Abstract
Aims
In theory, the magnitude of an in vivo drug–drug interaction arising from the inhibition of metabolic clearance can be predicted using the ratio of inhibitor concentration ([I]) to inhibition constant (Ki). The aim of this study was to construct a database for the prediction of drug–drug interactions from in vitro data and to evaluate the use of the various estimates for the inhibitor concentrations in the term [I]/Ki.
Methods
One hundred and ninety-three in vivo drug–drug interaction studies involving inhibition of CYP3A4, CYP2D6 or CYP2C9 were collated from the literature together with in vitro Ki values and pharmacokinetic parameters for inhibitors, to allow calculation of average/maximum systemic plasma concentration during the dosing interval and maximum hepatic input plasma concentration (both total and unbound concentration). The observed increase in AUC (decreased clearance) was plotted against the estimated [I]/Ki ratio for qualitative zoning of the predictions.
Results
The incidence of false negative predictions (AUC ratio > 2, [I]/Ki < 1) was largest using the average unbound plasma concentration and smallest using the hepatic input total plasma concentration of inhibitor for each of the CYP enzymes. Excluding mechanism-based inhibition, the use of total hepatic input concentration resulted in essentially no false negative predictions, though several false positive predictions (AUC ratio < 2, [I]/Ki > 1) were found. The incidence of true positive predictions (AUC ratio > 2, [I]/Ki > 1) was also highest using the total hepatic input concentration.
Conclusions
The use of the total hepatic input concentration of inhibitor together with in vitro Ki values was the most successful method for the categorization of putative CYP inhibitors and for identifying negative drug–drug interactions. However this approach should be considered as an initial discriminating screen, as it is empirical and requires subsequent mechanistic studies to provide a comprehensive evaluation of a positive result.
Keywords: CYP2C9, CYP2D6, CYP3A4, drug interactions, prediction
Introduction
Inhibition of CYP-mediated drug metabolism by a concomitantly administered second drug is one of the major causes of drug–drug interactions in humans and can lead to serious toxicities. The use of in vitro data to predict the inhibition potential of a drug is attractive because of the rapid and simple experimental procedures involved. Although there have been substantial technological advances in the conduct of in vitro studies, the interpretation of the parameters generated remains problematic due to lack of a quantitative framework for the relationship between in vitro and in vivo data on drug–drug interactions [1].
Biochemical principles [2] state that when the metabolism of a drug (substrate) is reversibly inhibited by a second drug (inhibitor), the metabolic intrinsic clearance (CLint) of substrate is decreased by a factor related to the inhibitor concentration available to the enzyme [I] and the inhibition constant, Ki (equation 1). The distinction between competitive and noncompetitive inhibition me-chanisms is not relevant when the substrate concentration is much lower than the Km value, the commonly encountered in vivo situation that results in linear kinetics.
where subscript I represents the value in the presence of inhibitor.
This theory, and the suitability of equation 1 to describe in vivo data, has been confirmed in several animal studies under well defined, steady state conditions for various levels of inhibition achieved by intravenous infusions, for example, in the decrease in clearance of diazepam caused by omeprazole [3], of theophylline by enoxacin and ciprofloxacin [4] and of antipyrine by ketoconazole and fluconazole [5].
In human in vivo interaction studies, drug plasma concentration profiles are determined in the presence and absence of inhibitor (after multiple oral dosing) and the degree of interaction is expressed as the increase in the area under the plasma concentration-time curve (AUC) of substrate. If the substrate is eliminated by a single metabolic pathway that is subject to inhibition, the AUC ratio of orally administered substrate in the presence and absence of inhibitor reflects the ratio of clearances, provided that the conditions of the ‘well-stirred’ liver model are assumed and that the inhibitor does not affect either the intestinal absorption or plasma protein binding of the substrate:
where Fa is the fraction absorbed from gut into the portal vein, D is the dose, and fuB is the unbound fraction in blood. Therefore the ratio of AUCs is dependent on the [I]/Ki ratio (equation 3) based on the assumptions mentioned above.
In recent years there has been much interest in the use of equation 3 to describe the degree of in vivo interaction between two drugs [1, 6–12]. Ki values can be readily obtained from in vitro studies using human liver microsomes. However, it is not normally possible to measure the inhibitor concentration available to the hepatic enzyme in vivo in humans. Predictions have been attempted using various [I] values in equation 3, including the plasma total or unbound concentration or hepatic input concentration of the inhibitor [6, 8, 10, 12, 13]. However, most of these studies have dealt with particular combinations of drugs with only one dosage regimen for inhibitor and a general agreement has not been reached as to which concentration should be used for [I] in equation 3 [1, 14].
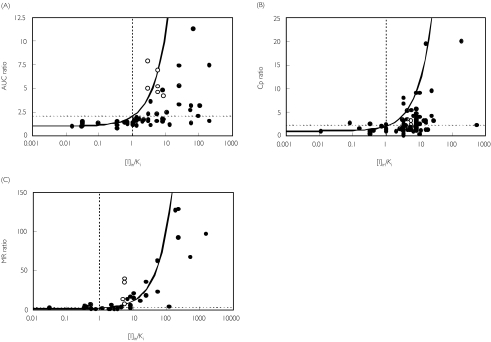
According to equation 3, interactions are regarded to be with low risk if the estimated [I]/Ki ratio is less than 0.1, and high risk if it is greater than 1. Based on a plot of AUC ratio against [I]/Ki(Figure 1), predictions can be categorized into four zones: true positives (AUC ratio > 2, [I]/Ki > 1), true negatives (AUC ratio < 2, [I]/Ki < 1), false positives (AUC ratio < 2, [I]/Ki > 1), or false negatives (AUC ratio > 2, [I]/Ki < 1). The threshold of two-fold increase in the AUC was selected based on a previous consensus report [1].
Figure 1.
Qualitative zoning for the prediction of drug–drug interactions involving CYP inhibition. The curve represents the theoretical curve based on equation 1. F-: false negative, T-: true negative, F+: false positive, T+: true positive
The aims of this study were 1) to extend previous analyses based on relatively small number of studies and to construct a database for the prediction of drug–drug interactions involving CYP inhibition from in vitro data, and 2) to evaluate the utility of the simple [I]/Ki ratio by using various inhibitor concentrations in vivo to designate qualitatively CYP inhibition interaction predictions into zones.
Methods
Data collection
Three hundred and twenty-one in vivo drug–drug interaction studies involving inhibition of the CYP enzymes 3A4/5, 2D6 and 2C9 were obtained from the literature and collated as shown in Table 1. The degree of interaction in each study was expressed as a fold increase in the AUC of the substrate. For the interactions involving CYP2D6, the ratio of the plasma concentration at a single time point and the metabolic ratio (urinary excretion ratio of parent/metabolite(s)) were also used as in vivo metrics. As summarized in Table 1, relatively few studies reported the inhibitor concentration (either as an average, maximum, minimum, or particular time point concentration) in the same subjects.
Table 1.
Numbers of studies in the drug-drug interaction database
| Category | Number of in vivro studies | Number of in vitro studies | ||||||
|---|---|---|---|---|---|---|---|---|
| CYP | Metric | S class | Total | With Cav | With Cmax | With other C | With same S | With unequivocal substitute S |
| 2D6 | AUC | 58 | 6 | 6 | 13 | 25 | 33 | |
| Cp | 89 | 0 | 0 | 21 | 40 | 49 | ||
| MR | 39 | 0 | 1 | 24 | 28 | 11 | ||
| 3A4 | AUC | BZ | 56 | 6 | 14 | 11 | 40 | 12 |
| TS | 31 | 13 | 1 | 3 | 19 | 11 | ||
| NIF | 14 | 1 | 3 | 0 | 0 | 0 | ||
| Other | 8 | 1 | 2 | 1 | 0 | 0 | ||
| 2C9 | AUC | 26 | 3 | 3 | 3 | 22 | 4 | |
BZ, benzodiazepines; TS, testosterone; NIF, nifedipine.
In vitro Ki values for the CYP inhibitors involved in the above studies were also collected from the literature. Often in vitro data were available for the same substrate as used in the in vivo interaction studies (Table 1), and when several human liver microsomal studies had been conducted, average values were used. For CYP2C9 and 2D6, in vitro data from alternative, well accepted substrates were used in the absence of data from the first choice substrate. For interactions involving CYP3A4, Ki values for the same substrate as in the in vivo study were available in about half of the studies (Table 1) and for others, probe(s) were selected that belong to the same substrate subgroup class (S) as that in the in vivo study [15] as indicated in Table 1.
Information on inhibitor pharmacokinetics in humans (oral clearance (CL/F), half-life (t1/2), and plasma unbound fraction (fu)) was obtained from the literature to calculate various concentrations as listed below.
