Abstract
Aims
To assess the feasibility of administering at the same time low doses of five probe drugs, metoprolol (25 mg), chlorzoxazone (250 mg), tolbutamide (250 mg), dapsone (100 mg) and caffeine (100 mg) to determine simultaneously the activities of CYP2D6, CYP2E1, CYP2C9, CYP3A4, CYP1A2, N-acetyltransferase-2 and xanthine oxidase.
Methods
Ten healthy young non-smoking males received the following drugs or combinations of drugs over a 5-week period: week 1) metoprolol; 2) tolbutamide; 3) caffeine, chlorzoxazone and dapsone; 4) caffeine, chlorzoxazone, dapsone and metoprolol; 5) caffeine, chlorzoxazone, dapsone, metoprolol and tolbutamide. The drugs were self-administered at bedtime and urine was collected for the following 8 h.
Results
Mean molar phenotypic ratios obtained after administering metoprolol (mean change of −11%) or tolbutamide (mean change of −0.3%) alone, were not significantly different from those obtained when other drugs were co-administered (P > 0.05). The mean within-subject coefficients of variation were 33%, 18%, 22%, 13%, 16%, 13% and 5% for CYP3A4, CYP2D6, CYP2C9, CYP2E1, CYP1A2, N-acetyltransferase 2 and xanthine oxidase metabolic ratios, respectively. No significant interactions (P > 0.5) were observed during the simultaneous administration of various combinations of the five probe drugs.
Conclusions
We propose that this cocktail, composed of five widely available drugs, constitutes a promising means of simultaneously determining the activities of the major CYP enzymes in large populations.
Keywords: caffeine, chlorzoxazone, cocktail, CYP1A2, CYP2C9, CYP2D6, CYP2E1, CYP3A4, cytochrome P450s, dapsone, metoprolol, N-acetyltransferase, tolbutamide
Introduction
Cytochrome P450 (CYP) enzymes are a superfamily of haemoproteins that are the terminal oxidases of the mono-oxygenase system. They are involved in the oxidative, reductive and peroxidative metabolism of numerous endogenous compounds as well as xenobiotics [1]. CYP enzymes have individual characteristics that are related to the size, lipophilicity, spatial configuration or ionization of the substrate at physiological pH [2]. However, a high degree of overlap exists and most substrates are metabolized by multiple CYP enzymes [3].
The six major human CYP enzymes involved in the metabolism of drugs are in order of importance CYP3A4, CYP2D6, CYP2C9, CYP2C19, CYP2E1 and CYP1A2 [1, 4]. CYP2D6, CYP2C9, CYP2C19 and CYP1A2 are polymorphically expressed leading to high interindividual variation in CYP enzyme activities [5]. Absence of functional CYP enzymes can result in an increased systemic exposure of their respective substrates, leading to a pharmacodynamic effect of increased intensity and/or duration in affected individuals (called poor metabolizers, PMs). An extreme example of the effect of the PM genotype on drug effect is a reported fluoxetine-related death in a CYP2D6-deficient child [6]. Similarly, an African-American woman with substantial toxic effects to phenytoin was found to eliminate phenytoin at a rate 17% of those of normal individuals resulting in a phenytoin elimination half-life of 13 days [7]. This woman was found to carry a CYP2C9*6 mutation. At the other end of the spectrum, over-expression of the CYP enzyme CYP2D6 in so-called ultrarapid metabolizers (UMs) can potentially result in therapeutic failure due to decreased systemic exposure following administration of normal therapeutic doses [8].
Optimal therapeutic effect can be achieved by individualising drug therapy to a patient's genotype/phenotype [9]. Extensive efforts have been directed towards developing genotyping methods for detecting specific DNA mutations that determine individual drug metabolizing capacity [9, 10]. However, the occurrence of rare, unrecognized or functionally uncharacterized mutations in the drug metabolizing enzyme gene(s) of an individual and the cost of screening could be limiting factors in genotyping large populations [11]. Additionally, not all mutations result in a substantial effect on phenotype [12].
Phenotyping itself involves the administration of an appropriate substrate that is selectively metabolized to a single metabolite by a given CYP enzyme. A metabolic ratio, i.e. the ratio of the concentration of the substrate to that of a metabolite(s) in urine, plasma or saliva is used as the index of CYP activity [13]. Although many CYP enzymes exhibit overlapping substrate specificity [13], for some substrates a single enzyme is exclusively or at least principally involved in a particular pathway of their metabolism [14]. For instance, metoprolol α-hydroxylation is carried out by CYP2D6 [15] and tolbutamide tolylmethylhydroxylation by CYP2C9 [16]. Phenotyping provides a measure of the activity of a given drug-metabolizing enzyme, and takes account of not only genetic but also environmental and physiological factors [17].
