Abstract
Medieval Black Death is believed to have killed up to one-third of the Western European population during the 14th century. It was identified as plague at this time, but recently the causative organism was debated because no definitive evidence has been obtained to confirm the role of Yersinia pestis as the agent of plague. We obtained the teeth of a child and two adults from a 14th century grave in France, disrupted them to obtain the pulp, and applied the new “suicide PCR” protocol in which the primers are used only once. There were no positive controls: Neither Yersinia nor Yersinia DNA were introduced in the laboratory. A negative result is followed by a new test using other primers; a positive result is followed by sequencing. The second and third primer pair used, coding for a part of the pla gene, generated amplicons whose sequence confirmed that it was Y. pestis in 1 tooth from the child and 19/19 teeth from the adults. Negative controls were negative. Attempts to detect the putative alternative etiologic agents Bacillus anthracis and Rickettsia prowazekii failed. Suicide PCR avoids any risk of contamination as it uses a single-shot primer—its specificity is absolute. We believe that we can end the controversy: Medieval Black Death was plague.
Keywords: plague, ancient DNA, paleomicrobiology
The medieval pandemic that later came to be called the “Black Death” killed an estimated 17–28 million Europeans (from 30 to 40% of the total population) between the years 1347 and 1351 (1). Black Death was first reported in Central Asia between 1339 and 1340, then in the Genoese city of Caffa, where Black Death Mongol cadavers were hurled as bacteriological weapons over the city walls in 1346 (2). Epidemics reached Europe when Genoese vessels docked in Messina, Genoa, and Marseilles in November 1347. Yersinia pestis is considered to be the most likely agent of Black Death, based mainly on historical clinical records of bubonic and pulmonary forms of the disease (3). However, as the high levels of mortality and transmissibility associated with the Black Death were not observed during the third plague pandemic (4, 5), alternative etiologies have been proposed. Moreover, the spread of transmission of the disease as well as the supposed densities of rats and fleas made unlikely for several authors the role of Y. pestis (5). The alternatives have included the anthrax agent Bacillus anthracis, the typhus agent Rickettsia prowazekii, Mycobacterium tuberculosis, and hemorrhagic fever (6–8). Confirmation of Y. pestis as the agent of the Black Death would end this controversy and improve our understanding of Y. pestis epidemics in light of the current reemergence of this infectious disease (9, 10).
PCR-based methods have led to the identification of microbial pathogens in ancient (11–15) and forensic (16) human remnants. However, these studies have had to employ exhaustive preventive measures to minimize the risk of false-positive results arising from contamination of samples with previously attempted amplification reactions (17, 18). In addition to this in vitro contamination, most ancient samples will have been exposed to, and probably colonized by, a natural microbial flora. To circumvent both these problems we have developed a strategy for the reliable detection of ancient pathogens in human remains. We have recently demonstrated the presence of DNA remnants from ancient systemic pathogens in skeletal dental pulp, a source of DNA free from natural contamination (14). Furthermore, we have designed for the present work a new PCR procedure, which we named suicide PCR, to detect microbial DNA. Therefore, we attempted to amplify the DNA of Y. pestis and two alternative putative agents, B. anthracis and R. prowazekii, in dental pulp removed from teeth collected from three skeletons of Black Death victims.
Materials and Methods
Excavation of a Patient Who Died of Black Death.
The 800-grave Saint-Côme and Saint-Damien site in Montpellier, Southern France, had been used as an extramural church cemetery during the 9th–17th centuries. Of the 800 graves excavated at this site, it seems likely that 4 were catastrophe graves because they contained multiple skeletons without shrouds. These four graves have been dated as having been dug between the 13th and late 14th centuries, because of their position on the top of a 13th century remblai, behind a wall dated from the second half of the 14th century. Dating of the different parts of this site was based on historical data, stratigraphy, the study of 7,059 ceramic remnants, and 14C data. This is the period when the Black Death swept through the Montpellier area, including the 1348 Black Death and further plague resurgences in 1361, 1374, 1375, 1385, and 1397 (19). These epidemics resulted in a sharp decrease in the Montpellier population, which fell from an estimated 9,500 homesteads in 1348 to only 1,000 homesteads in 1379 (19). No other catastrophe or epidemic was recorded in this area during this period. Therefore, we hypothesized that these grave skeletons were those of Black Death victims. One of these graves exhibited a multiple burial containing the skeletons of a man, a woman, and a child estimated to be 8–10 years old when he died. For further molecular investigation, 4 teeth were collected from the child's skull, 9 from the female's skull, and 10 from the male's skull.