Analyses
Various inhibitor concentrations were calculated for use in the analyses. For consistency these were estimated from literature pharmacokinetic parameters. Average systemic plasma concentration after repeated oral administration ([I]av), maximum systemic plasma concentration after repeated oral administration ([I]max), and maximum hepatic input concentration ([I]in) [13] were calculated as follows:
where D and τ are the dose and the dosing interval, respectively, of inhibitor used in the in vivo interaction study, k is the elimination rate constant, ka is the absorption rate constant, Fa is the fraction absorbed from gut into the portal vein, and Qh is the hepatic blood flow rate. The values of ka, Fa, Qh, and RB (blood-to-plasma concentration ratio) were assumed to be 0.1 min−1, 1, 1610 ml min−1, and 1, respectively. The corresponding unbound plasma concentrations after repeated oral administration (e.g. [I]av,u) was calculated by multiplying the value of [I] by fu.
The [I]/Ki ratio was calculated for each of the in vivo interaction studies using the various [I] values described above. For inhibitors with a metabolite that also inhibits the same CYP enzyme, the [I]/Ki ratio was calculated for both the parent drug and the metabolite and the values were added. The predictive performance using various [I] values was compared using the chi-squared test.
Simulations
In a typical interaction study, the input term greatly exceeds the systemic term in equation 6. In order to assess the importance of the assigned values for ka, Fa (these two parameters appear as a product in equation 6), and Qh, the relationship between the AUC ratio and 1/Ki was simulated based on the equations 3 and 6, using various values for ka × Fa (0.001, 0.01 and 0.1 min−1) and Qh (805, 1610 and 3220 ml min−1). For other parameters, values reported for quinidine were used (D = 268 nmol, τ = 24 h, and CL/F = 412 ml min−1).
Results and discussion
Of the 321 in vivo interaction studies collated from the literature, 193 provided information on the increase in substrate AUC after repeated oral administration of inhibitor (n = 58 for CYP2D6, n = 109 for CYP3A4, and n = 26 for CYP2C9). Of these 94 and 99 studies reported AUC ratios greater and lesser than 2, respectively. Data are listed in Tables 23–4 as AUC ratios together with in vitro Ki values and four [I]/Ki parameters, based on average systemic total drug plasma concentration ([I]av), average systemic unbound drug plasma concentration ([I]av,u), maximum systemic plasma concentration ([I]max), and maximum hepatic input concentration ([I]in). The latter represents the theoretical maximum drug concentration entering the liver, which is the sum of the hepatic artery and portal vein concentrations during the absorption process.
Table 2.
In vivo database for the interactions involving CYP2D6
| Substrate | Inhibitor | AUC ratio | Ki* (µm) | [I]av/ Ki | [I]av,u/ Ki | [I]max/ Ki | [I]in/ Ki | Reference |
|---|---|---|---|---|---|---|---|---|
| Flecainide | Amiodarone | 1.37 | 43 (20) | 0.11 | 0.0023 | 0.11 | 0.57 | 2.01 |
| Atenolol | Amitriptyline | 1.24 | 30 (12) | 0.13 | 0.0095 | 0.19 | 0.7 | 2.02 |
| Metoprolol | Amitriptyline | 1.44 | 30 (12) | 0.13 | 0.0095 | 0.19 | 0.7 | 2.02 |
| Propranolol | Chlorpromazine | 1.69 | 4.8 | 0.11 | 0.004 | 0.13 | 2.2 | 2.03 |
| Atenolol | Cimetidine | 1.07 | 40 | 0.1 | 0.081 | 0.24 | 1.3 | 2.04 |
| Metoprolol | Cimetidine | 1.61 | 40 | 0.1 | 0.081 | 0.24 | 1.3 | 2.04 |
| Propranolol | Cimetidine | 1.91 | 40 | 0.1 | 0.081 | 0.24 | 1.3 | 2.04 |
| Propranolol | Cimetidine | 1.94 | 40 | 0.1 | 0.081 | 0.24 | 1.3 | 2.05 |
| Propranolol | Cimetidine | 1.47 | 40 | 0.12 | 0.097 | 0.28 | 2 | 2.06 |
| Imipramine | Citalopram | 1.15 | 24 (14) | 0.015 | 0.0045 | 0.019 | 0.34 | 2.07 |
| (Desipramine) | Citalopram | 1.47 | 24 (14) | 0.015 | 0.0045 | 0.019 | 0.34 | 2.07 |
| Levomepromazine | Citalopram | 0.74 | 24 (14) | 0.015 | 0.0045 | 0.019 | 0.34 | 2.07 |
| Atenolol | Diltiazem | 1.07 | 150 | 0.0005 | 0.0001 | 0.001 | 0.03 | 2.08 |
| Metoprolol | Diltiazem | 1.33 | 150 | 0.0005 | 0.0001 | 0.001 | 0.03 | 2.08 |
| Propranolol | Diltiazem | 1.48 | 150 | 0.0005 | 0.0001 | 0.001 | 0.03 | 2.08 |
| Propranolol | Diltiazem | 1.33 | 150 | 0.0016 | 0.0003 | 0.003 | 0.091 | 2.09 |
| Propranolol | Verapamil | 1.42 | 16 | 0.0072 | 0.0007 | 0.013 | 1 | 2.09 |
| Metoprolol | Diphenhydramine | 1.61 | 10 | 0.056 | 0.012 | 0.076 | 1.2 | 2.10 |
| R(+)-carvedilol | Fluoxetine | 1.77 | 0.54 (0.58) | 0.7 | 0.042 | 0.81 | 8.1 | 2.11 |
| Desipramine | Fluoxetine | 4.80 | 0.54 (0.58) | 0.7 | 0.042 | 0.81 | 8.1 | 2.12 |
| Desipramine | Sertraline | 1.20 | 10 (24) | 0.0071 | 0.0001 | 0.01 | 1 | 2.12 |
| Desipramine | Fluoxetine | 7.43 | 0.54 (0.58) | 2.1 | 0.13 | 2.4 | 24 | 2.13 |
| Imipramine | Fluoxetine | 3.33 | 0.54 (0.58) | 2.1 | 0.13 | 2.4 | 24 | 2.13 |
| (Desipramine) | Fluoxetine | 5.31 | 0.54 (0.58) | 2.1 | 0.13 | 2.4 | 24 | 2.13 |
| Propafenone | Fluoxetine | 1.50 | 0.54 (0.58) | 0.7 | 0.042 | 0.81 | 8.1 | 2.14 |
| Ritonavir | Fluoxetine | 1.19 | 0.54 (0.58) | 2.1 | 0.13 | 2.3 | 13 | 2.15 |
| Tolterodine | Fluoxetine | 4.84 | 0.54 (0.58) | 0.7 | 0.042 | 0.81 | 8.1 | 2.16 |
| Desipramine | Fluvoxamine | 1.14 | 5.8 | 0.055 | 0.013 | 0.09 | 3.4 | 2.17 |
| Imipramine | Fluvoxamine | 3.63 | 5.8 | 0.055 | 0.013 | 0.09 | 3.4 | 2.17 |
| Metoprolol | Hydroxychloroquine | 1.65 | 66 | 0.025 | 0.015 | 0.025 | 1.1 | 2.18 |