Through the administration of a cocktail of drugs, it is possible to determine the activity of more than one enzyme at a time. Two-drug, three-drug, four-drug, as well as five-drug cocktails have been reported in the literature [14, 18–21]. However, drugs like debrisoquine [14], sparteine [21], and mephenytoin [14, 20, 21], which were included in many of the previously described cocktails, are neither approved by the FDA for clinical use nor are they available in Canada and some other countries [17]. Both midazolam and mephenytoin, established probes for determining CYP3A4 [22] and CYP2C19 [23] activities, respectively, can cause significant sedation, need clinical monitoring and require multiple blood sampling [24]. This makes the procedure expensive and impractical for large-scale genotyping. In addition, following prolonged storage, mephenytoin (a CYP2C19 substrate) enantiomers are unstable in urine samples. This may lead to significant increases in S/R-mephenytoin ratios in EMs and potential mis-assignment of phenotype [25]. Thus, the aim of this pilot study was to study the feasibility of administering a drug cocktail for CYP phenotyping (a) that consists of drugs which are available in Canada, (b) that assesses the activities of five of the most important human CYPs and (c) that can be self-administered at bedtime allowing for an overnight urine collection without the need for close medical supervision or blood sampling.
Methods
The Ethics Committee of Laval Hospital approved this study and all subjects provided written informed consent prior to entry.
Subjects
Ten healthy, young (mean age ± SD: 26 ± 4.83 years, range: 22–35 years; mean body weight ± SD: 73.06 ± 10.75 kg, range: 63.5–97.5 kg) male volunteers (nine Caucasians and one East Indian) were recruited from the research and hospital personnel. The good health of the participants was confirmed by the results of a questionnaire as well as by a physical and clinical examination (12 lead ECG, heart rate, blood pressure and blood biochemistry). Blood samples were also taken for the determination of glucose-6-phosphate dehydrogenase activity using the diagnostic kit provided by Sigma diagnostics® (St Louis, MO, USA). In all subjects, activities ranged from 7 to 13 U g−1 Hb at 30 °C. Subjects abstained from consuming alcohol, caffeinated beverages, charbroiled foods, grapefruit juice, and any prescription or over-the-counter medication including vitamins and herbal products for at least 48 h prior to each study period.
Administration of the drug cocktail
Each subject participated in five consecutive randomly assigned study arms consisting of the administration of various combinations of single oral subtherapeutic doses of metoprolol tartrate (Lopressor®, Novartis Pharma, ON), tolbutamide (Apo-tolbutamide®, Apotex Pharma, ON), caffeine (Wake-ups®, Adrem, ON), dapsone (Avlosulphon®, Wyeth-Ayerst, ON), and chlorzoxazone (Paraflex®, Ortho-McNeil, NJ): metoprolol 25 mg only; tolbutamide 250 mg only; a cocktail of caffeine (100 mg), dapsone (100 mg) and chlorzoxazone (250 mg); a cocktail of caffeine (100 mg), dapsone (100 mg), chlorzoxazone (250 mg) and metoprolol (25 mg); and a 5-drug cocktail consisting of caffeine (100 mg), dapsone (100 mg), chlorzoxazone (250 mg), metoprolol (25 mg), and tolbutamide (250 mg). Caffeine, chlorzoxazone and dapsone were given together, without testing individually, since the absence of a significant interaction between these three drugs has been previously reported [14]. In contrast, metoprolol and tolbutamide have not been co-administered previously with this drug combination and hence were studied individually before introduction into the cocktail. There was a washout period of at least 1 week between each study period. Each drug or cocktail was self-administered at bedtime. All subjects were questioned about any overt adverse effects potentially related to the administered drugs after each study period. Control urine was obtained immediately before the administration of the drugs and a complete overnight urine collection was done for 8 h following drug administration. Urine was collected into containers containing 1 g of ascorbic acid in order to stabilize the N-hydroxylamine metabolite of dapsone [26, 27]. The caffeine metabolite 5-acetylamino-6-formylamino-3-methyluracil (AFMU) can undergo spontaneous nonenzymatic degradation to 5-acetylamino-6-amino-3-methyluracil (AAMU) in urine [28]. Thus urinary pH was raised during sample analysis to promote this conversion [29] and hence, any spontaneous degradation of AFMU to AAMU that might have occurred during the storage phase was accounted for. The volume and pH of urine samples were measured and aliquots were stored at −20 °C until analysis.
