Abstract
Aims
To confirm the identity of the major metabolites of domperidone and to characterize the cytochrome P450s (CYPs) involved in their formation.
Methods
Human liver microsomes (HLMs) were used to characterize the kinetics of domperidone metabolism and liquid chromatography-mass spectrometry to identify the products. Isoform-specific chemical inhibitors, correlation analysis and expressed human CYP genes were used to identify the CYPs involved in domperidone oxidation.
Results
In HLMs, domperidone underwent hydroxylation to form 5-hydroxydomperidone (MIII) and N-dealkylation to form 2,3-dihydro-2-oxo-1H-benzimidazole-1-propionic acid (MI) and 5-chloro-4-piperidinyl-1,3-dihydro-benzimidazol-2-one (MII). The formation of all three metabolites (n = 4 HLMs) followed apparent Michaelis-Menten kinetics. The mean Km values for MI, MII and MIII formation were 12.4, 11.9, and 12.6 µm, respectively. In a panel of HLMs (n = 10), the rate of domperidone (5 µm and 50 µm) metabolism correlated with the activity of CYP3A (r > 0.94; P < 0.0001). Only ketoconazole (1 µm) (by 87%) and troleandomycin (50 µm) (by 64%) inhibited domperidone (5 µm) metabolism in HLMs. Domperidone (5 and 50 µm) hydroxylation and N-dealkylation was catalyzed by expressed CYP3A4 at a higher rate than the other CYPs. CYP1A2, 2B6, 2C8 and 2D6 also hydroxylated domperidone
Conclusions
CYP3A-catalyzed N-dealkylation and aromatic hydroxylation are the major routes for domperidone metabolism. The drug would be expected to demonstrate highly variable bioavailability due to hepatic, and possibly intestinal first-pass metabolism after oral administration. Increased risk of adverse effects might be anticipated during concomitant administration with CYP3A inhibitors, as well as decreased efficacy with inducers of this enzyme.
Keywords: cytochrome P450, domperidone, prokinetic, metabolites, hydroxylation
Introduction
The benzimidazole derivative domperidone is a dopamine D2 receptor antagonist that has been marketed in Europe and other countries as a prokinetic and antiemetic since 1978. It prevents nausea and vomiting associated with chemotherapy and antiparkinsonian drug therapy [1–3]. One of the more popular off-label uses for domperidone is for the stimulation of breast milk production in women. The recent restriction of cisapride in the United States and other countries due to drug-related pro-arrhythmic effects has left a gap in the therapeutic armamentarium for the treatment of upper gastrointestinal motility disorders. Since no new prokinetic drugs are available to replace cisapride, clinicians have relied on other agents such as metoclopramide and domperidone. Whereas metoclopramide and domperidone have similar therapeutic profiles, domperidone crosses the blood–brain barrier less readily and rarely causes the extrapyramidal side-effects that limit the use of metoclopramide, particularly in the elderly and paediatric populations [1–3]. Thus, domperidone appears to be a safer alternative to cisapride and metoclopramide.
The pharmacological similarity between domperidone and cisapride suggests that domperidone may have similar cardiotoxic effects to those seen with cisapride, as do other drugs like antihistamines such as terfenadine [4, 5]. Multiple cases of adverse cardiac effects associated with domperidone use, particularly after intravenous dosing, including QT interval prolongation, ventricular arrhythmia, and cardiac arrest at therapeutically recommended doses, have been reported. Drolet et al.[6] have shown that domperidone prolongs cardiac repolarization by blocking the rapid component (Ikr) of the delayed-rectifier potassium current in a concentration dependent manner, which may explain the drug-induced ventricular arrhythmias.
Domperidone is well absorbed after oral administration and extensively metabolized in humans, with less than 8% of the total dose appearing unchanged in urine and faeces [7]. On the basis of human excretion data, the major routes of domperidone metabolism include aromatic hydroxylation and oxidative N-dealkylation consistent with CYP450-mediated reactions [7]. Despite its use for more than two decades, the specific enzymes involved in domperidone metabolism have not been characterized. In the present study, we sought to confirm the identify of the major metabolites of domperidone and to characterize the CYP isoforms responsible for their formation.
Methods
Chemicals
Domperidone was purchased from Research Diagnostics Inc. (Pleasant Hill Road, Flanders, NJ). Ketoconazole, quinidine sulphate, troleandomycin, diethyldithiocarbamate, ticlopidine, glucose 6-phosphate (G6P), glucose 6-phosphate dehydrogenase (G6PD), β-NADP and magnesium chloride were purchased from Sigma Chemical Co. (St Louis, MO). Sulphaphenazole and furafylline were obtained from Ultrafine Chemicals (Manchester, England). Levallorphan and thioTEPA were obtained from United States Pharmacopeia Convention (Rockville, MD). Omeprazole was a generous gift from Dr Tommy Anderson (Clinical Pharmacology, Astra Hässle AB, Mölndal, Sweden). All other chemicals were of analytical grade appropriate for HPLC.
Microsomal preparations
Human liver microsomes (HLMs) used were obtained either from human liver tissues that were medically unsuitable for liver transplantation and processed as described in detail in previous publications [8, 9] or from a commercial source (BD Bioscience/Gentest, Bedford, MA). Gentest states that the human materials were prepared from human liver donor tissues that were obtained with informed consent in conformance with the guidelines promulgated by the USA Uniform Anatomical Gift Act (1987). The microsomal pellets were re-suspended in a reaction buffer (0.1 m potassium phosphate, 1.0 mm EDTA, 5.0 mm MgCl2, pH 7.4) to a protein concentration of 10 mg ml−1 (stock) and were kept at −80 °C until used. Protein concentrations were determined according to Bradford et al.[10] using bovine serum albumin as a standard. Baculovirus-insect cell expressed human P450s (1A2, 2A6, 2B6, 2C8, 2C9, 2C19, 2D6, 2E1, 3A4 and 3A5) were purchased from BD Bioscience/Gentest (Bedford, MA).
