were between 5 and 10% and between 7 and 15%, respectively. The limit of quantification was 4 ng ml−1 for both substances.
As clozapine is metabolized to norclozapine mainly by CYP1A2 [10–12], a caffeine phenotyping test [13] was performed in order to define the CYP1A2 metabolic status of the patient. Caffeine and its metabolite (paraxanthine) were measured by gas chromatography–mass spectrometry according to a method developed in our laboratory [Eap et al. unpublished data, available on request]. Intraday and interday coefficients of variation for the determination of caffeine and paraxanthine were between 2 and 7% and between 6 and 15%, respectively. The limit of quantification was 0.8 ng ml−1 for both substances.
Using a genomic DNA sample from this woman, we analysed the nucleotide sequence of the 5′-flanking region (from nucleotide −4078 to −870), the 7 exons and the exon-intron boundaries of the CYP1A2 gene. In order to sequence the 5′-flanking region of CYP1A2, a 3.2 kb fragment was first amplified by using a pair of CYP1A2-specific primers (forward primer: 5′-CAGGGACTTCT TGGATGCTTATGATGTCTC-3′; reverse primer: 5′-GGGTTGTAATGGCTGGTGTGGAGCTTCTGG-3′) and a TaKaRa Ex Taq™ kit (BioWhittaker, Verviers, Belgium), according to the manufacturer's instructions. Each of the 7 exons and their proximal flanking sequences were amplified according to a double step-PCR procedure, as described previously [14]. Nucleotide sequences were then determined by using an automated DNA sequencer (Model 373 A, Applied Biosystems, Foster City, USA) and the ABIPRISM Dye Terminator Cycle Sequencing Ready Reaction FS kit (Applied Biosystems) according to manufacturer's instructions. Both strands of each DNA fragment were sequenced. Approval for the study was obtained from the local Ethics Committee (Avis de la Commission d'Ethique de la Psychiatrie, University of Lausanne) and the patient gave her written informed consent for the phenotyping and genotyping tests, as well as for the possible publication of the data collected during this case study.
In order to estimate the frequency of this polymorphism, we re-examined the exon 6 of CYP1A2 in a previously studied population of 100 unrelated healthy volunteers of French Caucasian origin, using a PCR-SSCP strategy, as described elsewhere [14].
Results
Therapeutic drug monitoring of clozapine and norclozapine was first requested due to the suspicion of a clozapine overdose, despite the patient being administered a standard dose, i.e. 300 mg day−1, for 6 days. To achieve this dose, increasing amounts of the drug were given over a 7-week period.
The analysis revealed a high steady-state plasma concentration of clozapine (1296 ng ml−1) and of its metabolite (760 ng ml−1); (comedication: lactitol 20 ml day−1). Clozapine dose was then reduced to 100 mg day−1 because concentrations of clozapine above 1000 ng ml−1 increase the risk of adverse effects on the central nervous system, causing confusion, delirium and generalized seizures [15]. Two analyses performed 2 weeks and 2 months after the dose reduction showed a corresponding decrease in the plasma concentrations of clozapine and norclozapine (first determination: clozapine: 406 ng ml−1; norclozapine: 324 ng ml−1; comedication: clomethiazole 300 mg day−1, lactilol 20 ml day−1; second determination: clozapine: 475 ng ml−1; norclozapine: 205 ng ml−1; comedication: flurazepam 30 mg day−1, lactilol 20 ml day−1).
The caffeine phenotyping test revealed a slow CYP1A2 phenotype for this patient. Her plasma caffeine and paraxanthine concentrations measured 6 h after the oral intake of 200 mg of caffeine were 7.1 µg ml−1 and 1.4 µg ml−1, respectively, giving rise to a paraxanthine-caffeine ratio of 0.21, corresponding to a value of 0.32 ml min−1 kg−1 for systemic caffeine plasma clearance [13]. The patient was also taking clozapine 100 mg day−1 and flurazepam 30 mg day−1 at the time of the caffeine test. The paraxanthine-caffeine ratio is among the lowest found in the values (approximate range: 0.25–1.5) presented in a report that analysed retrospectively the caffeine clearance in four studies comprising a total of 78 subjects [13].