Drug and metabolite analysis
All test compounds and their respective metabolites were determined by modified HPLC/UV methods. Analysis was performed at ambient temperature using a Shimadzu HPLC system consisting of an automatic sample injector (model SIL-9 A), a programmable pump (model LC-600), a variable UV detector (model SPD-6 A), and an integrator (model Chromatopac C-R5A). Metoprolol was detected by a fluorescence detection (model Shimadzu RF-10Ax1).
Dapsone and metabolites
Dapsone and dapsone hydroxylamine were analyzed in urine using the method of May et al.[27] with minor modifications. Analysis was performed on a 5 µ Beckman® ultrasphere ODS column (4.6 × 250 mm) with UV detection (254 nm). The mobile phase was water : acetonitrile : glacial acetic acid : TEA (77 : 14 : 9 : 0.5% v/v) and was delivered at a flow rate of 1 ml min−1. Sulfamethoxazole (20 µl of a 1 mg ml−1 solution; internal standard) was added to urine samples (500 µl) prior to incubation for 1 h at 37 °C in the presence of 42 µl of 6 n HCl. Following neutralization with 42 µl of 6 n NaOH, samples were centrifuged and 45 µl of the supernatant was injected directly onto the HPLC system. The intraday variability was 3.1 and 2.3% after assessing 10 urine samples spiked with 4 nm of dapsone and dapsone hydroxylamine, respectively. Interday variability was 0.01% at 8 nm, 3.5% at 4 nm and 5.7% at 0.4 nm for dapsone, and 0.01% at 8 nm, 0.01% at 4 nm and 8.5% at 1 nm for dapsone hydroxylamine, respectively. Accuracy ranged from 98 to 104%.
Metoprolol and α–hydroxymetoprolol
Racemic metoprolol and its metabolite α–hydroxymetoprolol were analyzed using a HPLC method that was established and validated in our laboratory [30].
Tolbutamide and metabolites
Tolbutamide was analyzed by a modification of the method reported by Veronese and colleagues [31]. Analysis was performed on a Phenomenex® Luna 5µ phenylhexyl column (150 × 4.60 mm) and a Zorbax® phenyl column (250 × 4.60 mm) fitted in series and UV detection (230 nm). The mobile phase was 0.01 m sodium acetate buffer (pH 3) and acetonitrile (68 : 32% v/v) and was delivered at a flow rate of 1 ml min−1. Tolbutamide was extracted from 3 ml urine following the addition of 30 µl of 3 n HCl and 30 µg of chlorpropamide (internal standard) with two times 4 ml diethyl ether. The ether phases were combined, dried under a gentle stream of nitrogen and reconstituted in 100 µl mobile phase and an aliquot (50 µl) was injected onto the HPLC system. The method had an intraday CV of 2.5% at 1.23 nm tolbutamide. The CV for the interday variability was 7.7% at 0.06 nm, 5.7% at 0.6 nm and 4% at 1.23 nm. Accuracy ranged from 95% to 106%.
The hydroxymethyl- (CH2OH-) and carboxy- (COOH-tolbutamide) metabolites of tolbutamide were analyzed as reported previously with minor modifications [32]. The separation was performed on a 5 µ Beckman® ultrasphere ODS column (250 × 4.60 mm) with UV detection (254 nm). The mobile phase consisted of A: Sorensen phosphate buffer : acetonitrile (95 : 5% v/v; pH 7) containing 0.3% pic A (Waters®, Milford, MA), and B: Sorensen phosphate buffer : acetonitrile (60 : 40% v/v; pH 7) containing 0.3% pic A. The mobile phase was delivered at 1 ml min−1 and the following gradient was applied: 100%A for 14.9 min, 50%A/50%B at 15 min and transition to 100%B up to 35 min, 100%A at 36 min and re-equilibration for 10 min. Tolbutamide metabolites were extracted from 250 µl urine following the addition of chlorpropamide (internal standard) and 200 µl of 3 n HCl by mixing with two times 5 ml of diethyl ether : n-hexane (80 : 20 v/v). The combined organic layers were dried under a gentle steam of nitrogen and reconstituted in 200 µl mobile phase prior to injection of 50 µl. Ten urine samples spiked with 0.13 µm COOH-tolbutamide and 0.68 µm CH2OH-tolbutamide were analyzed to assess the intraday reproducibility. The CV was 5.6% for COOH- and 1.7% for CH2OH-tolbutamide, respectively. The CV for interday variability was 10.3% at 0.02 µm, 3.0% at 0.1 µm, 6.4% at 0.5 µm, 1.0% at 1.27 µm of COOH-tolbutamide. Accuracy ranged from 97 to 107%. Similarly, the CV of interday variability was 9.1% at 0.03 µm, 5.1% at 0.14 µm, 1.6% at 0.66 µm of CH2OH-tolbutamide. Accuracy ranged from 99 to 103%.