Incubation conditions
Based upon previous studies [8], preliminary experiments were performed to develop an HPLC method to measure the metabolites of domperidone and to optimize microsomal incubation conditions. Domperidone was dissolved in methanol, which was removed by evaporation prior to incubation with microsomes. The drug (10 µm) was prewarmed in 1.5 ml microfuge tubes for 5 min with 80 mm phosphate reaction buffer (pH 7.4) and a NADPH-regenerating system (1.3 mmβ-NADP, 3.3 mm G6P, 0.4 units ml−1, G6PD, and 3.3 mm MgCl2) at 37 °C. The reaction was initiated by adding human liver microsomal protein (1 mg ml−1) and the mixture was incubated for 30 min (final volume, 250 µl). Negative control incubations, from which the following were excluded: (a) domperidone; (b) a β-NADPH-generating system; or (c) HLMs (bovine serum albumin was used instead) were run in parallel. Reactions were terminated by placing tubes on ice and by the addition of 20 µl of 60% perchloric acid. After addition of internal standard (40 µl of 16 µm levallorphan) and centrifugation in an Eppendorf model 5415C microfuge (Brinkman Instruments, Westbury, NY) at 14000 r.p.m. for 5 min, an aliquot of the supernatant (40 µl) was injected onto the HPLC system. Three fluorescent chromatographic peaks (designated as M-I, M-II and M-III, respectively) were detected at retention times of about 6.0, 6.7, and 8.5 min, with the internal standard (levallorphan) at 9.6 min and domperidone at 14 min, respectively, following the assay of incubation mixtures that contain the substrate, cofactors and HLMs, but were not present in the negative control incubations (data not shown).
Identification of domperidone metabolites
Total ion count and single ion recordings (SIR) of domperidone and its metabolites were monitored by liquid chromatography-mass spectrometry (LC-MS). Metabolites were separated by a DuPont Zorbax SB-Phenyl 150 × 4.6 mm column and a mobile phase that consisted of 50 mm ammonium acetate/methanol/acetonitrile (50 : 25 : 25, v/v/v). The flow rate was 0.2 ml min−1. The mass spectrometer (PE/SciFx API100) was operated in positive ion mode and tuned for optimal resolution over the mass range by PE/SciEx proprietary LC2Tune program software. The exact conditions were: capillary voltage 5300 V, orifice voltage 30 V, and mass range of 150–450. Nitrogen was used as driving gas. The molecular weight data obtained from the MS were compared with the urinary metabolite profiles of domperidone previously reported in humans [7].
Linear conditions for incubation were then determined by incubating 10 µm domperidone at 37 °C with human liver microsomal proteins (0.025–1 mg ml−1) and a NADPH-generating system over different incubation times (0–60 min). A 30 min incubation time and a 0.25 mg ml−1 final protein concentration represented linear conditions and were used in the subsequent experiments.
HPLC assay for domperidone and its metabolites
An HPLC method with fluorescence detection was developed to measure domperidone and its metabolites. The HPLC system consisted of a Waters Assoc. model 510 pump (Milford, MA), a Waters Assoc. Model 717 autosampler, and a Hewlett Packard Model 1046 A Programmable Fluorescent Detector. Domperidone and metabolites were separated using a DuPont Zorbax SB-Phenyl 150 × 4.6 mm column and a mobile phase consisting of 80 mm monobasic potassium phosphate, 23% acetonitrile, 20% methanol and 0.114% triethylamine, pH 4.0 and delivered at a flow rate of 0.5 ml min−1. Fluorescent detection was carried out at an excitation wavelength of 282 nm and an emission wavelength of 328 nm.
Concentrations of metabolites were measured by standard curves obtained with domperidone, as authentic samples of the synthetic metabolite were not available to us. The possibility that the fluorescent intensity might differ between the metabolites and domperidone or between the metabolites themselves could not be excluded. Formation rates of metabolites normally presented as pmol min−1 mg−1 protein (or pmol min−1 pmol−1 P450) should be viewed more appropriately as apparent velocities (arbitrary unit min−1 mg−1 protein or pmol P450] where an arbitrary unit = 1000 × (metabolite peak area/internal standard peak area)/slope of the standard curve.
Kinetics studies of domperidone metabolism in HLMs
The enzyme kinetics of domperidone were characterized in four different HLMs. Domperidone (1–50 µm) was incubated in duplicate for 30 min at 37 °C at a protein concentration of 0.25 mg ml−1.
Correlation experiments in a panel of HLMs
Domperidone (5 and 50 µm) was incubated for 30 min at 37 °C with microsomes from 10 different human livers (0.25 mg protein ml−1) and a NADPH-generating system. Values for the activity of each CYP were provided by the supplier of the HLMs studied (see http://www.gentest.com) and are detailed in an earlier publication [9].