| Imipramine | Labetalol | 1.53 | 7 | 0.012 | 0.0062 | 0.026 | 5.4 | 2.19 |
| (Desipramine) | Labetalol | 2.27 | 7 | 0.012 | 0.0062 | 0.026 | 5.4 | 2.19 |
| Metoprolol | Mexiletine HCl | 1.82 | 30 | 0.095 | 0.035 | 0.13 | 1.5 | 2.20 |
| Metoprolol (R) | Omeprazole | 1.01 | 240 | 0.0003 | 0.0001 | 0.008 | 0.03 | 2.21 |
| Metoprolol (S) | Omeprazole | 1.02 | 240 | 0.0003 | 0.0001 | 0.03 | 2.21 | |
| Propranolol | Omeprazole | 1.02 | 240 | 0.0002 | 0.0001 | 0.004 | 0.015 | 2.22 |
| Desipramine | Paroxetine | 4.64 | 0.69 | 0.1 | 0.0051 | 0.16 | 5.6 | 2.23 |
| Imipramine | Paroxetine | 1.74 | 0.69 | 0.15 | 0.0076 | 0.24 | 8.4 | 2.24 |
| (Desipramine) | Paroxetine | 4.28 | 0.69 | 0.15 | 0.0076 | 0.24 | 8.4 | 2.24 |
| Metoprolol (R) | Paroxetine | 7.93 | 0.69 | 0.1 | 0.0051 | 0.13 | 2.8 | 2.25 |
| Metoprolol (S) | Paroxetine | 5.08 | 0.69 | 0.1 | 0.0051 | 2.8 | 2.25 | |
| Perphenazine | Paroxetine | 6.96 | 0.69 | 0.1 | 0.0051 | 0.16 | 5.6 | 2.26 |
| Desipramine | Paroxetine | 5.21 | 0.69 | 0.1 | 0.0051 | 0.16 | 5.6 | 2.27 |
| Desipramine | Sertraline | 1.37 | 10 (24) | 0.0065 | 0.0001 | 0.0091 | 1 | 2.27 |
| Propranolol | Propafenone | 2.13 | 0.55 | 19 | 0.95 | 30 | 93 | 2.28 |
| Desipramine | Quinidine sulphate | 7.50 | 0.082 | 5.5 | 0.72 | 16 | 210 | 2.29 |
| Imipramine | Quinidine sulphate | 1.54 | 0.082 | 5.5 | 0.72 | 16 | 210 | 2.29 |
| Encainide | Quinidine sulphate | 3.18 | 0.082 | 5.5 | 0.72 | 7.6 | 56 | 2.30 |
| Encainide | Quinidine sulphate | 11.40 | 0.082 | 5 | 0.64 | 7.5 | 66 | 2.31 |
| Metoprolol | Quinidine sulphate | 3.24 | 0.082 | 2.8 | 0.36 | 7.9 | 100 | 2.32 |
| Mexiletine | Quinidine sulphate | 1.32 | 0.082 | 5.5 | 0.72 | 7.6 | 56 | 2.33 |
| Propafenone | Quinidine sulphate | 2.70 | 0.082 | 4.1 | 0.54 | 6.2 | 55 | 2.34 |
| Desipramine | Ritonavir | 2.45 | 4.8 | 1.8 | 0.036 | 3.4 | 11 | 2.35 |
| Desipramine | Sertraline | 1.74 | 10 (24) | 0.02 | 0.0002 | 0.027 | 3 | 2.36 |
| Desipramine | Sertraline | 1.54 | 10 (24) | 0.02 | 0.0002 | 0.027 | 3 | 2.37 |
| Imipramine | Sertraline | 1.68 | 10 (24) | 0.02 | 0.0002 | 0.027 | 3 | 2.37 |
| (Desipramine) | Sertraline | 2.29 | 10 (24) | 0.02 | 0.0002 | 0.027 | 3 | 2.37 |
| Metoprolol | Verapamil | 1.33 | 16 | 0.0072 | 0.0007 | 0.013 | 1 | 2.38 |
Numbers in parentheses represent the values for metabolites.
References for Table 2
2.01Funck-Brentano et al. Clin Pharmacol Ther 1994; 55:256–69. 2.02Kirch et al. Br J Clin Pharmacol 1984; 17: 65S–8S. 2.03Vestal et al. Clin Pharmacol Ther 1979; 25: 19–24. 2.04Kirch et al. Klin Wochenschr 1982; 60: 1401–7. 2.05Reimann et al. Br J Clin Pharmacol 1981; 12: 785–90. 2.06Donn et al. J Clin Pharmacol 1984; 24: 500–8. 2.07Gram et al. Ther Drug Monit 1993; 15:18–24. 2.08Tateishi et al. Eur J Clin Pharmacol 1989; 36: 67–70. 2.09Hunt et al. Clin Pharmacol Ther 1990; 47: 584–91. 2.10Hamelin et al. Clin Pharmacol. Ther 2000; 67: 466–77. 2.11Graff et al. J Clin Pharmacol 2001; 41: 97–106. 2.12Preskorn et al. J Clin Psychopharmacol 1994; 14: 90–8. 2.13Bergstrom et al. Clin Pharmacol Ther 1992; 51: 239–48. 2.14Cai et al. Clin Pharmacol Ther 1999; 66: 516–21. 2.15Ouellet et al. Antimicrob Agents Chemother 1998; 42:3107–12.2.16Brynne et al. Br J Clin Pharmacol 1999; 48: 553–563. 2.17Spina et al. Ther Drug Monit 1993; 15: 243–6. 2.18Somer et al. Br J Clin Pharmacol 2000; 49: 549–54. 2.19Hermann et al. J Clin Pharmacol 1992; 32: 176–83. 2.20Sakamoto et al. Jpn J Clin Pharmacol Ther 1995; 26: 159–60. 2.21Andersson et al. Eur. J Clin Pharmacol 1991; 40: 61–5. 2.22Henry et al. Eur J Clin Pharmacol 1987; 33: 369–73. 2.23Brosen et al. Eur J Clin Pharmacol 1993; 44: 349–55. 2.24Albers et al. Psychiatry Res 1996; 59: 189–96. 2.25Hemeryck et al. Clin Pharmacol Ther 2000; 67:283–91. 2.26Ozdemir et al. Clin Pharmacol Ther 1997; 62:334–47. 2.27Alderman et al. J. Clin Psychopharmacol 1997; 17:284–91. 2.28Kowey et al. J Clin Pharmacol 1989; 29: 512–7. 2.29Brosen, Gram. Eur J. Clin Pharmacol 1989; 37: 155–60. 2.30Funck-Brentano et al. J Pharmacol Exp Ther 1989; 249: 134–42. 2.31Turgeon et al. J Pharmacol Exp Ther 1990; 255:642–9. 2.32Johnson, Burlew. Drug Metab Dispos 1996; 24:350–5. 2.33Turgeon et al. J Pharmacol Exp Ther 1991; 259:789–98. 2.34Funck-Brentano et al. Br J Clin Pharmacol 1989; 27: 435–44. 2.35von Moltke et al. J Pharm Sci 1998; 87: 1184–9. 2.36Zussman et al. Br J Clin Pharmacol 1995; 39: 550–1. 2.37Kurtz et al. Clin Pharmacol Ther 1997; 62: 145–56. 2.38Keech et al. Am J Cardiol 1986; 58: 551–2.
Table 3.
In vivo database for the interactions involving CYP3A4
| Substrate | Inhibitor | AUC ratio | Ki* (µm) | [I]av/Ki | [I]av,u/Ki | [I]max/Ki | [I]in/Ki | Reference |
|---|---|---|---|---|---|---|---|---|
| Midazolam | Azithromycin | 0.87 | 30 | 0.0046 | 0.0033 | 0.0056 | 0.71 | 3.01 |
| Midazolam | Azithromycin | 1.19 | 30 | 0.0092 | 0.0066 | 0.011 | 1.4 | 3.02 |
| Midazolam | Clarithromycin | 3.57 | 57 | 0.0087 | 0.0047 | 0.024 | 0.37 | 3.02 |
| Triazolam | Azithromycin | 1.03 | 30 | 0.0092 | 0.0066 | 0.01 | 0.71 | 3.03 |
| Triazolam | Clarithromycin | 5.06 | 57 | 0.017 | 0.0094 | 0.048 | 0.74 | 3.03 |
| Triazolam | Erythromycin | 3.65 | 77 | 0.0068 | 0.0011 | 0.035 | 0.56 | 3.03 |
| Midazolam | Azithromycin | 1.27 | 30 | 0.0092 | 0.0066 | 0.011 | 1.4 | 3.04 |
| Midazolam | Erythromycin | 3.81 | 170 | 0.0046 | 0.0004 | 0.016 | 0.25 | 3.04 |
| Alprazolam | Cimetidine | 1.58 | 140 | 0.034 | 0.028 | 0.081 | 0.56 | 3.05 |