Chlorzoxazone and metabolites
The analysis of 6-hydroxychlorzoxazone was by a previously reported method [33]. Separation was performed on a 5 µ Beckman® ultrasphere ODS column (4.6 × 250 mm) with UV detection (287 nm). The mobile phase was water : acetonitrile : glacial acetic acid (75 : 25 : 0.5%v/v) and was delivered at a flow rate of 1 ml min−1. Twenty µl of β-glucuronidase/arylsulphatase obtained from the juice of Helix pomatia (Roche Diagnostics®) and 500 µl 0.1 m acetate buffer (pH 5) were added to 50 µl urine and incubated at 37 °C for 18 h. Following incubation, phenacetin (internal standard), 300 µl modified tris/HCL buffer (0.28 m tris and 0.26 m HCl; pH 8.3) and 1020 µl water were added and the mixture was extracted twice with 4 ml of diethyl ether. The combined ether layers were dried under a gentle stream of nitrogen, reconstituted in 200 µl mobile phase and 30 µl of this solution was injected onto the HPLC. Raising the pH of the final solution before extraction with the modified tris/HCl buffer was found to decrease considerably interference on the chromatograms. Intraday variability was determined at 2.2 µm of 6-hydroxychlorzoxazone and the CV was 4.2%. The CV of interday variability was 12.4% at 0.44 µm, 3.5% at 1.3 µm and 0.8% at 2.16 µm. Accuracy ranged from 97 to 105%.
This method lacked sensitivity for the analysis of chlorzoxazone. Other groups have also expressed an inability to detect chlorzoxazone in urine samples following HPLC analysis [34, 35] and hence the samples were assayed for the amount of chlorzoxazone using ESI/LC/MS/MS as previously described [36]. The method used for the extraction of chlorzoxazone from urine was the same as that for its metabolite except that a 10-fold greater urine volume was used. The LC/MS/MS analysis was performed using a TSQ 7000 triple quad mass spectrometer (Thermo Finnigan, San Jose, CA). The inter- and intraday coefficients of variation were less than or equal to 5%.
Caffeine and metabolites
Caffeine and its metabolites, namely 1-methylxanthine (1X), 1-methylurate (1 U), 1,7-dimethylxanthine (17X), 1,7-dimethylurate (17 U), 5-acetylamino-6-amino-3-methyluracil (AAMU) were analyzed according to a previously reported HPLC method [29].
Data analysis
Molar metabolic/phenotypic ratios were calculated (Table 1). Data were expressed as mean ± SD (95% confidence intervals). A two-way Anova design was used to analyse two experimental factors, one associated with the subjects (factor subject), and a second one associated with the different study weeks, which were assigned within each subject. This second factor was analyzed as a repeated-measures factor. Different statistical models with different covariance structures were tested to obtain the one with the best fit as determined by the adjusted Akaike's information criterion (AICC). A variance component covariance structured model was selected. The univariate normality assumptions were verified with the Shapiro-Wilk tests and multivariate normality was verified with Mardia tests. All assumptions were fulfilled. The results were considered significant with P values < 0.05. All analyses were conducted using the statistical package SAS (SAS Institute Inc, Cary, NC, U.S.A).
Table 1.
List of probe substrates and urinary metabolic ratios used to measure CYP activities
| Isoform | Substrate | Metabolic ratio (MR) | Reference |
|---|---|---|---|
| CYP3A4 | Dapsone | Dapsone hydroxylamine/(dapsone+dapsone hydroxylamine) | May et al. [26], Frye et al. [14] |
| CYP2D6 | Metoprolol | Metoprolol/α-hydroxymetoprolol | Labbéet al. [46] |
| CYP2C9 | Tolbutamide | (COOH-tolbutamide+CH2OH-tolbutamide)/tolbutamide | Veronese et al. [31] |
| CYP2E1 (MR 2E1.1) | Chlorzoxazone | 6-hydroxychlorzoxazone/chlorzoxazone | |
| CYP2E1 (MR 2E1.2) | Chlorzoxazone | Oral dose/8 h 6-hydroxychlorzoxazone urinary excretion | Dreisbach et al. [34] |
| NAT2 | Caffeine | AAMU/(AAMU+1X+1U) | Hamelin et al.[29] |
| CYP1A2 | Caffeine | (AAMU+1X+1U)/17U | Hamelin et al. [29] |
| XO | Caffeine | 1U/(1U+1X) | Hamelin et al. [29] |
NAT2 = N-acetyltransferase 2, XO = xanthine oxidase.
