Abstract
Aims
To investigate the frequencies of CYP2C19*1, CYP2C19*2 and CYP2C19*3 alleles and CYP2C19 genotypes in a Tamilian population.
Methods
The study was conducted in 112 unrelated healthy human volunteers. DNA was extracted from leucocytes and analyzed by the PCR-RFLP protocol. The PCR product was digested with restriction enzymes (SmaI and BamH1) and then separated electrophoretically using polyacrylamide gel.
Results
The frequencies of the CYP2C19*1, *2 and *3 alleles were 0.598 [95% confidence interval (CI) 0.507, 0.689], 0.379 (95% CI, 0.350,0.407) and 0.022 (95% CI −0.005, 0.049), respectively. The distribution of CYP2C19*1/*1,*1/*2, *1/*3, *2/*2 and *2/*3 genotypes were 0.295 (95% CI, 0.210, 0.379), 0.580 (95% CI, 0.488, 0.671), 0.027 (95% CI −0.003, 0.057), 0.080 (95% CI 0.030, 0.130) and 0.018 (95% CI −0.006, 0.042), respectively.
Conclusions
The distribution of CYP2C19*1/*1 in the Tamilian population is lower than that in Caucasians, Africans and the North Indian population. The CYP2C19*1/*2 is significantly higher in Tamilians when compared with other populations. The CYP2C19*1/*3 allele, which was not reported in the North Indian and Caucasian populations has been identified in 2.7% of the Tamilian population.
Keywords: CYP2C19 polymorphism, genotyping, Indian, Tamilian
Introduction
CYP2C19 plays a major role in the metabolism of some clinically important drugs such as omeprazole, diazepam and proguanil [1]. CYP2C19*2 and CYP2C19*3 are the most frequently identified defective alleles in Orientals and Caucasian poor metabolizers (PM). The presence of the CYP2C19*2 allele leads to an aberrant splice site, whereas the CYP2C19*3 allele produces a premature stop codon [1]. The prevalence of PMs was reported to be 2–5% in Caucasians [2, 3], 4–8% in Africans [4] and 11–23% in Orientals [5–7].
In an earlier study in a North Indian population, the frequency of the CYP2C19*2 allele was reported to be 29.7% whereas CYP2C19*3 was absent [8]. North Indians are mainly Aryan in origin, whereas Tamilians are mainly Dravidians residing in Southern India [9]. The objective of this study was to estimate the frequencies of CYP2C19*2 and CYP2C19*3 alleles in the Tamilian population.
Methods
The study was conducted in 112 unrelated healthy Tamilian subjects residing in Tamilnadu and Pondicherry. There were 97 males and 15 females with a mean age (SD) of 31.4 (10.4) years. Institutional Ethics Committee approval and written informed consent of the volunteers were obtained.
About 5 ml of blood was collected from each volunteer and DNA was extracted from leucocytes. Genotyping of extracted crude DNA for CYP2C19*1, CYP2C19*2 and CYP2C19*3 alleles was performed by the PCR-RFLP method described by Sviri et al. [10]. PCR amplification of CYP2C19*2 was done using the forward and reverse primers: 5′-AATTACAACCAGAGCTTGGC-3′ and 5′-TATCACTTTCCATAAAAGCAAG-3′. The primers used for analysis of CYP2C19*3 mutant allele were: 5′-TATTATTATCTGTTAAC TAAT ATGA-3′ and 5′ACT TCAGGGCTTGGTCAATA-3′. The PCR products of CYP2C19*2 and CYP2C19*3 were digested with SmaI and BamH1 enzymes, respectively, and separated on a 6% polyacrylamide gel. Since the restriction site is absent in the mutant alleles (CYP2C19*2 and CYP2C19*3) the PCR products are not digested by restriction enzymes. In the CYP2C19*1 (wild-type) allele the restriction enzymes SmaI and BamH1 splice the 169 bp and 329 bp DNA fragments into 120 bp, 49 bp (Figure 1) and 233 bp, 96 bp, respectively. Positive and negative controls were always included in each PCR analysis and digestion of samples with restriction enzymes. Samples containing mutant alleles were reanalysed to ensure the accuracy of the method. There was 100% reproducibility.
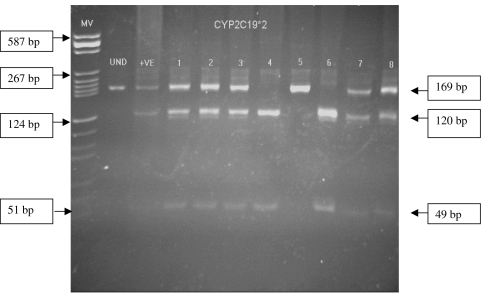
Figure 1.