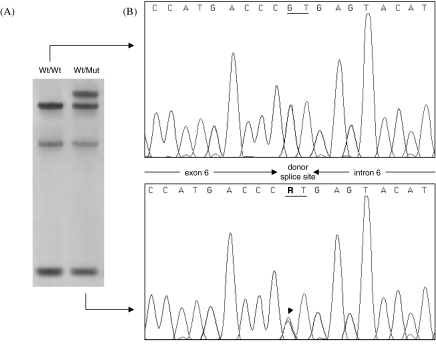
The sequencing of the 5′-flanking region, the 7 exons and their exon-intron junctions of the CYP1A2 gene identified only one heterozygous point mutation in the patient's DNA (Figure 1b). This polymorphism is located in the 5′ splice-site consensus sequence of intron 6, substituting the invariant GT dinucleotide with AT (mutation 3534G > A according to the CYP1A2 gene sequence published by Corchero et al. [16]). According to the current human CYP allele nomenclature, this novel allelic variant was termed CYP1A*7. Using the PCR-SSCP strategy previously described by Chevalier et al. [14], the mutation was clearly detected through an abnormal SSCP pattern (Figure 1a) and no other carrier of the 3534G > A substitution was identified in a control group comprising 100 unrelated French individuals, suggesting that the mutation is very rare in Caucasian populations.
Figure 1.
Detection by PCR-SSCP (a) and identification by direct sequencing (b) of the 3534G > A mutation in the CYP1A2 gene. A PCR fragment encompassing exon 6 of CYP1A2 and its proximal flanking sequences has been analysed by SSCP and direct sequencing as described in the Methods section. A DNA sample from the patient with a slow CYP1A2 activity (Wt/Mut) is compared with that from a subject with a homozygous wild-type genotype (Wt/Wt). The arrow indicates the position of the heterozygous G/A mutation in the donor splice site of intron 6. R; guanine or adenine.
Discussion
Our findings strongly suggest that the 3534G > A mutation in the CYP1A2 gene identified in the patient, although only present in a heterozygous state, is a likely explanation for the elevated plasma concentrations of clozapine and the low CYP1A2 activity as determined by the caffeine phenotyping test.
It is recognized that donor (GT) and acceptor (AG) splice-site consensus sequences are major determinants of accurate splicing [17] and therefore the observed G > A transition in the donor splice-site of CYP1A2 intron 6 is likely to affect normal splicing efficiency. Considering the absence of a potential alternative donor splice site in the immediate vicinity of the mutation, exon skipping is more likely to occur than activation of a cryptic splice-site [17]. Extensive studies of the consequences of mutations in splice-sites have shown that when a mutation of the 5′ splice-site leads to exon skipping, it is always the upstream exon immediately preceding the lesion that is removed from the subsequent mRNA transcript [17]. Exon 6 skipping could alter the CYP1A2 protein sequence whereby a missense mutation (Ser388Arg) and a 29-amino acid deletion (delThr389-Pro417) would occur, leading to a truncated protein with no retained activity. In vitro expression analysis of an allelic minigene, consisting of exons 5–7 of the mutant gene, would allow the confirmation of our hypothesis.
Variability in CYP1A2 activity is a major determinant of the large interindividual differences in plasma concentrations of clozapine [11]. In addition, the concentrations of clozapine and norclozapine measured in this patient are compatible with a heterozygous status, as they are in the upper range of those usually measured. Thus, in one study in which 24 out of 29 patients received clozapine at a dosage of 400 mg day−1, the mean ±SD (range) clozapine and norclozapine concentrations were 374 ± 233 ng ml−1, 84–1088 ng ml−1: 116 ± 65 ng ml−1, 25–272 ng ml−1, respectively [18]. In another study in which 61 patients received a fixed dose of 400 mg day−1 of clozapine for 6 weeks, the mean blood concentrations of clozapine were 598 ± 314 ng ml−1 (range: 111–1585 ng ml−1) [19]. To our knowledge, a homozygous CYP1A2 poor metabolizer status has never been described. However, phenocopying can occur when a potent CYP1A2 inhibitor such as fluvoxamine, an antidepressant, is given at a medium to high dose. In one patient clozapine and norclozapine concentrations were 2166 ng ml−1 and 615 ng ml−1 after receiving 400 mg day−1 of clozapine and 100 mg day−1 of fluvoxamine [20]. After fluvoxamine, plasma concentrations of both clozapine and norclozapine were increased, which suggests that CYP1A2 was also involved in the degradation of norclozapine [21]. This is in agreement with the present case report where elevated plasma concentrations were observed both for clozapine and its metabolite. In two other patients treated with 300 mg day−1 of clozapine plus 75 mg day−1 of fluvoxamine, and with 500 mg day−1 of clozapine plus 150 mg day−1 of fluvoxamine, the concentrations of clozapine were 1678 ng ml−1 and a maximum of 2911 ng ml−1, respectively [20].