Inhibition studies
Domperidone (5 µm) was incubated for 30 min at 37 °C with HLMs (0.25 mg ml−1) and a NADPH generating system in the absence (control) and presence of the following isoform-selective inhibitors: furafylline (10 µm) for CYP1A2, sulfaphenazole (50 µm) for CYP2C9, ticlopidine (5 µm) and omeprazole (10 µm) for 2C19, quinidine (1 µm) for CYP2D6, diethydithiocarbamate (50 µm) for CYP2E1 ketoconazole (1 µm) and troleandomycin (50 µm) for CYP3A, and ticlopidine (5 µm) and thioTEPA (50 µm) for CYP2B6. The concentrations of the inhibitors were chosen to be selective for the respective CYPs on the basis of published Ki values [8, 11–13]. Normally, domperidone was prewarmed for 5 min with or without the inhibitor in the presence of a NADPH-generating system, and then the reaction was initiated by adding HLMs. Incubation was for 30 min at 37 °C in a final incubation volume of 250 µl. However, troleandomycin, furafylline, thioTEPA and dietheyldithiocarbamate were pre-incubated with a NADPH regenerating system and HLMs for 15 min at 37 °C before initiation of the reaction by addition of domperidone and further incubation for 30 min. Inhibitors were dissolved in water where appropriate or in a suitable organic solvent (ethanol, methanol, or dimethyl sulfoxide) before being serially diluted with water to contain less than 0.1% of organic solvent in the final incubation volume.
Data from the above experiments suggested that ketoconazole was the most potent inhibitor of domperidone metabolism in HLMs. To estimate the Ki value for inhibition of domperidone metabolism by ketoconazole, we incubated domperidone (2–15 µm) with ketoconazole (0.1–0.75 µm) in HLMs.
Domperidone metabolism by recombinant human CYP450 isoforms
Domperidone (5 and 50 µm) was incubated with recombinant human CYP 1A2, 2A6, 2B6, 2C8, 2C9, 2C19, 2D6, 2E1, 3A4 or 3A5 (13–26 pmol P450/250 µl) and a NADPH-generating system at 37 °C for 30 min. All other incubation and HPLC assay conditions were the same as described for HLMs. The kinetic parameters for metabolism by CYP3A4 were determined by incubating domeperidone (1–50 µm) with the enzyme (26 pmol P450/250 µl) for 30 min.
Data analysis
Apparent kinetic parameters were obtained by fitting a single-site Michaelis Menten model to the data using nonlinear regression analysis software (GraphPad Prism Software inc, Version 3.02, San Diego, CA). Correlation analysis was performed using a Spearman's rank correlation test. P < 0.05 was considered statistically significant. Inhibition constants (Ki values) were estimated by fitting the model for competitive inhibition to the data using WinNonlin (Software Version 4.0; Pharsight, Mountain View, CA). All experiments were performed in duplicate. Data are presented as mean ± SD or as the mean of duplicate measurements.
Results
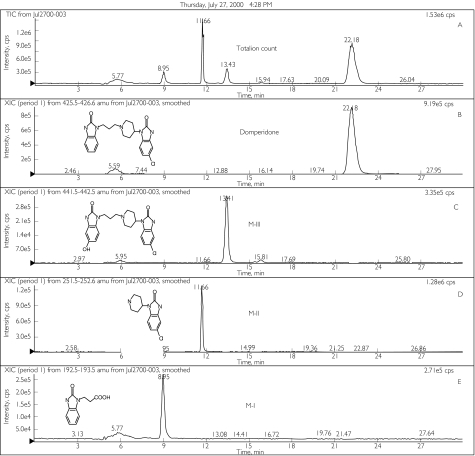
Three metabolite peaks were identified and confirmed by comparing the LC-MS fragmentation analyses (Figure 1) with previously reported structures obtained from a study in healthy subjects [7]. The metabolites named M-I, M-II and M-III corresponded to 2,3-dihydro-2-oxo-1H-benzimidazole-1-propionic acid, 5-chloro-4 - piperidinyl -1,3-dihydro- benzimidazol -2 - one, and 5-hydroxydomperidone, respectively (Figure 1). No other peaks were observed in any of the LC or LC-MS chromatograms.
Figure 1.
Total ion counts and selected ion-recording scans by liquid chromatography-mass spectrometry (LC-MS) for domperidone and its metabolites generated from HLMs. (A) represents total ion count (domperidone elutes at 22.2 min, M-III at 13.4 min, M-II at 11.7 min, and M-I at 8.9 min). Chromatograms B-E represent selected ion recording (SIR) scans for domperidone and its metabolites in HLMs incubates. SIRs for MW 426 m/z (B) corresponding to domperidone or 5-chloro-1-[1-[3-(2,3-dihydro-2-oxo-1H-benzimidazol-1-yl) propyl]-4-piperidinyl-1,3-dihydro-2H-benzimidazole-2-one, MW 442 m/z (C) corresponding to 5-hydroxydomperidone (MIII), and MW 252 m/z (D) corresponding to 5-chloro-4-piperidinyl-1,3-dihydro-benzimidazol-2-one (MII), and MW 193 m/z (E) corresponding to 2,3-dihydro-2-oxo-1H-benzimidazole-1-propionic acid (MI)
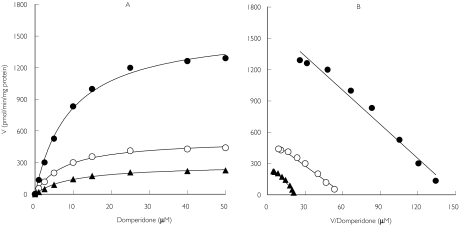
Representative kinetic plots of the apparent formation rates of metabolites as a function of domperidone concentration in HLMs are shown in Figure 2a. The corresponding Eadie-Hofstee plots revealed a linear relationship (Figure 2b). The Km values were similar for all three metabolites (mean 12.4 µm for M-I, 11.9 µm for M-II and 12.6 µm for M-III). The mean formation rate of metabolites is shown in Table 1. The mean formation rate for 5-hydroxydomperidone was 2.0- and 3.3-fold higher than those of the N-dealkylated products (M-I and M-II, respectively). A similar trend was observed with regard to the average apparent Vmax/Km ratio.