Recovery of DNA from Ancient Dental Pulp.
Twenty-three teeth were collected from Black Death skeletons and, as a negative control, four unerupted teeth were collected from ancient skeletons excavated from a medieval grave in Toulon, France. These skeletons showed no macroscopic signs of infectious disease and there was no anthropological evidence that these people could have died as a result of the plague. Teeth were thoroughly washed and longitudinally fractured (Fig. 1). Powdery remnants were scraped from the dental pulp cavities into sterile tubes for further DNA extraction. All manipulations of ancient teeth, including opening of the teeth and dental pulp collection, were performed in a laboratory located in a building where Y. pestis has never been introduced.
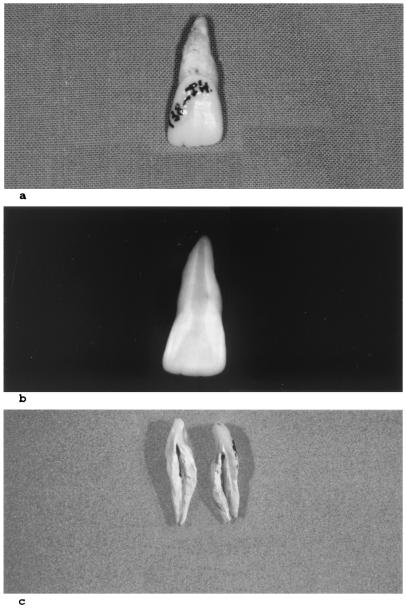
Figure 1.
Photography (a), radiography (b), and recovery (c) of ancient dental pulp from the skeleton of a 1348-Black Death child confirmed as having died from the Y. pestis plague.
Suicide PCR and Sequence Analysis of Ancient Y. pestis DNA.
Dental pulp DNA was extracted by using a standard protocol (20). To avoid any possibility of molecular contamination of the ancient DNA with amplicons resulting from previous investigations into ancient plague in our laboratory (14), DNA extracted from the dental pulp was submitted to suicide PCR amplifications using primers targeting Y. pestis genetic sequences not previously targeted in our laboratory. No positive controls were used at any step of the processing of ancient material. Our suicide PCR methodology permits primer pairs to be used only once, but multiple sets of primers could be tested until an amplicon of the expected size is yielded. This amplicon then has to be sequenced to confirm its identity. In a first set of experiments, only teeth collected from the child's skull were tested by suicide PCR. PCR amplifications were performed in 25-μl reaction volumes containing 5 ng of DNA in 10 mM Tris⋅HCl (pH 8.3), 50 mM KCl, 1.5 mM MgCl2, 200 μM 2′-deoxynucleotide and 5′-triphosphate, 0.2 mg of BSA, 2 μM each of the oligonucleotide primers, and 2 units of Taq DNA polymerase. A first set of primers INV.D (5′-AATGCTTATTTACCTGCACT-3′) and INV.R (5′-TCAAAAGGGGTTTTTATA-3′) targeting an IS200-like element within the gene env (21) was incorporated under the following conditions: initial denaturation of 95°C for 90 s was followed by 40 cycles consisting of denaturation (20 s at 95°C), primer annealing (20 s at 58°C), and elongation (30 s at 72°C). A second set of primers YP11D (5′-CTATGCCATATATTGGACTTGC-3′) and YP10R (5′-GAGCCGGATGTCTTCTCACG-3′) corresponding to nucleotides 854–1,001 of the pla gene (GenBank accession no. M27820) then was tested under the following conditions: Initial denaturation for 90 s at 95°C was followed by 40 cycles consisting of denaturation for 30 s at 95°C, primer annealing for 30 s at 52°C, and elongation for 90 s at 72°C, followed by postelongation for 4 min at 72°C. The pla sequence has been demonstrated to be