Results
All 10 participants completed the study. None of the subjects reported undesirable effects during any of the study periods. There were no statistically significant differences (P > 0.05) in the average urine output between the 5 study weeks: (week 1: 662 ± 377 ml, week 2: 619 ± 402 ml, week 3: 637 ± 337 ml, week 4: 597 ± 298 ml, week 5: 609 ± 377 ml). Urinary pH did not change between treatments (week 1: 6.29 ± 0.37, week 2: 6.17 ± 0.38, week 3: 6.28 ± 0.36, week 4: 6.35 ± 0.43, week 5: 6.48 ± 0.41; P > 0.05) and there were no interfering peaks during the analysis of the samples by HPLC or LC-MS/MS following the administration of the drug cocktails. The average molar metabolic/phenotypic ratios for metoprolol (changed by −11.3%) and tolbutamide (changed by −0.3%) did not change significantly from administration of the drugs alone compared with when they were administered as part of the five-drug cocktail (Tables 2, P > 0.05). Furthermore, there were no significant differences for any metabolic ratio between the different study periods as illustrated in Figures 1–5 and Table 2.
Table 2.
Metabolic ratios (MR) (mean ± SD [95% confidence intervals on differences*]) following the administration of probe substrates alone or in combination
| Enzyme | n | Metoprolol alone | Tolbutamide alone | 3 drug | 4 drug | 5 drug | P value |
|---|---|---|---|---|---|---|---|
| CYP3A4 | 10 | ND | ND | 0.32 ± 0.15[week 3-4: -0.09, 0.23] | 0.25 ± 0.16[week 4-5: -0.23, 0.10] | 0.32 ± 0.13[week 3-5: -0.16, 0.17] | 0.5 |
| CYP2D6 EM | 8 | 1.02 ± 1.21 | ND | ND | 0.90 ± 1.12 | 0.93 ± 1.21 | |
| CYP2D6 PM | 2 | 255.98 ± 21.65[week 1-4: -110, 122] | ND | ND | 255.58 ± 82.96(week 4-5: -116, 115] | 227.07 ± 63.04[week 1-5: -110, 122] | 0.98 |
| CYP2C9 EM | 9 | ND | 1383.6 ± 616.89 | ND | ND | 1369.06 ± 398.2 | 0.98 |
| CYP2C9 PM | 1 | ND | 53.21 | ND | 141[week 1-5: -110, 122][week 2-5: -662, 670] | ||
| CYP2E1 (MR 2E1.1) | 10 | ND | ND | 41306 ± 35123[week 3-4: -22397, 46003] | 29503 ± 21450[week 4-5: -44310, 25867] | 38724 ± 27430[week 3-5: -31618, 36782] | 0.65 |
| CYP2E1 (MR 2E1.2) | 10 | ND | ND | 1.11 ± 0.37[week 3-4: -0.48, 0.58] | 1.06 ± 0.53[week 4-5: -0.63, 0.42] | 1.16 ± 0.47[week 3-5: -0.58, 0.47] | 0.87 |
| NAT2 | 10 | ND | ND | 0.49 ± 0.16[week 3-4: -0.20, 0.08] | 0.55 ± 0.13[week 4-5: -0.12, 0.17] | 0.52 ± 0.09[week 3-5: -0.18, 0.11] | 0.56 |
| CYP1A2 | 10 | ND | ND | 6.44 ± 2.96[week 3-4: -2.52, 2.77] | 6.31 ± 1.79[week 4-5: -2.29, 3] | 5.96 ± 2.04[week 3-5: -2.17, 3.13] | 0.89 |
| XO | 10 | ND | ND | 0.56 ± 0.04[week 3-4: -0.03, 0.08] | 0.53 ± 0.05[week 4-5: -0.07, 0.04] | 0.55 ± 0.05[week 3-5: -0.05, 0.06] | 0.5 |
Tukey adjustment for multiple comparisons, NAT2 = N-acetyltransferase 2, XO = xanthine oxidase, EM = extensive metabolizer, PM = poor metabolizer, ND = not done, P value is for the week effect, 3 drug = cocktail of caffeine, chlorzoxazone and dapsone, 4 drug = caffeine, chlorzoxazone, dapsone and metoprolol, 5 drug = caffeine, chlorzoxazone, dapsone, metoprolol and tolbutamide.