| Triazolam | Cimetidine | 1.54 | 99 | 0.048 | 0.039 | 0.11 | 0.8 | 3.05 |
| Alprazolam | Cimetidine | 1.73 | 140 | 0.028 | 0.023 | 0.067 | 0.38 | 3.06 |
| Triazolam | Cimetidine | 2.20 | 99 | 0.04 | 0.033 | 0.096 | 0.54 | 3.06 |
| Midazolam | Cimetidine | 2.02 | 180 | 0.022 | 0.0089 | 0.052 | 0.29 | 3.07 |
| Midazolam | Ranitidine | 1.66 | 170 | 0.0027 | 0.0012 | 0.011 | 0.17 | 3.07 |
| Midazolam | Cimetidine | 1.35 | 180 | 0.018 | 0.0071 | 0.074 | 0.56 | 3.08 |
| Midazolam | Ranitidine | 1.23 | 170 | 0.0027 | 0.0012 | 0.011 | 0.17 | 3.08 |
| Nifedipine | Cimetidine | 1.60 | 140 | 0.028 | 0.023 | 0.24 | 1.8 | 3.09 |
| Nifedipine | Ranitidine | 1.22 | 170 | 0.0027 | 0.0023 | 0.011 | 0.17 | 3.09 |
| Nifedipine | Cimetidine | 2.01 | 140 | 0.023 | 0.018 | 0.19 | 1.4 | 3.10 |
| Nifedipine | Ranitidine | 1.06 | 170 | 0.0055 | 0.0047 | 0.022 | 0.35 | 3.10 |
| Nimodipine | Cimetidine | 1.74 | 140 | 0.028 | 0.023 | 0.067 | 0.38 | 3.11 |
| Nimodipine | Ranitidine | 0.99 | 170 | 0.0027 | 0.0023 | 0.022 | 0.35 | 3.11 |
| Nisoldipine | Cimetidine | 1.49 | 140 | 0.028 | 0.023 | 0.067 | 0.38 | 3.12 |
| Nitrendipine | Cimetidine | 2.41 | 140 | 0.027 | 0.022 | 0.057 | 0.38 | 3.13 |
| Triazolam | Cimetidine | 1.55 | 99 | 0.048 | 0.039 | 0.11 | 0.8 | 3.14 |
| Triazolam | Cimetidine | 1.32 | 99 | 0.048 | 0.039 | 0.11 | 0.8 | 3.15 |
| Midazolam | Clarithromycin | 7.00 | 57 | 0.017 | 0.0094 | 0.048 | 0.74 | 3.16 |
| Lovastatin | Cyclosporin | 1.89 | 2 | 0.059 | 0.0041 | 0.11 | 3.7 | 3.17 |
| Buspirone | Diltiazem | 5.50 | 65 | 0.0024 | 0.0005 | 0.0047 | 0.14 | 3.18 |
| Buspirone | Verapamil | 3.40 | 16 | 0.0047 | 0.0005 | 0.0087 | 0.68 | 3.18 |
| Cyclosporin | Diltiazem | 1.49 | 63.0 | 0.0033 | 0.0007 | 0.0084 | 0.29 | 3.19 |
| Cyclosporin | Diltiazem | 1.57 | 63.0 | 0.0025 | 0.0006 | 0.0063 | 0.22 | 3.20 |
| Cyclosporin | Ketoconazole | 4.39 | 0.37 | 1.2 | 0.012 | 6.2 | 66 | 3.20 |
| Lovastatin | Diltiazem | 3.57 | 65 | 0.0032 | 0.0007 | 0.0081 | 0.28 | 3.21 |
| Midazolam | Diltiazem | 3.75 | 54 | 0.0029 | 0.0003 | 0.0056 | 0.17 | 3.22 |
| Midazolam | Verapamil | 2.92 | 13 | 0.0061 | 0.0003 | 0.011 | 0.87 | 3.22 |
| Nifedipine | Diltiazem | 2.22 | 65 | 0.0012 | 0.0003 | 0.0023 | 0.07 | 3.23 |
| Nifedipine | Diltiazem | 3.11 | 65 | 0.0036 | 0.0008 | 0.007 | 0.21 | 3.23 |
| Simvastatin | Diltiazem | 4.82 | 65 | 0.0032 | 0.0007 | 0.0081 | 0.28 | 3.24 |
| Triazolam | Diltiazem | 3.38 | 54 | 0.0029 | 0.0006 | 0.0056 | 0.17 | 3.25 |
| Triazolam | Diltiazem | 2.30 | 54 | 0.0029 | 0.0006 | 0.0056 | 0.17 | 3.26 |
| Alprazolam | Erythromycin | 2.47 | 120 | 0.005 | 0.0008 | 0.018 | 0.28 | 3.27 |
| Buspirone | Erythromycin | 5.90 | 110 | 0.0075 | 0.0012 | 0.027 | 0.41 | 3.28 |
| Buspirone | Itraconazole | 19.20 | 0.91 (2.2) | 0.26 | 0.0005 | 0.32 | 9.9 | 3.28 |
| Cerivastatin | Erythromycin | 1.21 | 110 | 0.0075 | 0.0012 | 0.027 | 0.41 | 3.29 |
| Cyclosporin | Erythromycin | 2.22 | 66 | 0.0079 | 0.0013 | 0.022 | 0.33 | 3.30 |
| Cyclosporin | Erythromycin | 2.15 | 66 | 0.016 | 0.0025 | 0.044 | 0.66 | 3.31 |
| Felodipine | Erythromycin | 2.49 | 110 | 0.005 | 0.0008 | 0.014 | 0.21 | 3.32 |
| Midazolam | Erythromycin | 4.42 | 170 | 0.0046 | 0.0004 | 0.016 | 0.25 | 3.33 |
| Quinidine | Erythromycin | 1.19 | 110 | 0.005 | 0.0008 | 0.014 | 0.21 | 3.34 |
| Quinidine | Itraconazole | 2.58 | 0.91 (2.2) | 0.13 | 0.0003 | 0.19 | 9.8 | 3.34 |
| Simvastatin | Erythromycin | 6.20 | 110 | 0.0075 | 0.0012 | 0.023 | 0.41 | 3.35 |
| Simvastatin | Verapamil | 4.60 | 16 | 0.0047 | 0.0005 | 0.0087 | 0.68 | 3.35 |
| Triazolam | Erythromycin | 2.06 | 120 | 0.0042 | 0.0007 | 0.015 | 0.23 | 3.36 |
| Quinidine | Felodipine | 1.07 | 11 | 0.0003 | <0.0001 | 0.0005 | 0.15 | 3.37 |
| Quinidine | Nifedipine | 1.15 | 17 | 0.0047 | 0.0002 | 0.022 | 0.21 | 3.37 |
| Cyclosporin | Fluconazole | 1.84 | 40 | 0.54 | 0.48 | 0.69 | 1.55 | 3.38 |
| Midazolam | Fluconazole | 3.60 | 6 | 3.6 | 1.6 | 4.6 | 10 | 3.39 |
| Midazolam | Itraconazole | 6.64 | 0.92 (6.3) | 0.18 | 0.0004 | 0.26 | 19 | 3.39 |
| Rifabutin | Fluconazole | 1.82 | 12 | 1.8 | 1.6 | 2.3 | 5.2 | 3.40 |
| Triazolam | Fluconazole | 1.63 | 1.8 | 2.9 | 2.6 | 3.8 | 8.5 | 3.41 |
| Triazolam | Fluconazole | 2.05 | 1.8 | 5.9 | 5.2 | 7.6 | 17 | 3.41 |
| Triazolam | Fluconazole | 4.42 | 1.8 | 12 | 10 | 15 | 34 | 3.41 |
| Triazolam | Fluconazole | 2.46 | 1.8 | 5.9 | 5.2 | 7.6 | 17 | 3.42 |
| Triazolam | Terbinafine | 0.81 | 100 | 0.012 | <0.0001 | 0.02 | 0.55 | 3.42 |
| Alprazolam | Fluoxetine | 1.34 | 65 (10) | 0.088 | 0.0053 | 0.1 | 0.27 | 3.43 |
| Alprazolam | Fluoxetine | 1.26 | 65 (10) | 0.059 | 0.0035 | 0.063 | 0.12 | 3.44 |
| Triazolam | Fluoxetine | 1.02 | 26 (5.4) | 0.14 | 0.0085 | 0.16 | 0.61 | 3.45 |
| Alprazolam | Fluvoxamine | 1.96 | 9.2 | 0.035 | 0.0080 | 0.057 | 2.2 | 3.46 |
| Buspirone | Fluvoxamine | 2.35 | 11 | 0.029 | 0.0066 | 0.048 | 1.8 | 3.47 |
| Quinidine | Fluvoxamine | 1.41 | 11 | 0.029 | 0.0066 | 0.048 | 1.8 | 3.48 |
| Triazolam | Isradipine | 0.80 | 17 | 0.0001 | <0.0001 | 0.0003 | 0.043 | 3.49 |
| Triazolam | Mibefradil | 8.97 | 2.5 | 0.25 | 0.001 | 0.39 | 2.8 | 3.49 |
| Alprazolam | Itraconazole | 2.66 | 0.95 (2.2) | 0.26 | 0.0005 | 0.37 | 19 | 3.50 |
| Astemizole | Itraconazole | 2.77 | 0.91 (2.2) | 0.52 | 0.001 | 0.63 | 20 | 3.51 |
| Atorvastatin | Itraconazole | 3.64 | 0.91 (2.2) | 0.26 | 0.0005 | 0.38 | 20 | 3.52 |
| Buspirone | Itraconazole | 14.50 | 0.91 (2.2) | 0.26 | 0.0005 | 0.38 | 20 | 3.53 |