specific for Y. pestis (22, 23) and not to be present in the related subspecies Yersinia pseudotuberculosis (24). Further experiments included 19 teeth collected in two adult skeletons and four negative controls. Suicide PCR incorporated PCR primers YP12D (5′-CAGCAGGATATCAGGAAACA-3′) and YP11R (5′-GCAAGTCCAATATATGGCATAG-3′; reverse complement of primer YP11D) corresponding to the nucleotides 728–876 of the pla gene under the same experimental conditions as described above for YP11D/YP10R primers. When incorporated with the alternative etiologic agent R. prowazekii DNA, the YP11D/YP10R primer pair yielded a 280-bp product whose sequence disclosed no homology with sequences deposited in GenBank (data not shown). Amplified fragments were sequenced by using cycle sequencing and dye terminator methodologies using the Ampli-Taq cycle sequencing kit (Perkin–Elmer) and an automatic ABI Prism 377 DNA Sequencer (Applied Biosystems). The sequence was aligned with that of modern pla by using clustal (25). PCR detection of R. prowazekii was attempted with primer pair Rp1 (5′-CTCCTCTTACACTTGGTG-3′) and Rp2 (5′-ACCTGCTTGTAAATTTAAAG-3′) corresponding to nucleotides 528–718 of the R. prowazekii ompB gene (GenBank accession no. AF123718). PCR conditions were as above except that the annealing temperature was 53°C. PCR detection of B. anthracis was attempted as previously described (26).
Results
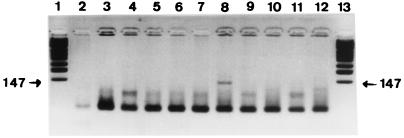
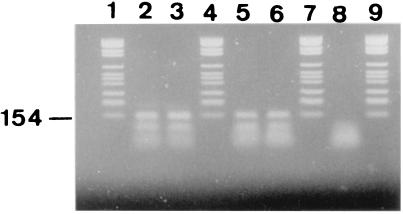
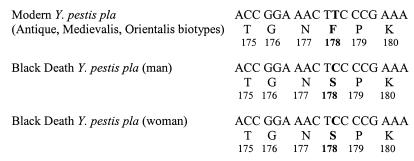
All ancient teeth were carefully fractured and exhibited a large dental pulp chamber, as expected from their radiographical examination. Ancient negative and test teeth yielded powdery remnants of a white or brown color (no correlation was found between the remnants' color and the success rate of DNA amplifications). A single round of 40 cycles of amplification targeting an IS200-like element within env failed to yield an amplicon from any DNA extracts derived from ancient dental pulp and the sterile control water. However, amplification product of the expected 147-bp size corresponding to positions 854–1,001 of the pla sequence (27) was obtained from 1/4 child teeth (Fig. 2) and amplification product of the expected 148-bp size corresponding to positions 728–876 of the pla sequence (27) was obtained from 19/19 adult teeth, but not in the negative control teeth or in PCR negative controls (Fig. 3). The sequence derived from the PCR product obtained from the child teeth shared 100% identity with that of the modern Y. pestis pla sequence in GenBank, whereas a Phe178 → Ser178 mutation was noted at the end of the hydrophobic domain for the PCR products recovered from adult teeth (Fig. 4). These sequences are specific for Y. pestis; particularly, it is not present in Y. pseudotuberculosis, which has been demonstrated to be the same genetic species as Y. pestis. No amplification was obtained from any specimen by using the primer pairs designed to detect R. prowazekii and B. anthracis.
Figure 2.