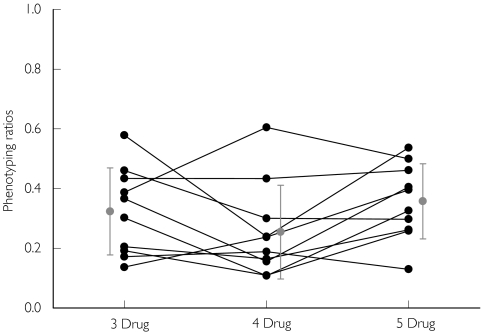
Figure 1.
CYP3A4 phenotyping ratios (dapsone hydroxylamine/(dapsone + dapsone hydroxylamine)) obtained following the administration of dapsone in various drug combinations (3 drug: caffeine, dapsone and chlorzoxazone; 4 drug: caffeine, dapsone, chlorzoxazone and metoprolol; 5 drug: caffeine, dapsone, chlorzoxazone, metoprolol and tolbutamide). Individual values (•), Mean ± SD ( )
)
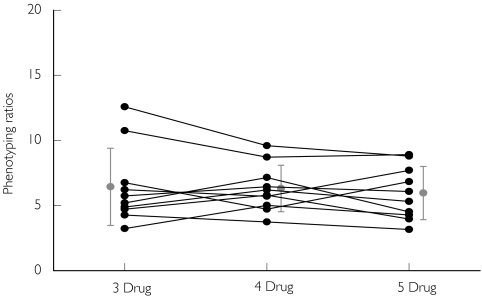
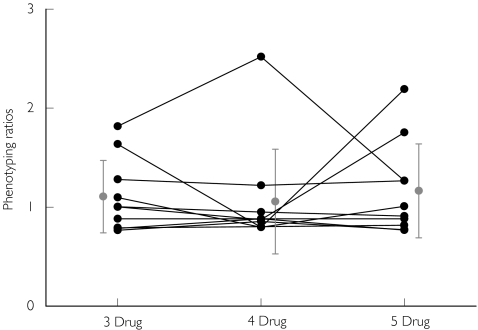
Figure 5.
CYP1A2 phenotyping ratios ((AAMU +1X +1 U)/17 U) obtained following the administration of caffeine in various drug combinations (3 drug: caffeine, chlorzoxazone and dapsone; 4 drug: caffeine, chlorzoxazone, dapsone and metoprolol; 5 drug: caffeine,chlorzoxazone, dapsone, metoprolol and tolbutamide). Individual values (•), Mean ± SD ( )
)
Discussion
The phenotyping probes used in this pilot study were selected based on a number of criteria, including (a) safety (at the doses administered); (b) drugs approved and/or easily available in Canada; (c) possibility of assessing the activities of the most common CYP enzymes; (d) feasibility of self-administration and practicality of large population phenotyping; (e) no or minimal interactions between co-administered drugs. The proposed cocktail was comprised of subtherapeutic doses of five drugs, namely dapsone, metoprolol, tolbutamide, chlorzoxazone and caffeine, which are all readily available in Canada. The results from this pilot study were encouraging as the cocktail was self-administered, well tolerated by the volunteers with no reported adverse effects, clinical surveillance was not necessary, and significant drug/drug interactions were absent.
This and previous cocktails may be questioned as to the specificity of some of the probe drugs used. In particular, although dapsone has been employed as a probe drug for CYP3A4 in previous investigations [14, 26], its use has recently been criticised. First, the contribution of the fractional metabolic clearance of dapsone to dapsone hydroxylamine accounted for only 23% (range: 11–36%) of the oral clearance of the drug [27], although differences in the rate of N-hydroxylation accounted for 94% of the interindividual variation in this parameter. Second, it has been argued that the involvement of several CYP enzymes in the N-hydroxylation of dapsone may make this drug less useful as a probe for CYP3A4. Furthermore a series of in vitro studies using human liver microsomes have shown discrepant data. At a concentration of 100 µm, Fleming and coworkers [37] illustrated the importance of CYP3A4 in the metabolism of dapsone. However, Gill and coworkers [38] showed that both CYP3A4 and CYP2C9 catalyzed this reaction at that same substrate concentration. In contrast, others have reported, that CYP2C9 was the major and CYP2C8 a minor contributor to the dapsone N-hydroxylation at a concentration of 4 µmin vitro, whereas both CYP3A4 and CYP2E1 were found not to be involved [39]. The kinetics of dapsone hydroxylamine formation were biphasic (Vmax/Km values of 9.3 and 32.5 ml min−1 mg−1 protein) in three human liver microsomal preparations [40] supporting the evidence that at least two enzymes catalyze the reaction.