The values of caffeine clearance measured in this patient (0.32 ml min−1) are also compatible with a heterozygous status for an inactivating CYP1A2 mutation. Thus, in other studies, systemic caffeine plasma clearance was found to vary from 0.3 to 3.3 ml min−1 kg−1 [13]. Again, when fluvoxamine is given as a comedication, a value of 0.03 ml min−1 kg−1 for systemic caffeine plasma clearance was obtained, indicative of an almost complete inhibition of CYP1A2 activity corresponding to a homozygous CYP1A2 poor metabolizer status [22].
In conclusion, this is the first report of a polymorphism affecting a splice-site consensus sequence of the CYP1A2 gene. This mutation could cause abnormal RNA splicing and, thereby, lead to the expression of a truncated protein lacking catalytic activity.
Acknowledgments
This work was supported by the Centre Hospitalier Régional et Universitaire de Lille, France.
References
- 1.Landi MT, Sinha R, Lang NP, Kadlubar FF. Human cytochrome P4501A2, Chapter 16. IARC Sci Publications. 1999;148:173–195. [PubMed] [Google Scholar]
- 2.Butler MA, Lang NP, Young JF, et al. Determination of CYP1A2 and NAT2 phenotypes in human populations by analysis of caffeine urinary metabolites. Pharmacogenetics. 1992;2:116–127. doi: 10.1097/00008571-199206000-00003. [DOI] [PubMed] [Google Scholar]
- 3.Welfare MR, Aitkin M, Bassendine MF, Daly AK. Detailed modelling of caffeine metabolism and examination of the CYP1A2 gene: lack of a polymorphism in CYP1A2 in Caucasians. Pharmacogenetics. 1999;9:367–375. doi: 10.1097/00008571-199906000-00012. [DOI] [PubMed] [Google Scholar]
- 4.Tantcheva-Poór I, Zaigler M, Rietbrock S, Führ U. Estimation of cytochrome P-450 CYP1A2 activity in 863 healthy Caucasians using a saliva-based caffeine test. Pharmacogenetics. 1999;9:131–144. [PubMed] [Google Scholar]
- 5.Chida M, Yokoi T, Fukui T, Kinoshita M, Yokota J, Kamataki T. Detection of three genetic polymorphisms in the 5′-flanking region and intron 1 of human CYP1A2 in the Japanese population. Jpn J Cancer Res. 1999;90:899–902. doi: 10.1111/j.1349-7006.1999.tb00832.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nakajima M, Yokoi T, Mizutani M, Kinoshita M, Funayama M, Kamataki T. Genetic polymorphism in the 5′-flanking region of human CYP1A2 gene: effect on the CYP1A2 inducibility in humans. J Biochem. 1999;125:803–808. doi: 10.1093/oxfordjournals.jbchem.a022352. [DOI] [PubMed] [Google Scholar]
- 7.Sachse C, Brockmoller J, Bauer S, Roots I. Functional significance of a C→A polymorphism in intron 1 of the cytochrome P450 CYP1A2 gene tested with caffeine. Br J Clin Pharmacol. 1999;47:445–449. doi: 10.1046/j.1365-2125.1999.00898.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Han XM, Ou-Yang DS, Lu PX, et al. Plasma caffeine metabolite ratio (17X/137X) in vivo associated with G-2964A and C734A polymorphisms of human CYP1A2. Pharmacogenetics. 2001;11:429–435. doi: 10.1097/00008571-200107000-00006. [DOI] [PubMed] [Google Scholar]
- 9.Bondesson U, Lindström LH. Determination of clozapine and its N-demethylated metabolite in plasma by use of gas chromatography-mass spectrometry with single ion detection. Psychopharmacology. 1988;95:472–475. doi: 10.1007/BF00172957. [DOI] [PubMed] [Google Scholar]