Agarose gel stained with ethidium bromide showing the 147-bp amplified Y. pestis pla fragment obtained from ancient DNA after suicide PCR. Products resulting from DNA extracted from the dental pulp tissues collected from the ancient child skeletons (lanes 3, 5, 8, and 10), negative control dental pulp tissues (lanes 4, 9, 11, and 12), and mock extraction controls (lanes 2, 6, and 7) are shown. Lanes 1 and 13 are molecular weight marker fragments. The measured molecular weight of the amplicon is indicated in the margin.
Figure 3.
Agarose gel stained with ethidium bromide showing the 148-bp amplified Y. pestis pla fragment obtained from ancient DNA after suicide PCR. Products resulting from DNA extracted from the dental pulp tissues collected from the ancient adult male skeleton (lanes 2 and 3), the ancient adult female skeleton (lanes 5 and 6) and negative control dental pulp tissues (lane 8) are shown. Lanes 1, 4, 7, and 9 are molecular weight marker fragments. The molecular weight of the marker is indicated in the margin.
Figure 4.
Partial sequence of the Y. pestis pla gene determined in two adult Medieval Black Death victims. The mutation identified in codon 178 and the resulting amino acid substitution are in bold.
Discussion
Field and laboratory contaminations of ancient human remnants limit the interpretation of molecular biological diagnosis of ancient infectious diseases. Field contamination of specimens can result from environmental flora contamination and hand-transmitted contamination from one ancient specimen to another. Ancient specimens currently used in paleomicrobiology, particularly bone specimens, are at risk of environmental contamination. We previously have proposed dental pulp as a convenient material on which to base PCR detection of ancient septicemic pathogens for reasons including its durability, natural sterility, and involvement in septicemic diseases (14). This material enabled us to perform the first detection of Y. pestis DNA in 16th–18th century plague skeletons. Interestingly, plague teeth were used as amulets during plague epidemics, being swallowed in an attempt to prevent the Black Death (28).
Laboratory contamination of specimens may result from previously extracted target DNA or PCR-amplified DNA. The absence of a previous or concurrent extraction of the target DNA in the laboratory is the only certain way to prevent DNA contamination. This therefore implies that positive controls should never be used during the molecular detection of an ancient pathogen. The omission of any positive control during the processing of ancient specimens is the first requirement in achieving a zero contamination goal. The suicide PCR amplification procedure we developed further ensures the absence of amplicon contamination in the laboratory of the ancient material, as PCR primers are designed to hybridize to targets outside genomic regions previously targeted in the laboratory. The primers having been used only once, no previous contamination could occur. Furthermore, as amplification products can only have been derived from the sample DNA, a second PCR can, if necessary, be used to increase amplicon yields to a level adequate for direct sequence determination. When the first trial is negative, another primer pair should be designed because a primer pair should be used only once even if negative. The suicide PCR amplification procedure therefore ensures a 100% positive predictive value for any sequence derived from suicide PCR products. The sensitivity in this experience is probably low, but our goal was the specificity. The question of how many different experiments have to be performed before an ancient specimen is considered to be negative needs further evaluation. The availability of complete microbial genomes, including the ongoing Y. pestis sequencing project, should permit an almost limitless source of new hybridization sites for suicide PCR amplification primers. The suicide PCR approach does not permit the accumulation of comparable sequence data in the same laboratory. Such collection could, however, be obtained through a network of laboratories. However, such a technique is very helpful when a basic question could be resolved only by PCR and when specimens are rare.