| Cerivastatin | Itraconazole | 1.15 | 0.91 (2.2) | 0.26 | 0.0005 | 0.38 | 20 | 3.54 |
| Felodipine | Itraconazole | 6.34 | 0.91 (2.2) | 0.26 | 0.0005 | 0.38 | 20 | 3.55 |
| Lovastatin | Itraconazole | 22.10 | 0.91 (2.2) | 0.26 | 0.0005 | 0.38 | 20 | 3.56 |
| Lovastatin | Itraconazole | 15.40 | 0.91 (2.2) | 0.13 | 0.0003 | 0.19 | 9.8 | 3.57 |
| Midazolam | Itraconazole | 7.97 | 0.92 (6.3) | 0.18 | 0.0004 | 0.26 | 19 | 3.58 |
| Midazolam | Itraconazole | 5.75 | 0.92 (6.3) | 0.089 | 0.0002 | 0.13 | 9.7 | 3.59 |
| Midazolarn | Itraconazole | 10.80 | 0.92 (6.3) | 0.36 | 0.0007 | 0.52 | 39 | 3.60 |
| Midazolam | Ketoconazole | 15.90 | 0.088 | 5 | 0.025 | 26 | 270 | 3.60 |
| Quinidine | Itraconazole | 2.42 | 0.91 (2.2) | 0.26 | 0.0005 | 0.38 | 20 | 3.61 |
| Simvastatin | Itraconazole | 18.60 | 0.91 (2.2) | 0.26 | 0.0005 | 0.38 | 20 | 3.62 |
| Triazolam | Itraconazole | 27.10 | 0.16 (0.2) | 2.2 | 0.0044 | 3.2 | 110 | 3.63 |
| Triazolam | Ketoconazole | 22.40 | 0.022 | 41 | 0.41 | 210 | 2200 | 3.63 |
| Alprazolam | Ketoconazole | 3.98 | 0.061 | 15 | 0.15 | 40 | 400 | 3.64 |
| Triazolam | Ketoconazole | 13.70 | 0.022 | 41 | 0.41 | 110 | 1100 | 3.64 |
| Cyclosporin | Ketoconazole | 5.15 | 0.36 | 2.5 | 0.025 | 6.7 | 67 | 3.65 |
| Cyclosporin | Ketoconazole | 5.31 | 0.36 | 1.2 | 0.012 | 6.2 | 66 | 3.66 |
| Midazolam | Ketoconazole | 16.00 | 0.088 | 10 | 0.051 | 28 | 280 | 3.67 |
| Nisoldipine | Ketoconazole | 24.40 | 0.099 | 4.5 | 0.045 | 23 | 240 | 3.68 |
| Tacrolimus | Ketoconazole | 2.88 | 8 | 0.056 | 0.0006 | 0.28 | 3 | 3.69 |
| Triazolam | Ketoconazole | 9.16 | 0.022 | 31 | 0.31 | 110 | 1100 | 3.70 |
| Alprazolam | Nefazodone | 1.41 | 1.2 | 0.98 | 0.0089 | 2 | 24 | 3.71 |
| Triazolam | Nefazodone | 3.90 | 1.2 | 0.98 | 0.0089 | 2 | 24 | 3.72 |
| Cyclosporin | Nifedipine | 0.88 | 10 | 0.008 | 0.0003 | 0.037 | 0.36 | 3.73 |
| Cyclosporin | Verapamil | 1.45 | 24 | 0.0036 | 0.0004 | 0.0066 | 0.51 | 3.73 |
| Quinidine | Nifedipine | 0.98 | 17 | 0.0071 | 0.0003 | 0.023 | 0.21 | 3.74 |
| Nifedipine | Quinidine sulphate | 1.37 | 160 | 0.019 | 0.0025 | 0.029 | 0.26 | 3.74 |
| Midazolam | Ranitidine | 1.66 | 170 | 0.0027 | 0.0012 | 0.011 | 0.17 | 3.75 |
| Alprazolam | Ritonavir | 2.48 | 0.065 | 71 | 1.4 | 120 | 340 | 3.76 |
| Midazolam | Roxithromycin | 1.47 | 110 | 0.061 | 0.0012 | 0.19 | 0.26 | 3.77 |
| Midazolam | Saquinavir | 5.14 | 3.5 | 0.12 | 0.0012 | 0.15 | 32 | 3.78 |
| Quinidine | Verapamil | 1.47 | 16 | 0.0047 | 0.0005 | 0.0087 | 0.68 | 3.79 |
| Quinidine | Verapamil | 1.50 | 16 | 0.0071 | 0.0007 | 0.013 | 1 | 3.79 |
*Numbers in parentheses represent the values for metabolites.
References for Table 3
3.01 Pourbaix et al. Int J Clin Pharmacol Ther Toxicol 1985; 23: 447–51. 3.02 Friedman et al. J Clin Pharmacol 1988; 28: 228–33. 3.03 Kirch et al. Arch Toxicol Suppl 1984; 7:256–9. 3.04 Azie et al. Clin Pharmacol Ther 1998; 64: 369–77. 3.05 Tateishi et al. J Clin Pharmacol 1989; 29: 994–7. 3.06 Soons et al. Clin Pharmacol Ther 1991; 50: 394–403. 3.07 Olbricht et al. Clin Pharmacol Ther 1997; 62: 311–21. 3.08 Asberg et al. Eur J Clin Pharmacol 1999; 55: 383–7. 3.09 Foradori et al. Transplant Proc 1998; 30: 1685–7. 3.10 Khan et al. Br J Clin Pharmacol 1991; 32: 519–22. 3.11 Varhe et al. Clin Pharmacol Ther 1996; 59: 369–75. 3.12 Lamberg et al. Clin Pharmacol Ther 1998; 63: 640–5.3.13 Yasui et al. Clin Pharmacol Ther 1996; 59: 514–9. 3.14 Muck et al. Eur J Clin Pharmacol 1998; 53: 469–73. 3.15 Gupta et al. Br J Clin Pharmacol 1989; 27: 475–81. 3.16 Bailey et al. Clin Pharmacol Ther 1996; 60: 25–33. 3.17 Kivisto et al. Clin Pharmacol Ther 1997;62: 348–54. 3.18 Damkier et al. Br J Clin Pharmacol 1999; 48: 829–38. 3.19 Kosuge et al. Br J Clin Pharmacol 1997; 43: 367–72. 3.20 Kantola et al. Clin Pharmacol Ther 1998; 64:177–82. 3.21 Bailey et al. Clin Pharmacol Ther 1993; 53: 354–9. 3.22 Canafax et al. Transplantation 1991; 51: 1014–8. 3.23 Trapnell et al. Ann Intern Med 1996; 124: 573–6. 3.24 Varhe et al. Br J Clin Pharmacol 1996; 41: 319–23. 3.25 Phillips et al. J Clin Psychopharmacol 1986; 6: 297–9. 3.26 Lasher et al. Psychopharmacology(Berl) 1991; 104: 323–7.3.27 Greenblatt et al. Clin Pharmacol Ther 1992; 52:479–86. 3.28 Wright et al. Pharmacotherapy 1992; 12: 103–6. 3.29 Fleishaker, Hulst. Eur J Clin Pharmacol 1994; 46: 35–9. 3.30 Lamberg et al. Eur J Clin Pharmacol 1998; 54:761–6. 3.31 Backman et al. Clin Pharmacol Ther 1999; 66:401–7. 3.32 Lefebvre et al. Br J Clin Pharmacol 1997; 43: 319–22. 3.33 Yasui et al. Psychopharmacology (Berl) 1998; 139:269–73. 3.34 Kantola et al. Clin Pharmacol Ther 1998; 64: 58–65. 3.35 Kivisto et al. Pharmacol Toxicol 1999; 84: 94–7. 3.36 Kantola et al. Eur J Clin Pharmacol 1999; S4: 851–5. 3.37 Jalava et al. Clin Pharmacol Ther 1997; 61: 410–5. 3.38 Neuvonen, Jalava. Clin Pharmacol Ther 1996; 60:54–61.3.39 Kivisto et al. Br J Clin Pharmacol 1998; 46: 49–53. 3.40 Kaukonen et al. Clin Pharmacol Ther 1997; 62:510–7. 3.41 Neuvonen et al. Clin Pharmacol Ther 1998; 63:332–41. 3.42 Varhe et al. Clin Pharmacol Ther 1994; 56: 601–7 3.43 Batman et al. J Heart Lung Transplant 1991; 10:351–8. 3.44 Gomez et al. Clin Pharmacol Ther 1995; 58: 15–9. 3.45 Greenblatt et al. Clin Pharmacol Ther 1998; 64:237–47.3.46 Heinig et al. Eur J Clin Pharmacol 1999; 55: 57–60 3.47 Floren et al. Clin Pharmacol Ther 1997; 62: 41–9 3.48 von Moltke et al. J Pharmacol Exp Ther 1996; 276:370–9. 3.49 Tortorice et al. Ther Drug Monit 1990; 12: 321–8. 