- 10.Pirmohamed M, Williams D, Madden S, Templeton E, Park BK. Metabolism and bioactivation of clozapine by human liver in vitro. J Pharmacol Exp Ther. 1995;272:984–990. [PubMed] [Google Scholar]
- 11.Bertilsson L, Carrillo JA, Dahl ML, et al. Clozapine disposition covaries with CYP1A2 activity determined by a caffeine test. Br J Clin Pharmacol. 1994;38:471–473. doi: 10.1111/j.1365-2125.1994.tb04385.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Aitchison KJ, Jann MW, Zhao JH, et al. Clozapine pharmacokinetics and pharmacodynamics studied with CYP1A2-null mice. J Psychopharmacol. 2000;14:353–359. doi: 10.1177/026988110001400403. [DOI] [PubMed] [Google Scholar]
- 13.Führ U, Rost KL. Simple and reliable CYP1A2 phenotyping by the paraxanthine/caffeine ratio in plasma and in saliva. Pharmacogenetics. 1994;4:109–116. doi: 10.1097/00008571-199406000-00001. [DOI] [PubMed] [Google Scholar]
- 14.Chevalier D, Cauffiez C, Allorge D, et al. Five novel natural allelic variants – 951A>C, 1042G>A (D348N), 1156A>T (I386F), 1217G>A (C406Y) and 1291C>T (C431Y) – of the human CYP1A2 gene in a French Caucasian population. Hum Mutat. 2001;17:355–356. [PubMed] [Google Scholar]
- 15.Freeman DJ, Oyewumi LK. Will routine therapeutic drug monitoring have a place in clozapine therapy? Clin Pharmacokinet. 1997;32:93–100. doi: 10.2165/00003088-199732020-00001. [DOI] [PubMed] [Google Scholar]
- 16.Corchero J, Kimura S, Gonzalez FJ. Organization of the CYP1A cluster on human chromosome 15: implications for gene regulation. Pharmacogenetics. 2001;11:1–6. doi: 10.1097/00008571-200102000-00001. [DOI] [PubMed] [Google Scholar]
- 17.Krawczak M, Reiss J, Cooper DN. The mutational spectrum of single base-pair substitutions in mRNA splice junctions of human genes: causes and consequences. Hum Genet. 1992;90:41–54. doi: 10.1007/BF00210743. [DOI] [PubMed] [Google Scholar]
- 18.Perry PJ, Miller DD, Arndt SV, Cadoret RJ. Clozapine and norclozapine plasma concentrations and clinical response of treatment-refractory schizophrenic patients. Am J Psychiatry. 1991;148:231–235. doi: 10.1176/ajp.148.2.231. [DOI] [PubMed] [Google Scholar]
- 19.Liu HC, Chang WH, Wei FC, Lin SK, Jann MW. Monitoring of plasma clozapine levels and its metabolites in refractory schizophrenic patients. Ther Drug Monit. 1996;18:200–207. doi: 10.1097/00007691-199604000-00015. [DOI] [PubMed] [Google Scholar]
- 20.Hiemke C, Weigmann H, Härtter S, Dahmen N, Wetzel H, Müller H. Elevated levels of clozapine in serum after addition of fluvoxamine. J Clin Psychopharmacol. 1994;14:279–281. [PubMed] [Google Scholar]
- 21.Wetzel H, Anghelescu I, Szegedi A, et al. Pharmacokinetic interactions of clozapine with selective serotonin reuptake inhibitors: differential effects of fluvoxamine and paroxetine in a prospective study. J Clin Psychopharmacol. 1998;18:2–9. doi: 10.1097/00004714-199802000-00002. [DOI] [PubMed] [Google Scholar]
- 22.Bender S, Eap CB. Very high cytochrome P4501A2 activity and nonresponse to clozapine. Arch Gen Psychiatry. 1998;55:1048–1050. doi: 10.1001/archpsyc.55.11.1048. [DOI] [PubMed] [Google Scholar]