In this study, suicide PCR was applied to the detection of Y. pestis DNA in dental pulp collected from three Black Death skeletons. The Black Death killed up to 90% of the Medieval population of the Montpellier area within 30 years (19). The human remnants we obtained in a catastrophe grave were dated to be from the 14th century. Therefore, any remnant of this period has a 90% probability of being related to Black Death, and the fact that the three skeletons we studied were lying in a common catastrophe grave enhances this probability. Suicide PCR allowed the amplification of a pla sequence previously shown to be specific to Y. pestis among several Yersinia spp. and to be homologous in several Y. pestis isolates from different sources and different countries (22, 23). Further experiments including ancient materials may confirm whether the point mutation we noted indicates a real heterogeneity within Y. pestis. If confirmed, this result could be highly significant because no sequence divergence was found at any of six gene loci analyzed in 36 Y. pestis strains representing the global diversity of this species (29). The fact that Y. pestis pla sequence was independently amplified in three different individuals reinforced the conclusions of our work. Therefore, suicide PCR results gave certainty that Y. pestis was present in the dental pulp of these persons and was the likely cause of death. The fact that two alternative etiologic agents, R. prowazekii and B. anthracis were not detected reinforced this conclusion. This result indicates that the plague had authentically been recognized as a unique morbid entity as early as the Middle Ages, and suggests that medieval descriptions of Black Death can be regarded as true descriptions of plague epidemics. The French medieval physician Gui de Chaulliac was appointed dean of the oldest French medical university in Montpellier by Pope Clément VI in 1347, when the Black Death was sweeping over Southern France. He himself suffered and survived the plague. His original description of Black Death epidemics was lost, but printed copies were preserved and are still available for historical research (30) This description indicates that Black Death swept from Marseille to Avignon (100 km) within 1 month and that it presented in both bubonic and pulmonary forms.
The application of a suicide PCR method may also resolve the controversy regarding the etiology of the Great Plague in Athens during the Peloponnesian war of 430–425 B.C. As described by Thucydides, it has been proposed as the Ebola virus (31), the agent of the epidemic typhus R. prowazekii (7), B. antracis (32), or Y. pestis (3). It may also confirm the Y. pestis etiology of the Justinian plague (3) and could further be applied to the identification of Y. pestis biotype-specific sequences to test the theory of Y. pestis biotypes being responsible for the three pandemics of plague identified from historical records over the past 2 millenia (3). The suicide PCR amplification procedure and the imminent availability of the annotated Y. pestis genome sequencing will offer the opportunity to test this hypothesis. We propose that suicide PCR should become a standard in PCR technology to completely avoid contamination of material with previously amplified sequences.
Acknowledgments
We thank J.-C. Hélas and C. Arlaud for their contribution in anthropological investigation of the Saint-Côme and Saint-Damien site.
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. M27820, AF123718, and AF292645).
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.220225197.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.220225197
References
- 1.Ziegler P. The Black Death. Wolfeboro Falls, NH: Alan Sutton Publishing Inc.; 1991. [Google Scholar]
- 2.Derbes V J. J Am Med Assoc. 1966;19:179–182. [Google Scholar]
- 3.Perry R D, Fetherston J D. Clin Microbiol Rev. 1997;10:35–66. doi: 10.1128/cmr.10.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yersin A. Ann Inst Pasteur. 1894;8:428–430. [Google Scholar]