Clinically, dapsone peak plasma concentrations range from 8 to 13 µm[41]. However, liver concentrations of lipophilic amine drugs have been reported to be several fold (10–130 times) higher than the plasma concentrations [6, 42–44]. Animal data suggest that this is also the case for dapsone [44, 45]. Therefore, there is evidence that liver concentrations of dapsone could be much greater than the plasma concentrations, leading to significant involvement of CYP3A4 in its metabolism.
In light of this evidence, we and others [14, 26] have used dapsone as a probe for CYP3A4. The dapsone metabolic ratios obtained for the 10 subjects in our study were in the range of the values previously reported for extensive metabolisers [26] and did not vary significantly by administering dapsone in various combinations (Figure 1 and Table 2).
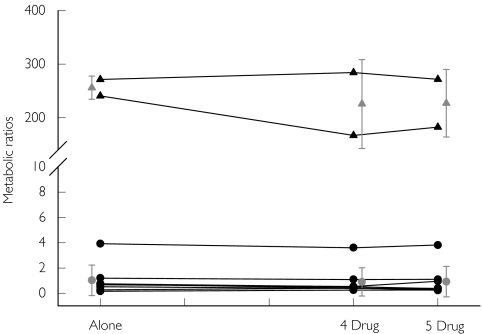
Metoprolol has been extensively employed as a probe drug for CYP2D6 [30, 46]. Its hydroxylation is exclusively mediated by CYP2D6 and the partial metabolic clearance of metoprolol to α-hydroxymetoprolol is about 200-fold smaller in poor metabolizers than in extensive metabolizers [47]. The molar metabolic ratio of metoprolol/α-hydroxymetoprolol was bimodally distributed with an antimode of 1.02 [15]. The metoprolol : α-hydroxymetoprolol ratios obtained in the current study were in the range of those reported in the literature [46]. Two subjects were found to be poor metabolizers of metoprolol (Figure 2 and Table 2).
Figure 2.
CYP2D6 metabolic ratios (metoprolol/a-hydroxy-metoprolol) obtained following the administration of metoprolol in various drug combinations (alone: metoprolol only 4 drug: metoprolol, caffeine, chlorzoxazone and dapsone; 5 drug: metoprolol, caffeine,chlorzoxazone, dapsone and tolbutamide). EMs-Individual values (•), PMs-Individual values (▵), EMs (Mean ± SD) ( ), PMs (Mean ± SD) (
), PMs (Mean ± SD) ( )
)
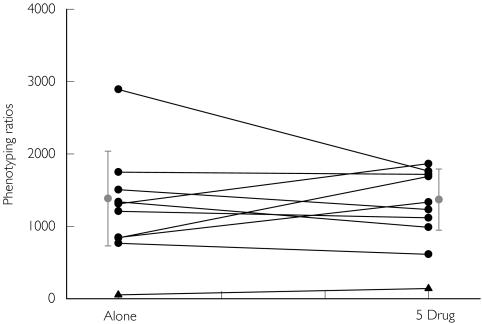
In humans, the elimination of tolbutamide occurs mainly (85%) along a single pathway, with the initial and rate-limiting step being tolylmethylhydroxylation to form CH2OH-tolbutamide [48], catalyzed by CYP2C9 [10, 49]. CH2OH-tolbutamide is further oxidized by alcohol and aldehyde dehydrogenases to form COOH-tolbutamide. Veronese and coworkers [31] proposed a molar phenotypic ratio of (CH2OH-tolbutamide + COOH-tolbutamide) : tolbutamide as an index of CYP2C9 activity (Tables 1 and 2; MR 2C9.1). The ratio following the administration of 500 mg tolbutamide to 106 healthy subjects ranged from 324 to 3033 with one poor metabolizer subject having a value of 159 [50]. Values for the tolbutamide metabolic ratio found in the current study were similar, and we identified one poor metabolizer (Figure 3 and Table 2).
Figure 3.