Gel electrophoresis photograph showing the analysis of CYP2C19*2. Note: 1, 2, 3, 7 and 8 are heterozygous (CYP2C19*1/*2), 5 is homozygous (CYP2C19*2/*2), 4 and 6 are wild genotype (CYP2C19*1/*1). UND – undigested; + ve is heterozygous positive control
The data were analysed by Fisher's exact test. P < 0.05 was considered statistically significant. The observed genotype frequencies of CYP2C19 were compared with expected genotype frequencies according to the Hardy–Weinberg law.
Results
CYP2C19*1 was the most frequently identified allele in the Tamilian population [0.598, 95% confidence interval (CI) 0.507, 0.689, Table 1]. There were 33 subjects with the CYP2C19*1/*1 genotype (0.295, CI 0.210, 0.379) and 68 subjects were heterozygous having a CYP2C19*1/*2 or CYP2C19*1/*3 genotype (0.607, CI 0.517, 0.697). CYP2C19*2 was the most frequently identified mutant allele (0.379, CI 0.350, 0.407). Nine subjects were homozygous for the CYP2C19*2 allele (0.080, CI 0.029, 0.130). Five subjects possessed the CYP2C19*3 allele (0.022, CI −0.005, 0.049), of whom three were heterozygous for CYP2C19*1 and two for the CYP2C19*2. None of the subjects was homozygous for the CYP2C19*3 mutant allele. The observed CYP2C19 genotype frequencies were consistent with Hardy–Weinberg expectations (data not shown).
Table 1.
The genotype and allele frequencies of CYP2C19 in a Tamilian population compared with other populations
| Frequencies of CYP2C19 | |||||
|---|---|---|---|---|---|
| CYP2C19 alleles and genotype | Tamilians (n = 112) | North Indians†(n = 121) | Orientals‡(n = 367) | Caucasians§(n = 837) | Africans*(n = 251) |
| 2C19*1 | 0.598a | 0.703 | 0.610 | 0.860 | 0.814 |
| (0.507, 0.689) | (0.622, 0.784) | (0.561, 0.659) | (0.837, 0.883) | (0.766, 0.862) | |
| 2C19*2 | 0.379a | 0.297 | 0.305 | 0.140 | 0.179 |
| (0.350, 0.407) | (0.216, 0.378) | (0.258, 0.352) | (0.117, 0.163) | (0.132, 0.226) | |
| 2C19*3 | 0.022b | 0 | 0.085 | 0 | 0.007 |
| (−0.005, 0.049) | (0.057, 0.113) | (0.003, 0.017) | |||
| 2C19*1/*1 | 0.295c | 0.479 | 0.384 | 0.745 | 0.661 |
| (0.210, 0.379) | (0.390, 0.568) | (0.335, 0.433) | (0.716, 0.774) | (0.603, 0.719) | |
| 2C19*1/*2 | 0.580d | 0.446 | 0.367 | 0.230 | 0.298 |
| (0.488, 0.671) | (0.358, 0.534) | (0.318, 0.416) | (0.201, 0.258) | (0.242, 0.354) | |
| 2C19*1/*3 | 0.027e | 0 | 0.084 | 0.005 | 0.007 |
| (−0.003, 0.057) | (0.057, 0.113) | (0.001, 0.009) | (−0.003, 0.017) | ||
| 2C19*2/*2 | 0.080f | 0.074 | 0.087 | 0.022 | 0.027 |
| (0.030, 0.130) | (0.028, 0.120) | (0.059, 0.115) | (−0.012, 0.031) | (0.007, 0.047) | |
| 2C19*2/*3 | 0.018 | 0 | 0.065 | 0 | 0.004 |
| (−0.006, 0.042) | (0.040, 0.090) | (−0.003, 0.011) | |||
| 2C19*3/*3 | 0 | 0 | 0.010 | 0 | 0 |
| (0, 0.02) | |||||
Values given in parentheses are 95% confidence intervals; n = number of subjects;
Reference 8;
Compiled data of Orientals (Japanese, Chinese and Thais) calculated from references 5–7;
Compiled data of Caucasians (European Americans, Turks, Germans) calculated from references 2 and 3;
Reference 4.
Tamilians vs Caucasians P < 0.0001, Africans P < 0.0001;
Tamilians vs Orientals P < 0.03, Caucasians P < 0.001;
Tamilians vs North Indians P < 0.005, Orientals P < 0.0001, Caucasians P < 0.0001;
Tamilians vs North Indians P < 0.05, Orientals P < 0.0001, Caucasians P < 0.0001, Africans P < 0.0001;
Tamilians vs Orientals P < 0.03, Caucasian P < 0.03.
Tamilians vs Caucasians P < 0.002, Africans P < 0.05.