3.50 Greenblatt et al. Clin Pharmacol Ther 2000; 67:335–41. 3.51 Edwards et al. Clin. Pharmacol Ther 1987; 41: 68–73. 3.52 Zimmermann et al. Arzneimittelforschung 1996; 46:213–7. 3.53 Yeates et al. Int J Clin Pharmacol Ther 1996; 34:400–5. 3.54 Backman et al. Int J Clin Pharmacol Ther 1995; 33:356–9. 3.55 Olkkola et al. Clin Pharmacol Ther 1993; 53: 298–305. 3.56 Backman et al. Eur J Clin Pharmacol 1994; 46: 551–5. 3.57 Elliott et al. Eur J Anaesthesiol 1984; 1: 245–51. 3.58 Fee et al. Clin Pharmacol Ther 1987; 41: 80–4. 3.59 Elwood et al. Br J Clin Pharmacol 1983; 15: 743–5. 3.60 Olkkola et al. Anesth Analg 1996; 82: 511–6. 3.61 Backman et al. Br J Clin Pharmacol 1994; 37:221–5. 3.62 Olkkola et al. Clin Pharmacol Ther 1994; 55: 481–5. 3.63 Palkama et al. Clin Pharmacol Ther 1999; 66: 33–9. 3.64 Greenblatt et al. Clin Pharmacol Ther 1998; 64: 278–85. 3.65 Cox et al. Biopharm Drug Dispos 1986; 7: 567–75. 3.66 Abernethy et al. Psychopharmacology 1983; 80:275–8. 3.67 Mousa et al. Clin Pharmacol Ther 2000; 67: 267–74. 3.68 Varhe et al. Br J Clin Pharmacol 1996; 42: 465–70. 3.69 Tsunoda et al. Clin Pharmacol Ther 1999; 66: 461–71. 3.70 Muck et al. Eur J Clin Pharmacol 1992; 42: 325–8. 3.71 van Harten et al. Clin Pharmacol Ther 1988; 43:332–41. 3.72 Backman et al. Eur J Clin Pharmacol 1998; 54: 53–8. 3.73 Freeman et al. Br J Clin Pharmacol 1987; 23: 776–8. 3.74 Damkier et al. Eur J Clin Pharmacol 1999; 55: 451–6. 3.75 Greene et al. J Clin Psychopharmacol 1995; 15:399–408. 3.76 Bowles et al. J Clin Pharmacol 1993; 33: 727–31. 3.77 Gorski et al. Clin Pharmacol Ther 1998; 64:133–43. 3.78 Barbhaiya et al. J Clin Psychopharmacol 1995; 15:320–6. 3.79 Ahonen et al. Br J Clin Pharmacol 1995; 40: 270–2.
Table 4.
In vivo database for the interactions involving CYP2C9
| Substrate | Inhibitor | AUC ratio | Ki(µm) | [I]av/Ki | [I]av,u/Ki | [I]max/Ki | [I]in/Ki | Reference |
|---|---|---|---|---|---|---|---|---|
| S-Warfarin | Amiodarone | 2.11 | 95 | 0.033 | 0.0014 | 0.033 | 0.23 | 4.01 |
| S-Warfarin | Amiodarone | 1.27 | 95 | 0.024 | 0.001 | 0.025 | 0.31 | 4.02 |
| Phenytoin | Amiodarone | 1.40 | 95 | 0.016 | 0.0006 | 0.016 | 0.21 | 4.03 |
| S-Warfarin | Benzbromarone | 2.15 | 0.0085 | 550 | 0.55 | 960 | 1400 | 4.04 |
| S-Warfarin | Bucolome | 3.29 | 20 | 1.9 | 0.19 | 2.4 | 5.3 | 4.05 |
| S-Warfarin | Fluconazole | 2.84 | 7.5 | 6.2 | 5.5 | 8 | 17 | 4.06 |
| Tolbutamide | Fluconazole | 2.09 | 16 | 0.73 | 0.65 | 0.94 | 2 | 4.07 |
| Phenytoin | Fluconazole | 1.75 | 7.5 | 3.2 | 2.8 | 4.1 | 8.6 | 4.08 |
| Phenytoin | Fluconazole | 1.33 | 7.5 | 6.2 | 5.5 | 8 | 17 | 4.09 |
| Losartan | Fluconazole | 1.69 | 13 | 1.8 | 1.6 | 2.4 | 5 | 4.10 |
| Losartan | Fluconazole | 1.27 | 13 | 1.8 | 1.6 | 2.4 | 5 | 4.11 |
| Fluvastatin | Fluconazole | 1.84 | 13 | 1.8 | 1.6 | 2.4 | 5 | 4.12 |
| Glimepiride | Fluconazole | 2.38 | 13 | 1.8 | 1.6 | 2.4 | 5 | 4.13 |
| Diclofenac | Fluvastatin | 1.25 | 0.18 | 0.32 | 0.0032 | 7.1 | 31 | 4.14 |
| Tolbutamide | Fluvastatin | 1.23 | 0.25 | 0.23 | 0.0032 | 5.1 | 22 | 4.15 |
| Tolbutamide | Ketoconazole | 1.77 | 8.1 | 0.65 | 0.0065 | 3.3 | 3.5 | 4.16 |
| Phenprocoumon | Lornoxicam | 1.13 | 12 | 0.1 | 0.001 | 0.24 | 0.22 | 4.17 |
| R-Acenocoumarol | Lornoxicam | 1.06 | 10 | 0.12 | 0.0012 | 0.28 | 0.26 | 4.18 |
| S-Warfarin | Miconazole | 4.72 | 0.5 | 0.54 | 0.054 | 0.75 | 33 | 4.19 |
| Phenytoin | Sertraline | 1.19 | 33 | 0.013 | 0.0001 | 0.018 | 1.1 | 4.20 |
| Tolbutamide | Sertraline | 1.19 | 33 | 0.005 | <0.0001 | 0.0065 | 1.1 | 4.21 |
| Tolbutamide | Sulphamethizole | 1.62 | 75 | 0.0012 | 0.004 | 3.1 | 4.22 | |
| Tolbutamide | Sulphaphenazole | 5.28 | 0.32 | 220 | 70 | 320 | 530 | 4.23 |
| Tolbutamide | Sulphaphenazole | 3.10 | 0.32 | 220 | 70 | 320 | 530 | 4.24 |
| S-Warfarin | Sulphinpyrazone | 1.70 | 230 | 0.018 | 0.0004 | 0.042 | 0.15 | 4.25 |
| S-Warfarin | Sulphinpyrazone | 1.93 | 230 | 0.018 | 0.0004 | 0.042 | 0.15 | 4.26 |
References for Table 4
4.01 O’Reilly et al. Clin Pharmacol Ther 1987; 42: 290–4. 4.02 Heimark et al. Clin Pharmacol Ther 1992; 51:398–407. 4.03 Nolan et al. Clin Pharmacol Ther 1989; 46: 43–50. 4.04 Takahashi et al. Clin Pharmacol Ther 1999; 66:569–81. 4.05 Takahashi et al. Drug Metab Dispos 1999; 27: 1179–86. 4.06 Black et al. Drug Metab Dispos 1996; 24: 422–8. 4.07 Lazar, Wilner. Rev Infect Dis 1990; 12: S327–33. 4.08 Blum et al. Clin Pharmacol Ther 1991; 49: 420–5. 4.09 Touchette et al. Br J Clin Pharmacol 1992; 34: 75–8. 4.10 Kazierad et al. Clin Pharmacol Ther 1997; 62:417–25. 4.11 Kaukonen et al. Eur J Clin Pharmacol 1998; 53:445–9. 4.12 Kantola et al. Eur J Clin Pharmacol 2000; 56: 225–29. 4.13 Niemi et al. Clin Pharmacol Ther 2001; 69: 194–200 4.14 Transon et al. Clin Pharmacol Ther 1995; 58: 412–17. 4.15 Appel et al. Am J Cardiol 1995; 76: 29A–32A. 4.16 Krishnaiah et al. Br J Clin Pharmacol 1994; 37: 205–7. 4.17 Masche et al. Eur J Clin Pharmacol 1999; 54: 857–64. 4.18 Masche et al. Eur J Clin Pharmacol 1999; 54: 865–68. 4.19 O’Reilly et al. Clin Pharmacol Ther 1992; 51: 656–67. 4.20 Rapeport et al. J Clin Psychiatry 1996; 57: Suppl 1 24–8. 4.21 Tremaine et al. Clin Pharmacokinet 1997; 32: SuppI 1 31–6. 4.22 Lumholtz et al. Clin Pharmacol Ther 1975; 17: 731–4. 4.23 Veronese et al. Clin Pharmacol Ther 1990; 47: 403–11. 4.24 Back et al. Eur J Clin Pharmacol 1988; 34: 157–63. 4.25 Toon et al. Clin Pharmacol Ther 1986; 39: 15–24. 4.26 O’Reilly. Circulation 1982; 65: 202–7.