Figure 2.
Representative kinetic plots for the metabolism domperidone by HLMs. (A) Michaelis-Menten plots of apparent formation rates (V) of domperidone metabolites vs domperidone concentrations. (B) The corresponding Eadie-Hofstee plot (V of metabolite vs V/domperidone concentrations). Each data point represents the mean of duplicate measurements. MIII (•), MII (▴), MI (○)
Table 1.
Kinetic parameters for the formation of domperidone metabolites in HLMs and baculovirus insect cell CYP3A4
| MI | MII | MIII | |||||||
|---|---|---|---|---|---|---|---|---|---|
| HLMS | Vmax | Km | Vmax/Km | Vmax | Km | Vmax/Km | Vmax | Km | Vmax/Km |
| HL014/99 | 7415 | 15.3 | 485 | 4252 | 14.0 | 304 | 13312 | 15.7 | 848 |
| HL09 | 1428 | 14.5 | 98.5 | 959 | 12.0 | 79.9 | 4259 | 15.5 | 275 |
| HLD | 592 | 13.3 | 44.5 | 606 | 12.9 | 46.9 | 1183 | 11.5 | 103 |
| HL8 | 488 | 6.5 | 75.0 | 261 | 8.5 | 30.6 | 1453 | 7.7 | 189 |
| Mean | 2481 | 12.4 | 176 | 1519 | 11.9 | 115 | 5051 | 12.6 | 354 |
| SD | 3316 | 4.0 | 207 | 1844 | 2.4 | 127 | 5679 | 3.8 | 337 |
| CYP3A4 | 3.4 | 3.0 | 1.13 | 1.58 | 3.12 | 0.5 | 6.53 | 3.6 | 1.81 |
Kinetic parameters were estimated by fitting formation rates vs. substrate concentrations to a one-site binding equation using a nonlinear regression analysis (see Data analysis). Vmax, pmol min−1 mg−1 protein (or pmol min−1 pmol−1 P450); Km, µm; and Vmax/Km, µl min−1 mg−1 protein. MI corresponds to 2,3-dihydro-2-oxo-1H-benzimidazole-1-propionic acid, MII to 5-chloro-4-piperidinyl-1,3-dihydro-benzimidazol-2-one and MIII to 5-hydroxydomperidone.
Correlations between the apparent formation rates of each domperidone metabolite determined in microsomes from 10 different human liver donors and their isoform-specific CYP-catalytic activity (or total CYP content) were tested. There was a significant correlation (Spearman r > 0.93; P < 0.0002) between the apparent formation rates of M-I and M-II, M-I and M-III, and M-II and M-III.
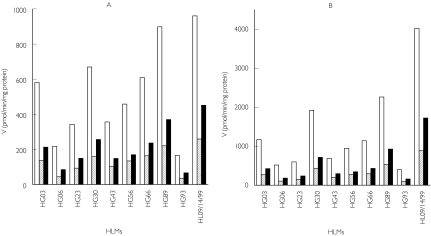
The apparent formation rates of domperidone metabolites showed high interindividual variability among the livers tested (Figure 3). The formation rates of metabolites from 5 µm and 50 µm domperidone were respectively, MI, 173 ± 144 (range: 11.9–489) and 305 ± 230 (range: 20–714); MII, 83 ± 48 (range: 20–163) and 163 ± 89 (range: 38–285); and MIII, 40 ± 26 (range: 3.6–84) and 36 ± 16 (range 3.6–70). In each HLMs tested, M-III was formed consistently at the highest apparent rate at both domperidone concentrations.
Figure 3.
Domperidone metabolism by a panel of characterized HLMs. Incubations from 5 µm (A) and 50 µm (B) domperidone are shown. Data are mean apparent formation rates of domperidone metabolites (pmol min−1 mg−1 protein) of duplicate incubations. MI (▪), MII ( ), MIII (□)
), MIII (□)
The apparent formation rates of all three metabolites showed significant correlation with the activity of CYP3A-catalayzed testosterone 6β-hydroxylation (r ≥ 0.95; P < 0.0001). There was also significant correlation with the activity of CYP2B6-mediated S-mephenytoin N-demethylation (r ≥ 0.75; P < 0.05) and total CYP (r ≥ 0.67; P < 0.05). The significant correlation between the activity of CYP2B6 and domperidone metabolism may be linked to the significant correlation found between the activity of CYP3A and CYP2B6 (Spearman r = 0.72; p = 0.02) [9]. Domperidone metabolism is not inhibited by thioTEPA, a specific inhibitor of CYP2B6 [11]. The activity of other CYP isoforms, FMO, cytochrome b5 or oxidoreductase did not correlate with the apparent metabolite formation rates.