Discussion
In our Tamilian population, the CYP2C19*1 allele (wild type) frequency was significantly less when compared with Caucasians and Africans (Table 1). The most common mutant allele in Tamils was CYP2C19*2. Its frequency was significantly higher compared with Caucasians and Africans (Table 1) but not significantly different from Orientals and North Indians [5–8]. The presence of the CYP2C19*3 allele was not reported in North Indians [8], whereas in our study five Tamilians had this allele.
CYP2C19*1/*1 genotype was less frequent in Tamilians compared with Caucasians, Africans and North Indians [Table 1]. In Caucasians, Africans and North Indians, most of the extensive metabolizers (EMs) were homozygous for the wild type allele [3–5, 9], whereas in Tamilians most of the EMs were heterozygous for the wild type allele. The majority of Tamilian poor metabolizers were homozygous for the CYP2C19*2 mutant allele, which is similar to other ethnic populations. The prevalence of PMs in Tamilians was significantly higher compared with Caucasians and Africans (Table 1).
In conclusion, the polymorphism of CYP2C19 in the Tamilian population shows differences to that in Caucasians and North Indians.
Acknowledgments
The research project is funded by Indian Council of Medical Research, New Delhi, India and INSERM, Paris, France (ICMR Ref.No.50/6/2000-BMS dated 11/12/2001). We gratefully acknowledge the technical assistance provided by Mr R. Balakrishnan, Mr Rajan and Ms Mala.
References
- 1.Desta Z, Zhao X, Shin JG, Flockhart DA. Clinical significance of the cytochrome P450 2C19 genetic polymorphism. Clin Pharmacokinet. 2002;41:913–958. doi: 10.2165/00003088-200241120-00002. [DOI] [PubMed] [Google Scholar]
- 2.Goldstein JA, Ishizaki T, Chiba K, de Morais SM, Bell D, Krahn PM, et al. Frequencies of the defective CYP2C19 alleles responsible for the mephenytoin poor metabolizer phenotype in various Oriental, Caucasian, Saudi Arabian and American black populations. Pharmacogenetics. 1997;7:59–64. doi: 10.1097/00008571-199702000-00008. [DOI] [PubMed] [Google Scholar]
- 3.Aynacioglu AS, Sachse C, Bozkurt A, Kortunay S, Nacak M, Schroder T, et al. Low frequency of defective alleles of cytochrome P450 enzymes 2C19 and 2D6 in the Turkish population. Clin Pharmacol Ther. 1999;66:185–192. doi: 10.1053/cp.1999.v66.100072001. [DOI] [PubMed] [Google Scholar]
- 4.Herrlin K, Massele AY, Jande M, Alm C, Tybring G, Abdi YA, et al. Bantu Tanzanians have a decreased capacity to metabolize omeprazole and mephenytoin in relation to their CYP2C19 genotype. Clin Pharmacol Ther. 1998;64:391–401. doi: 10.1016/S0009-9236(98)90070-4. [DOI] [PubMed] [Google Scholar]
- 5.Kubota T, Chiba K, Ishizaki T. Genotyping of S-mephenytoin 4′-hydroxylation in an extended Japanese population. Clin Pharmacol Ther. 1996;60:661–666. doi: 10.1016/S0009-9236(96)90214-3. [DOI] [PubMed] [Google Scholar]
- 6.de Morais SM, Goldstein JA, Xie HG, Huang SL, Lu YQ, Xia H, et al. Genetic analysis of the S-mephenytoin polymorphism in a Chinese population. Clin Pharmacol Ther. 1995;58:404–411. doi: 10.1016/0009-9236(95)90053-5. [DOI] [PubMed] [Google Scholar]
- 7.Tassaneeyakul W, Tawalee A, Tassaneeyakul W, Kukongviriyapan V, Blaisdell J, Goldstein JA, et al. Analysis of the CYP2C19 polymorphism in a North-eastern Thai population. Pharmacogenetics. 2002;12:221–225. doi: 10.1097/00008571-200204000-00006. [DOI] [PubMed] [Google Scholar]
- 8.Lamba JK, Dhiman RK, Kohli KK. CYP2C19 genetic mutations in North Indians. Clin Pharmacol Ther. 2000;68:328–335. doi: 10.1067/mcp.2000.109365. [DOI] [PubMed] [Google Scholar]
- 9.Roland J, Berton L. Atlas of the Languages and Ethnic Communities of South Asia. New Delhi: Sags Publications; 1993. [Google Scholar]
- 10.Sviri S, Shpizen S, Leitersdorf E, Levy M, Caraco Y. Phenotypic-genotypic analysis of CYP2C19 in the Jewish Israeli population. Clin Pharmacol Ther. 1999;65:275–282. doi: 10.1016/S0009-9236(99)70106-2. [DOI] [PubMed] [Google Scholar]