- 5.Slack P. Trans R Soc Trop Med Hyg. 1989;83:461–463. doi: 10.1016/0035-9203(89)90247-2. [DOI] [PubMed] [Google Scholar]
- 6.Twigg G. The Black Death. A Biological Reappraisal. London: Batsford; 1984. [Google Scholar]
- 7.Weiss E. In: Encyclopedia of Microbiology. Lederberg J, editor. Vol. 3. San Diego: Academic; 1992. pp. 585–610. [Google Scholar]
- 8.Scott S, Duncan C J, Duncan S R. Ann Hum Biol. 1996;23:1–21. doi: 10.1080/03014469600004232. [DOI] [PubMed] [Google Scholar]
- 9.Ramalingaswani V. Nat Med. 1995;1:1237–1239. doi: 10.1038/nm1295-1237. [DOI] [PubMed] [Google Scholar]
- 10.Galimand M, Guiyoule A, Gerbaud G, Rasoamanana B, Chanteau S, Carniel E, Courvalin P. N Engl J Med. 1997;337:677–680. doi: 10.1056/NEJM199709043371004. [DOI] [PubMed] [Google Scholar]
- 11.Taubenberger J K, Reid A H, Krafft A E, Bijwaard K E, Fanning T G. Science. 1997;275:1793–1796. doi: 10.1126/science.275.5307.1793. [DOI] [PubMed] [Google Scholar]
- 12.Taylor G M, Goyal M, Legge A J, Shaw R J, Young D. Microbiology. 1999;145:899–904. doi: 10.1099/13500872-145-4-899. [DOI] [PubMed] [Google Scholar]
- 13.Rafi A, Spigelman M, Standford J, Lemma E, Donoghue H, Zias J. Lancet. 1994;343:1360–1361. [PubMed] [Google Scholar]
- 14.Drancourt M, Aboudharam G, Signoli M, Dutour O, Raoult D. Proc Natl Acad Sci USA. 1998;95:12637–12640. doi: 10.1073/pnas.95.21.12637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kolman C J, Centurion-Lara A, Lukehart S A, Owsley D W, Tuross N. J Infect Dis. 2000;180:2060–2063. doi: 10.1086/315151. [DOI] [PubMed] [Google Scholar]
- 16.Jackson P J, Hugh-Jones M E, Adair D M, Green G, Hill K K, Kuske C R, Grinberg L M, Abramova F A, Keim P. Proc Natl Acad Sci USA. 1998;95:1224–1229. doi: 10.1073/pnas.95.3.1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kwok S, Higuchi R. Nature (London) 1989;339:237–238. doi: 10.1038/339237a0. [DOI] [PubMed] [Google Scholar]
- 18.Ward R, Stringer C. Nature (London) 1997;388:226. doi: 10.1038/40746. [DOI] [PubMed] [Google Scholar]
- 19.Combes J. In: Histoire de Montpellier. Cholvy G, editor. Toulouse, France: Privat; 1984. p. 82. [Google Scholar]
- 20.Pääbo S. Proc Natl Acad Sci USA. 1989;86:1939–1943. doi: 10.1073/pnas.86.6.1939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Simonet M, Riot B, Fortineau N, Berche P. Infect Immun. 1996;64:375–379. doi: 10.1128/iai.64.1.375-379.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hinnebusch J, Schwan T G. J Clin Microbiol. 1993;31:1511–1514. doi: 10.1128/jcm.31.6.1511-1514.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Campbell J, Lowe J, Walz S, Ezzell J. J Clin Microbiol. 1993;31:758–759. doi: 10.1128/jcm.31.3.758-759.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bercovier H, Mollaret H H, Alonso J M, Brault J, Fanning G R, Steigerwalt A G, Brenner D J. Curr Microbiol. 1980;4:225–229. [Google Scholar]
- 25.Higgins D G, Sharp P M. Gene. 1989;73:237–244. doi: 10.1016/0378-1119(88)90330-7. [DOI] [PubMed] [Google Scholar]
- 26.Patra G, Sylvestre P, Ramisse V, Thérasse J, Guesdon J-L. FEMS Immunol Med Microbiol. 1996;15:223–231. doi: 10.1111/j.1574-695X.1996.tb00088.x. [DOI] [PubMed] [Google Scholar]
- 27.Sodeinde O A, Goguen J D. Infect Immun. 1989;57:1517–1523. doi: 10.1128/iai.57.5.1517-1523.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mollaret H H, Brossolet J. Jaarbock 1965. Antwerp, Belgium: Koninklijk Museum voor Schone Kunsten; 1965. pp. 3–112. [Google Scholar]
- 29.Achtman M, Zurth K, Morelli G, Torrea G, Guiyoule A, Carniel E. Proc Natl Acad Sci USA. 1999;96:14043–14048. doi: 10.1073/pnas.96.24.14043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Enselme J. Rev Lyon Med. 1969;17:697–710. [PubMed] [Google Scholar]
- 31.Olson P E, Hames C S, Benenson A S, Genovese E N. Emerg Infect Dis. 1996;2:155–156. doi: 10.3201/eid0202.960220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McSherry J, Kilpatrick R. J R Soc Med. 1992;85:713. [PMC free article] [PubMed] [Google Scholar]