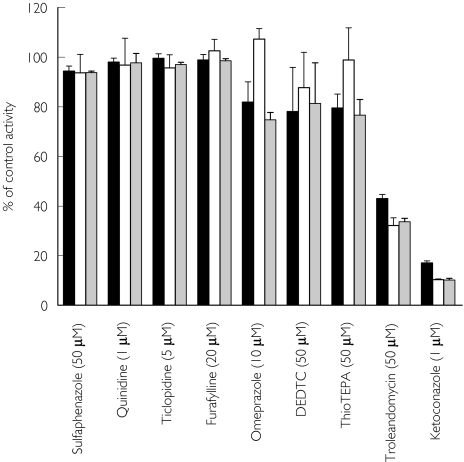
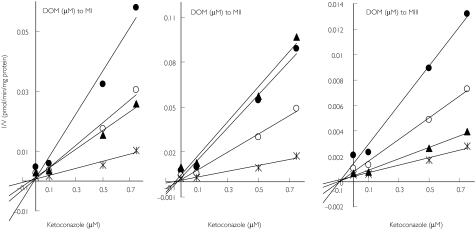
The effect of selective inhibitors of CYPs on domperidone metabolism in HLMs is shown in Figure 4. Ketoconazole was the most potent inhibitor of M-I, M-II and M-III formation (by > 80%) followed by troleandomycin (by 56–68%). Figure 5 demonstrates the inhibition of domperidone metabolism by ketoconazole in HLMs and Ki values were less than 0.05 µm.
Figure 4.
Inhibition of domperidone (5 µm) metabolism by HLMs. The selective inhibitors used were furafylline (10 µm) for CYP1A2, thioTEPA (50 µm) for CYP2B6, sulfaphenazole (50 µm) for CYP2C9, omeprazole (10 µm) and ticlopidine (5 µm) for CYP2C19, quinidine (1 µm) for CYP2D6, diethyldithiocarbamate (DEDTC, 50 µm) for CYP2E1 and ketoconazole (1 µm) and troleandomycin (50 µm) for CYP3A. Inhibition data are expressed as percent control activity remaining (mean ± s.d. of 3 different experiments in duplicate). MIII (▪), MII (□), MI ( )
)
Figure 5.
Dixon plots for the inhibition of domperidone metabolism by ketoconazole in HLMs. Domperidone (2.5, 5, 10 and 15 µm) was incubated at 37 °C for 30 min with HLMs (0.25 mg ml−1) and a NADPH-generating system without or with different concentrations of ketoconazole (0.1, 0.5 and 0.75 µm). 2.5 (•), 5 (○), 10 (▴), 15 ( )
)
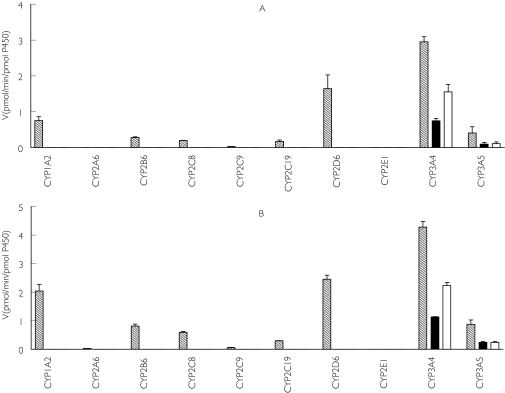
Figure 6 shows the apparent formation rates of M-I, M-II and M-III at low (5 µm, Figure 6a) and high (50 µm, Figure 6b) substrate concentrations following incubation with expressed human CYPs. CYP3A4 produced the highest rates of metabolite formation (M-I, 1.55 ± 0.21 and 2.24 ± 0.11, and M-II, 0.75 ± 0.07 and 1.11 ± 0.012 pmol min−1 pmol−1 P450 at 5 and 50 µm domperidone, respectively). CYP3A5 metabolized domperidone at a much lower rate (less than 0.25 pmol product min−1 pmol−1 P450) (Figure 6a and 6b). The metabolism to M-III was also catalyzed by CYP3A4 at the highest rate (2.95 ± 0.14 and 4.28 ± 0.195 pmol product min−1 pmol−1 P450 at 5 and 50 µm domperidone, respectively) followed by CYP2D6 (1.64 ± 0.38 vs 2.46 ± 0.14 pmol min−1 pmol−1 P450) and CYP1A2 (0.75 ± 0.11 vs 2.04 ± 0.24 pmol min−1 pmol−1 P450) (Figure 6a and 6b).
Figure 6.
Domperidone metabolism by expressed human CYP isoforms. Apparent formation rates of metabolites (pmol min−1 pmol−1 P450) from 5 µm (A) and 50 µm (B) domperidone generated by a panel of expressed CYPs are shown. The data are expressed as mean ± SD (n = 3 different experiments in duplicate). MIII ( ), MII (▪), MI (□)
), MII (▪), MI (□)
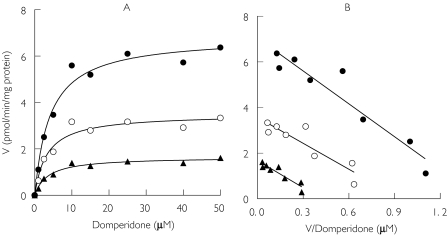
In Figure 7, the kinetics of the formation of MI, MII and MIII from domperidone by expressed CYP3A4 are similar to those observed in HLMs (Figure 2 and Table 1). Formation of MIII by CYP3A4 was more rapid than that of MI and MII. Accordingly, the apparent Vmax/Km of M-III in CYP3A4 was 1.6- and 3.6-fold higher than that of M-I and M-II, respectively, essentially mirroring the findings in HLMs. The estimated Km values for the three domperidone metabolites formed by expressed CYP3A4 were similar to each other, although 3.5- to 4-fold lower than those obtained in HLMs (Table 1). The difference in the in vitro intrinsic clearances down the three routes is a reflection of differences in Vmax, rather than Km.
Figure 7.