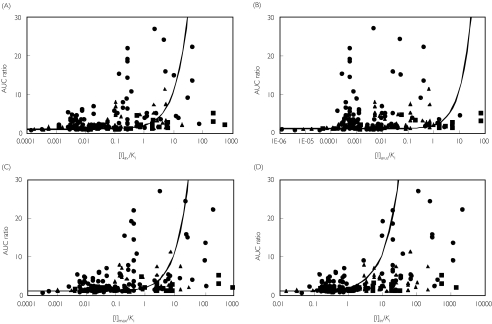
In Figure 2, the observed values of AUC ratio are plotted against the values of [I]/Ki ratio estimated using each of the four [I] above. The average degree of in vivo interaction was largest for CYP3A4 (4.5-fold), intermediate for CYP2D6 (2.6-fold) and smallest for CYP2C9 (2.0-fold). In vitro Ki values differed over five orders of magnitude. The use of [I]av (panel A) gave an approximate description of the data with 78% and 22% of the inhibitors with [I]/Ki ratios below and above 1, respectively. Table 5 provides a breakdown of the number of studies within the true positives (AUC ratio > 2, [I]/Ki > 1), true negatives (AUC ratio < 2, [I]/Ki < 1), false positives (AUC ratio < 2, [I]/Ki > 1), or false negatives (AUC ratio > 2, [I]/Ki < 1) zones.
Figure 2.
Relationship between the observed AUC ratio and the various [I]/Ki ratios for drug-drug interactions involving CYP3A4 (•), CYP2D6 (▴) or CYP2C9 (▪). The curves represent the theoretical curves based on equation 1 using average systemic total drug plasma concentration ([I]av– panel A), average systemic unbound drug plasma concentration ([I]av,u– panel B), maximum systemic plasma concentration ([I]max– panel C), and maximum hepatic input concentration ([I]in– panel D)
Table 5.
Summary of the predictions of drug-drug interactions based on the various values for [I]. The studies were designated according to the qualitative zoning shown in Figure 1
| CYP3A4 | CYP2D6 | CYP2C9 | ||||||||||
| [I]av | [I]av,u | [I]max | [I]in | [I]av | [I]av,u | [I]max | [I]in | [I]av | [I]av,u | [I]max | [I]in | |
| True positive | 16 | 5 | 17 | 36 | 10 | 0 | 10 | 15 | 6 | 4 | 6 | 8 |
| True negative | 31 | 31 | 30 | 24 | 33 | 36 | 33 | 13 | 12 | 11 | 9 | 6 |
| False positive | 2 | 2 | 3 | 9 | 3 | 0 | 3 | 23 | 5 | 5 | 8 | 11 |
| False negative | 23 | 34 | 22 | 3 | 5 | 15 | 5 | 0 | 3 | 5 | 3 | 1 |
| Total | 72 | 72 | 72 | 72 | 51 | 51 | 51 | 51 | 26 | 25 | 26 | 26 |
Using [I]av,u (panel B) shifted all the points to the left compared with [I]av, resulting in substantial underprediction for most of the studies, whereas the use of [I]max (panel C) appears little different from [I]av. In contrast, the points shifted to the right using [I]in (panel D), leading to a more convincing zoning of negatives and positives.
Among the inhibitors involved in CYP3A4 interaction studies, macrolide antibiotics (erythromycin, clarithromycin, etc.) and some of the calcium channel blockers (diltiazem, verapamil and mibefradil) are reported to be mechanism-based inhibitors of CYP3A4 that cause irreversible inhibition by forming an inactive complex with the enzyme [16–18]. In addition, paroxetine has recently been reported to be a mechanism-based inhibitor of CYP2D6 [19]. Since equation 1 is only applicable for reversible inhibition, the interactions involving mechanism-based inhibitors were identified. Once these are excluded, there is a marked improvement in zoning with almost no false negative predictions (see Figure 3 using [I]in).
Figure 3.
Identifying drug-drug interaction studies involving either reversible (closed symbols) or mechanism-based inhibition (open symbols). CYP3A4 (•), CYP2D6 (▴), CYP2C9
A comparison of the success of qualitative zoning of the inhibition predictions (excluding mechanism-based inhibitions) according to the four [I] values are summarized in Table 5. Results based on [I]av and [I]max were similar to each other for all of the CYP enzymes, with a high proportion of ‘true’ predictions (the sum of ‘true positives’ and ‘true negatives’). The incidence of false negative prediction was largest using the [I]av,u and smallest using the [I]in for any of the CYP enzymes, a difference that was statistically significant (P < 0.001). These findings are consistent with a previous study [14], which showed a high possibility of false negative predictions based only on the systemic concentration of inhibitor. The use of [I]in resulted in the highest incidence of true positive predictions (Table 5), though the number of false positive predictions was also the highest of the four values for [I]. There was a significant difference between the use of [I]in and the other three [I] values (P < 0.001).
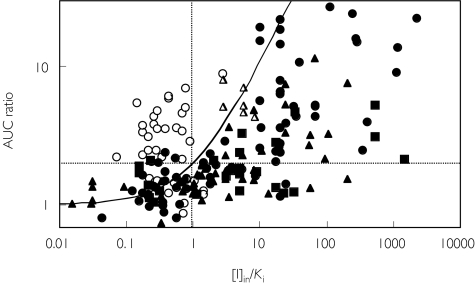
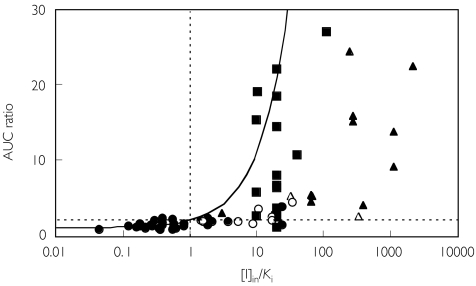
The inhibitors used in the interaction studies involving CYP3A4 and CYP2D6 are identified in Figures 4 and 5, respectively. Most of the interactions involving major effects on CYP3A4 were caused by azoles, particularly itraconazole and ketoconazole. For CYP2D6 interactions, more than half of the studies involved quinidine or SSRI antidepressants (fluoxetine, fluvoxamine, etc.) as inhibitors.
Figure 4.
Data from studies involving CYP3A inhibitors. Fluconazole (n = 7) (○), itraconazole (n = 17) (▪), ketoconazole (n = 11) (▴), HIV protease inhibitors (n = 2) (▵), others (n = 35)
Figure 5.
Data from studies involving CYP2D6 inhibitors. Quinidine (n = 7) (▴), citalopram (n = 3) (□), fluoxetine (n = 8) (▪), fluvoxamine (n = 2) (○), sertraline (n = 6) (▵), others (n = 25) (•)
Figure 6 shows the comparison of the three in vivo metrics (AUC ratio, Cp ratio and MR ratio) used in CYP2D6 interaction studies. All three gave similar patterns as illustrated by [I]in.
Figure 6.
Comparison of the use of three in vivo metrics for interactions involving CYP2D6 inhibition. The ordinates show the observed ratio of AUC (A), plasma concentration at a single time point (B), or metabolic ratio (C) for the substrate in the presence and absence of inhibitor. Open symbols represent studies involving paroxetine.