Kinetic plots for domperidone metabolism in expressed human CYP3A4. (A) Michaelis-Menten plots of apparent formation rates of metabolites vs domperidone concentrations. (B) The corresponding Eadie-Hofstee plots are displayed. Each data point represents the mean of duplicate measurements. The derived kinetic parameters are shown in Table 1. MIII (•), MII (▴), MI (○)
Discussion
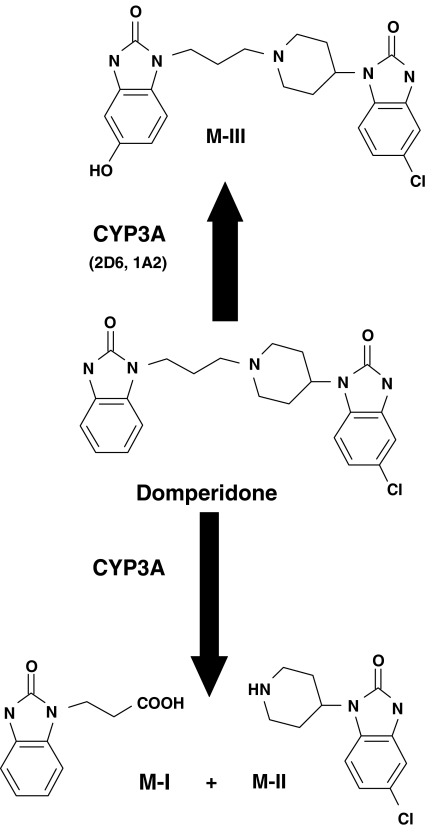
In the present study, characterization of domperidone metabolism in vitro is reported. Hydroxylation to 5-hydroxydomperidone (M-III) and N-dealkylation at the piperidine nitrogen to form both 5-chloro-4-piperidinyl-1,3-dihydro-benzimidazol-2-one (M-II) and 2,3-dihydro-2-oxo-1H-benzamidazole-1-propionic acid (M-I) were the major routes of domperidone metabolism. We have demonstrated that CYP3A is the major catalyst of domperidone N-dealkylation and hydroxylation. The proposed human metabolic routes of domperidone and the CYPs involved are summarized in Figure 8.
Figure 8.
Proposed human metabolism of domperidone and the forms of CYP involved. M-I is 2,3-dihydro-2-oxo-1H-benzimidazole-1-propionic acid, M-II is 5-chloro-4-piperidinyl-1,3-dihydro-benzimidazol-2-one and M-III is 5-hydroxydomperidone
In vivo animal and human metabolite excretion profiles (urinary and faecal) following oral administration of 14C-labelled domperidone [7] suggest that N-dealkylation and hydroxylation are the principal routes of domperidone oxidation. Our data agree with those findings. According to Meuldermans et al.[7] approximately 37 and 23% of domperidone dose was excreted as the 5-hydroxydomperidone and N-dealkylated metabolite (M-I), respectively, within the first 72 h. On the basis of the mean apparent in vitro intrinsic clearance (Vmax/Km) (Table 1), 5-hydroxylation of domperidone accounts for 55–67% of domperidone oxidation and N-dealkylation for 33–45%, values that are comparable with the in vivo human data. However, our results should be interpreted carefully because the lack of availability of authentic metabolite standards precluded precise estimation of their rates of formation and thus of their in vitro intrinsic clearances. Nevertheless, it is reasonable to suggest that the overall clearance of domperidone in vivo can be predicted from the aggregates of 5-hydroxylation and N-dealkylation reactions since these pathways are the major routes of domperidone oxidation in vitro (present data) and in vivo[7] and are catalyzed by the same enzyme system.
Several lines of evidence suggest that the N-dealkylation and hydroxylation of domperidone are predominantly catalyzed by CYP3A. First, domperidone oxidation to all three metabolites exhibited a monophasic kinetic pattern, consistent with the involvement of a single enzyme. Second, apparent formation rates of domperidone metabolites correlated significantly with each other and with that of CYP3A activity in HLMs. Third, formation of all three domperidone metabolites was potently inhibited by the CYP3A specific inhibitors ketoconazole and troleandomycin and, finally expressed-human CYP3A4 catalyzed domperidone N-dealkylation and hydroxylation at a higher rate than the other CYPs tested. Although CYP1A2 and CYP2D6 also generated 5-hydroxydomperidone, their activities were small compared with that of CYP3A4 and furthermore, we found no evidence for the involvement of these isoforms in our inhibition or correlation studies in HLMs. A statistically significant correlation between domperidone metabolism and CYP2B6 activity was observed, but there is little other evidence to suggest that this isoform plays an important role. This observed correlation may be due to the significant relationship shown previously between CYP3A and CYP2B6 in the same HLMs [9]. Together, these data provide strong evidence that CYP3A is the major enzyme responsible for domperidone metabolism, and the contribution of other isoforms, if any, is minimal.
It is well established now that CYP3A4/5 is the predominant CYPs in both small intestinal epithelium and the liver and plays a major role in the presystemic elimination of many drugs [14–17]. Total plasma clearance of domperidone after intravenous administration is 701 ml min−1[18], and since renal or biliary excretion of the unchanged drug is minimal (< 8%), the elimination of domperidone is a reflection of metabolism. Hekants et al.[18] reported that the systemic availability of domperidone after oral administration to healthy volunteers is very low (14% in fasted and 24% after a meal). The predicted bioavailability of domperidone is 37% based on an estimated hepatic extraction ratio of 0.63 after IV dosing [3, 18], which suggests that gut-wall metabolism occurs. An intestinal availability between 0.4 and 0.6 would explain the lower than predicted bioavailability.