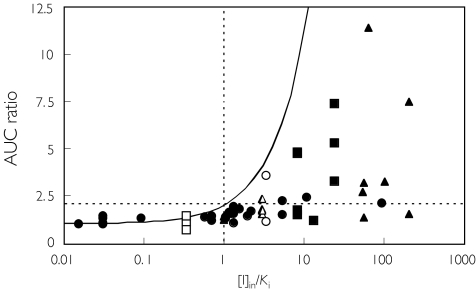
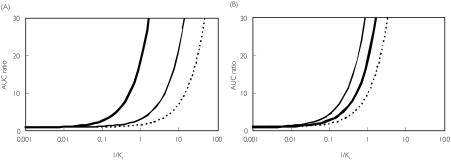
The use of [I]in relies on an input term for the hepatic portal vein plasma concentration calculated from the inhibitor absorption rate constant, the fraction absorbed, the dose and the hepatic blood flow (equation 6). One possible limitation of this equation is the use of theoretical maximum values of 0.1 min−1 and 1 for the inhibitor ka and Fa, respectively [8]. Therefore the effect of lowering the values of the inhibitor ka × Fa product was simulated (see Figure 7A). Since the input term (the second term in equation 6) is larger than [I]av (as illustrated for quinidine), the simulated AUC ratio was affected largely by the value of ka (or indeed Fa). This results in a shift of the theoretical line to the right. Quinidine represents an intermediate case where the input term exceeds the systemic term by 37-fold. Within the database the mean differential observed is 73-fold (87% studies had an input term of more than 10 times the systemic term). Although it is generally not easy to obtain accurate values for ka (or, to a lesser extent Fa), estimations of these values for each inhibitor would provide more precise predictions. In contrast the hepatic blood flow term appears far less important, since a four-fold difference in the Qh value resulted in only a small difference in the simulated value of AUC ratio (Figure 7B). The dose of inhibitor is the major determinant of the [I]in.
Figure 7.
Simulation of the effects of changing the rate of absorption of the inhibitor (A) or liver blood flow (B). The AUC ratio was calculated as 1 + [I]in/Ki and plotted against 1/Ki. The following parameter values for inhibition by quinidine were used, D = 268 nmol, τ = 24 h, & CL/F = 412 ml min−1 in equations 4 and 6 A— : ka Fa = 0.1 min−1, —: ka Fa = 0.01 min−1, …: ka Fa = 0.001 min−1B— : Qh = 1610 ml min−1, —: Qh = 805 ml min−1, …: Qh = 3220 ml min−1
In conclusion, qualitative zoning of CYP inhibition interactions can be achieved from the [I]/Ki ratio using in vitro kinetic parameters and the hepatic input concentration of inhibitor. True negatives can be identified and, in contrast to the use of other values for [I], false negatives are eliminated. True positives are also predicted well and, although the incidence of false positive predictions is quite high, the use of hepatic input concentration is recommended. This approach would be particularly valuable in drug screening, where false negative predictions need to be avoided. However, it is important to appreciate that the present analysis is empirical, and must be regarded as an initial step in the prediction of CYP inhibition interactions, since it ignores the interactions related to gut metabolism and specific substrate/inhibitor properties. The likely importance of these individual characteristics is evident from the number of true positives that are quantitatively over-predicted in this global analysis. Factors such as the role of hepatic uptake transporters, the existence of more than one metabolic/elimination pathway, the influence of multisite kinetics for CYP3A4/5 and the nonlinear kinetics of substrates, will require consideration in addition to the [I]/Ki ratio in order to progress the prediction of CYP inhibition interactions towards a quantitative basis.
Acknowledgments
HSB was financially supported by a Bristol Myers Squibb studentship. The authors would like to thank Dr Rene H. Levy for permission to use the Drug Interaction Database of the University of Washington to obtain some of the published articles used in this data base and Dr Aleksandra Galetin for valuable discussion.
References
- 1.Tucker GT, Houston JB, Huang S-M. Optimizing drug development: Strategies to assess drug metabolism/transporter interaction potential – toward a consensus. Br J Clin Pharmacol. 2001;52:107–17. doi: 10.1046/j.0306-5251.2001.temp.1441.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Segel IH. Enzyme KineticsBehaviour and Analysis of Rapid Equilibrium and Steady State Enzyme Systems. New York: Wiley & Sons Inc; 1975. [Google Scholar]
- 3.Zomorodi K, Houston JB. Effect of omeprazole on diazepam disposition in the rat: in vitro and in vivo studies. Pharm Res. 1995;12:1642–6. doi: 10.1023/a:1016241000480. [DOI] [PubMed] [Google Scholar]
- 4.Davis JD, Aarons L, Houston JB. Relationship between enoxacin and ciprofloxacin plasma concentrations and theophylline disposition. Pharm Res. 1994;11:1424–8. doi: 10.1023/a:1018991822440. [DOI] [PubMed] [Google Scholar]
- 5.Ervine CM, Matthew DE, Brennan B, Jezequel SG, Humphrey MJ, Houston JB. Comparison of the steady-state pharmacokinetics of fluconazole and ketoconazole and their relative effects on cytochrome P450 activity in rats: use of antipyrine and a steady-state infusion approach to assess plasma concentration-response relationships. Drug Metab Dispos. 1996;24:211–5. [PubMed] [Google Scholar]
- 6.von Moltke LL, Greenblatt DJ, Cotreau-Bibbo MM, Duan SX, Harmatz JS, Shader RI. Inhibition of desipramine hydroxylation in vitro by serotonin-reuptake-inhibitor antidepressants, and by quinidine and ketoconazole: a model system to predict drug interactions in vivo. J Pharmacol Exp Ther. 1994;268:1278–83. [PubMed] [Google Scholar]
- 7.Bertz RJ, Granneman GR. Use of in vitro and in vivo data to estimate the likelihood of metabolic pharmacokinetic interactions. Clin Pharmacokin. 1997;32:210–58. doi: 10.2165/00003088-199732030-00004. [DOI] [PubMed] [Google Scholar]
- 8.Ito K, Iwatsubo T, Kanamitsu S, Ueda K, Suzuki H, Sugiyama Y. Prediction of pharmacokinetic alterations caused by drug–drug interactions: Metabolic interaction in the liver. Pharmacol Rev. 1998;50:387–411. [PubMed] [Google Scholar]
- 9.Davit B, Reynolds K, Yuan R, et al. FDA evaluations using in vitro metabolism to predict and interpret in vivo metabolic drug–drug interactions: impact on labeling. J Clin Pharmacol. 1999;39:899–910. doi: 10.1177/00912709922008515. [DOI] [PubMed] [Google Scholar]
- 10.Lin JH. Sense and nonsense in the prediction of drug–drug interactions. Curr Drug Metab. 2000;1:305–31. doi: 10.2174/1389200003338947. [DOI] [PubMed] [Google Scholar]
- 11.Rodrigues AD, Winchell GA, Dobrinska MR. Use of in vitro drug metabolism data to evaluate metabolic drug–drug interactions in man: the need for quantitative databases. J Clin Pharmacol. 2001;41:368–73. doi: 10.1177/00912700122010212. [DOI] [PubMed] [Google Scholar]
- 12.Yao C, Levy RH. Inhibition-based metabolic drug–drug interactions: predictions from in vitro data. J Pharm Sci. 2002;91:1923–35. doi: 10.1002/jps.10179. [DOI] [PubMed] [Google Scholar]
- 13.Kanamitsu S, Ito K, Sugiyama Y. Quantitative prediction of in vivo drug–drug interactions from in vitro data based on physiological pharmacokinetics: Use of maximum unbound concentration of inhibitor at the inlet to the liver. Pharm Res. 2000;17:336–43. doi: 10.1023/a:1007509324428. [DOI] [PubMed] [Google Scholar]
- 14.Ito K, Chiba K, Horikawa M, et al. Which concentration of the inhibitor should be used to predict in vivo drug interactions from in vitro data? AAPS Pharm Sci. 2002;4:article 25. doi: 10.1208/ps040425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kenworthy KE, Bloomer J, Clarke SE, Houston JB. CYP3A4 drug interactions. correlation of ten in vitro probes substrates. Br J Clin Pharmacol. 1999;48:716–27. doi: 10.1046/j.1365-2125.1999.00073.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Periti P, Mazzei T, Mini E, Novelli A. Pharmacokinetic drug interactions of macrolides. Clin Pharmacokinet. 1992;23:106–31. doi: 10.2165/00003088-199223020-00004. [DOI] [PubMed] [Google Scholar]
- 17.Ma B, Prueksaritanont T, Lin JH. Drug interactions with calcium channel blockers. Possible involvement of metabolite-intermediate complexation with CYP3A. Drug Metab Dispos. 2000;28:125–30. [PubMed] [Google Scholar]
- 18.Prueksaritanont T, Ma B, Tang C, et al. Metabolic interactions between mibefradil and HMG-CoA reductase inhibitors: an in vitro investigation with human liver preparations. Br J Clin Pharmacol. 2000;49:87–90. doi: 10.1046/j.1365-2125.1999.00903.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bertelsen KM, Venkatakrishnan K, Von Moltke LL, Obach RS, Greenblatt DJ. Apparent mechanism-based inhibition of human CYP2D6 in vitro by paroxetine: comparison with fluoxetine and quinidine. Drug Metab Dispos. 2003;31:289–93. doi: 10.1124/dmd.31.3.289. [DOI] [PubMed] [Google Scholar]