In conclusion, we have shown that domepridone is mainly N-dealkylated and hydroxylated by CYP3A. Other studies suggest that domperidone is a substrate for P-glycoprotein [21–23]. These and our data on CYP3A provide the basis on which to predict drug interactions involving domperidone. Several case reports implicate domperidone in cardiac QT-interval prolongation [24–32], and in vitro the drug has been shown to block the rapid component of the delayed rectifier potassium current [6]. For a number of QTc-interval prolonging drugs that include cisapride, terfenadine and pimozide, metabolic drug interactions are important risk factors for cardiac toxicity. Therefore, it may be anticipated that CYP3A inhibitors (e.g. macrolide antibiotics, azole antifungals and certain HIV-1 protease inhibitors) would increase the risk of cardiac adverse effects from domperidone. It is also possible that concomitant administration of inhibitors of P-glycoprotein might increase domperidone-induced extrapyramidal side-effects by enhancing the accumulation of domperidone in the CNS, via diminished P-glycoprotein activity in the blood brain barrier. Conversely, patients may be placed at greater risk for therapeutic failure by concomitant administration of inducers of CYP3A (e.g. rifampicin). Thus domperidone should be used cautiously in patients receiving other drugs known to alter CYP3A or P-glycoprotein activity. Whether domperidone itself alters the pharmacokinetics of co-administered drugs through metabolic inhibition remains unknown. Recently, Obach et al. reported that high domperidone concentrations inhibit human aldehyde oxidase by about 90%[33], although the clinical relevance of this finding remains to be determined.
Acknowledgments
The study was funded by the National Institute of General Medical Sciences grants RO1-GM56898-01, U-01 G61373 and T32 G56898, Bethesda, MD, USA
References
- 1.Brogden RN, Carmine AA, Heel RC, Speight TM, Avery GS. Domperidone. A review of its pharmacological activity, pharmacokinetics and therapeutic efficacy in the symptomatic treatment of chronic dyspepsia and as an antiemetic. Drugs. 1982;24:360–400. doi: 10.2165/00003495-198224050-00002. [DOI] [PubMed] [Google Scholar]
- 2.Prakash A, Wagstaff AJ. Domperidone. A review of its use in diabetic gastropathy. Drugs. 1998;56:429–45. doi: 10.2165/00003495-199856030-00011. [DOI] [PubMed] [Google Scholar]
- 3.Barone JA. Domperidone: a peripherally acting dopamine2-receptor antagonist. Ann Pharmacother. 1999;33:429–40. doi: 10.1345/aph.18003. [DOI] [PubMed] [Google Scholar]
- 4.Woosley RL, Chen Y, Freiman JP, Gillis RA. Mechanism of the cardiotoxic actions of terfenadine. JAMA. 1993;269:1532–6. [PubMed] [Google Scholar]
- 5.Cubeddu LX. QT prolongation and fatal arrhythmias. a review of clinical implications and effects of drugs. Am J Ther. 2003;10:452–7. doi: 10.1097/00045391-200311000-00013. [DOI] [PubMed] [Google Scholar]
- 6.Drolet B, Rousseau G, Daleau P, Cardinal R, Turgeon J. Domperidone should not be considered a no-risk alternative to cisapride in the treatment of gastrointestinal motility disorders. Circulation. 2000;102:1883–5. doi: 10.1161/01.cir.102.16.1883. [DOI] [PubMed] [Google Scholar]
- 7.Meuldermans W, Hurkmans R, Swysen E, et al. On the pharmacokinetics of domperidone in animals and man III. Comparative study on the excretion and metabolism of domperidone in rats, dogs and man. Eur J Drug Metab Pharmacokinet. 1981;6:49–60. doi: 10.1007/BF03189515. [DOI] [PubMed] [Google Scholar]
- 8.Desta Z, Kerbusch T, Soukhova N, Richard E, Ko JW, Flockhart DA. Identification and characterization of human cytochrome P450 isoforms interacting with pimozide. J Pharmacol Exp Ther. 1998;285:428–37. [PubMed] [Google Scholar]
- 9.Ward BA, Gorski JC, Jones DR, Hall SD, Flockhart DA, Desta Z. The cytochrome P4502B6 (CYP2B6) is the main catalyst of efavirenz primary and secondary metabolism. Implication for HIV/AIDS therapy and utility of efavirenz as a substrate marker of CYP2B6 catalytic activity. J Pharmacol Exp Ther. 2003;306:287–300. doi: 10.1124/jpet.103.049601. [DOI] [PubMed] [Google Scholar]
- 10.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–54. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- 11.Rae JM, Soukhova NV, Flockhart DA, Desta Z. Triethylenethiophosphoramide is a specific inhibitor of cytochrome P450 2B6: implications for cyclophosphamide metabolism. Drug Metab Dispos. 2002;30:525–30. doi: 10.1124/dmd.30.5.525. [DOI] [PubMed] [Google Scholar]
- 12.Ko JW, Desta Z, Soukhova NV, Tracy T, Flockhart DA. In vitro inhibition of the cytochrome P450 (CYP450) system by the antiplatelet drug ticlopidine: potent effect on CYP2C19 and CYP2D6. Br J Clin Pharmacol. 2000;49:343–51. doi: 10.1046/j.1365-2125.2000.00175.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Richter T, Murdter TE, Heinkele G, et al. Potent mechanism-based inhibition of human CYP2B6 by clopidogrel and ticlopidine. J Pharmacol Exp Ther. 2004;308:189–97. doi: 10.1124/jpet.103.056127. [DOI] [PubMed] [Google Scholar]
- 14.Hall SD, Thummel KE, Watkins PB, et al. Molecular and physical mechanisms of first-pass extraction. Drug Metab Dispos. 1999;27:161–6. [PubMed] [Google Scholar]
- 15.Thummel KE, Wilkinson GR. In vitro and in vivo drug interactions involving human CYP3A. Annu Rev Pharmacol Toxicol. 1998;38:389–430. doi: 10.1146/annurev.pharmtox.38.1.389. [DOI] [PubMed] [Google Scholar]
- 16.Thummel KE, O'Shea D, Paine MF, et al. Oral first-pass elimination of midazolam involves both gastrointestinal and hepatic CYP3A-mediated metabolism. Clin Pharmacol Ther. 1996;59:491–502. doi: 10.1016/S0009-9236(96)90177-0. [DOI] [PubMed] [Google Scholar]
- 17.Gorski JC, Jones DR, Haehner-Daniels BD, Hamman MA, O'Mara EM, Jr, Hall SD. The contribution of intestinal and hepatic CYP3A to the interaction between midazolam and clarithromycin. Clin Pharmacol Ther. 1998;64:133–43. doi: 10.1016/S0009-9236(98)90146-1. [DOI] [PubMed] [Google Scholar]
- 18.Heykants J, Hendriks R, Meuldermans W, Michiels M, Scheygrond H, Reyntjens H. On the pharmacokinetics of domperidone in animals and man. IV. The pharmacokinetics of intravenous domperidone and its bioavailability in man following intramuscular, oral and rectal administration. Eur J Drug Metab Pharmacokinet. 1981;6:61–70. doi: 10.1007/BF03189516. [DOI] [PubMed] [Google Scholar]
- 19.Schuetz EG. Lessons from the CYP3A4 promoter. Mol Pharmacol. 2004;65:279–81. doi: 10.1124/mol.65.2.279. [DOI] [PubMed] [Google Scholar]
- 20.Kuehl P, Zhang J, Lin Y, et al. Sequence diversity in CYP3A promoters and characterization of the genetic basis of polymorphic CYP3A5 expression. Nat Genet. 2001;27:383–91. doi: 10.1038/86882. [DOI] [PubMed] [Google Scholar]
- 21.Schinkel AH, Wagenaar E, Mol CA, van Deemter L. P-glycoprotein in the blood–brain barrier of mice influences the brain penetration and pharmacological activity of many drugs. J Clin Invest. 1996;97:2517–24. doi: 10.1172/JCI118699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tsujikawa K, Dan Y, Nogawa K, et al. Potentiation of domperidone-induced catalepsy by a P-glycoprotein inhibitor, cyclosporin A. Biopharm Drug Dispos. 2003;24:105–14. doi: 10.1002/bdd.343. [DOI] [PubMed] [Google Scholar]
- 23.Dan Y, Murakami H, Koyabu N, Ohtani H, Sawada Y. Distribution of domperidone into the rat brain is increased by brain ischaemia or treatment with the P-glycoprotein inhibitor verapamil. J Pharm Pharmacol. 2002;54:729–33. doi: 10.1211/0022357021778880. [DOI] [PubMed] [Google Scholar]
- 24.Bruera E, Villamayor R, Roca E, Barugel M, Tronge J, Chacon R. Q-T interval prolongation and ventricular fibrillation with i.v. domperidone. Cancer Treat Report. 1986;70:545–6. [PubMed] [Google Scholar]
- 25.Cameron HA, Reyntjens AJ, Lake-Bakaar G. Cardiac arrest after treatment with intravenous domperidone. Br. Med J (Clin Res Ed) 1985. p. 160. [DOI] [PMC free article] [PubMed]
- 26.Giaccone G, Bertetto O, Calciati A. Two sudden deaths during prophylactic antiemetic treatment with high doses of domperidone and methylprednisolone. Lancet. 1984;ii:1336–7. doi: 10.1016/s0140-6736(84)90841-9. [DOI] [PubMed] [Google Scholar]
- 27.Joss RA, Goldhirsch A, Brunner KW, Galeazzi RL. Sudden death in cancer patient on high-dose domperidone. Lancet. 1982;i:1019. doi: 10.1016/s0140-6736(82)92016-5. [DOI] [PubMed] [Google Scholar]
- 28.Love EM, Yin JA, Delamore IW. Cardiotoxicity of intravenous domperidone. Lancet. 1985;ii:676. doi: 10.1016/s0140-6736(85)90050-9. [DOI] [PubMed] [Google Scholar]
- 29.Osborne RJ, Slevin ML, Hunter RW, Hamer J. Cardiac arrhythmias during cytotoxic chemotherapy: role of domperidone. Hum Toxicol. 1985;4:617–26. doi: 10.1177/096032718500400608. [DOI] [PubMed] [Google Scholar]
- 30.Osborne RJ, Slevin ML, Hunter RW, Hamer J. Cardiotoxicity of intravenous domperidone. Lancet. 1985;ii:385. doi: 10.1016/s0140-6736(85)92515-2. [DOI] [PubMed] [Google Scholar]
- 31.Quinn N, Parkes D, Jackson G, Upward J. Cardiotoxicity of domperidone. Lancet. 1985;ii:724. doi: 10.1016/s0140-6736(85)92959-9. [DOI] [PubMed] [Google Scholar]
- 32.Roussak JB, Carey P, Parry H. Cardiac arrest after treatment with intravenous domperidone. Br Med J (Clin Res Ed) 1984;289:1579. doi: 10.1136/bmj.289.6458.1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Obach RS, Huynh P, Allen MC, Beedham C. Human liver aldehyde oxidase: inhibition by 239 drugs. J Clin Pharmacol. 2004;44:7–19. doi: 10.1177/0091270003260336. [DOI] [PubMed] [Google Scholar]