CYP2C9 phenotyping ratios ((COOH-tolbutamide + CH2OH-tolbutamide)/tolbutamide) obtained following the administration of tolbutamide in various drug combinations (alone: tolbutamide only, 5 drug: caffeine, chlorzoxazone, dapsone, metoprolol and tolbutamide). EM subjects (•), PM subject (▴), Mean ± SD ( )
)
Chlorzoxazone has been used as a probe drug for assessing CYP2E1 activity in the past [34, 35], CYP2E1 is predominantly responsible for its metabolism to 6-hydroxychlorzoxazone [51]. In previous studies, HPLC analysis was not sufficiently sensitive to detect chlorzoxazone in urine samples [34, 35]. Dreisbach and coworkers [34] measured the 8 h urinary excretion of 6-hydroxychlorzoxazone following an oral dose of 500 mg and used a 0–8 h urinary molar ratio of chlorzoxazone dose : 6-hydroxychlorzoxazone, which ranged from 1.43 to 2.24. Frye and coworkers [35] demonstrated that a plasma ratio of 6-hydroxychlorzoxazone : chlorzoxazone 2 and 4 h after dosing is a better index of CYP2E1 activity. However, given that the aim of our study was to evaluate the feasibility of a cocktail that could be self-administered by subjects followed by an overnight urine collection, thus avoiding blood sampling, we employed the urinary dose ratio. The values obtained were similar to those reported by Dreisbach and coworkers [34] (Figure 4 and Table 2). We detected chlorzoxazone in all but two samples from two individuals using ESI/LC/MS/MS and hence, we also used the 8 h urinary 6-hydroxychlorzoxazone : chlorzoxazone molar ratio (Table 2). Neither ratio varied significantly between the different study phases.
Figure 4.
CYP2E1 urinary phenotyping ratios (oral dose/6-hydroxychlorzoxazone excreted in urine) obtained following the administration of chlorzoxazone in various drug combinations (3 drug: caffeine, chlorzoxazone and dapsone; 4 drug: caffeine, chlorzoxazone, dapsone and metoprolol; 5 drug: caffeine, chlorzoxazone, dapsone, metoprolol and tolbutamide). Individual values (•), mean ± SD ( )
)
Caffeine has been extensively employed as a probe drug for N-acetyltransferase-2, CYP1A2 and xanthine oxidase activities [19, 29]. The results obtained in the current study by using various ratios were in agreement with previously reported results (Figure 5 and Table 2).
The between-subject coefficients of variation were 51, 211, 53, 42, 33, 23, and 9% for CYP3A4, CYP2D6, CYP2C9, CYP2E1, CYP1A2, N-acetyltransferase 2 and xanthine oxidase activities, respectively. The variation in CYP2D6 activity was particularly high due to the presence of two poor metabolizers among the 10 subjects. The within-subject coefficients of variation were 33, 18, 22, 13, 16, 13 and 5% for CYP3A4, CYP2D6, CYP2C9, CYP2E1, CYP1A2, N-acetyltransferase 2 and xanthine oxidase activities, respectively. Hence, the between-subject variation was around 2–12 times higher than that within-subjects. Both types of variation were similar to those reported previously [14, 20, 52].
CYP2C19 is another important polymorphic CYP enzyme. When we started this project, the simultaneous assessment of CYP2C19 and CYP2C9 in the same cocktail did not appear feasible due to substrate overlap. Hence, we did not include a probe substrate to measure CYP2C19 activity in our cocktail. However, this has been recently attempted with warfarin as a probe for CYP2C9 and omeprazole as a probe drug for CYP2C19 [53]. Thus, we feel that our cocktail could benefit from incorporation of an appropriate CYP2C19 probe.
Only 10 subjects were studied in the present investigation and a larger number are needed to validate unequivocally the absence of clinically meaningful interactions between the substrates used. However, the current report reflects a successful pilot study of the feasibility of giving a cocktail of drugs that can be self-administered and therefore convenient for population studies, that is devoid of significant adverse drug effects and drug/drug interactions, that contains easily obtainable drugs and that allows activities of the major CYP enzymes involved in drug metabolism to be determined.
Acknowledgments
This study was supported by a grant from the Canadian Institutes of Health Research and the Quebec Heart Institute. A. Sharma was the recipient of a joint PhD scholarship from the Canada's Research Based Pharmaceutical Companies and Canadian Institutes of Health Research. The work was presented in part at the American Society of Clinical Pharmacology and Therapeutics 102nd annual meeting in Orlando, Florida, 6–10 March 2001.
The authors wish to thank Mr Serge Simard (biostatistician, Laval Hospital) for statistical analysis. Generous gifts of dapsone hydroxylamine from Jacobus Pharmaceuticals, Princeton, NJ and of metoprolol and a-hydroxymetoprolol from Astra-Zeneca, Molndal, Sweden, were much appreciated. We also thank Ms Sylvie Plante at Shire BioChem Inc. for the ESI/LC/MS/MS analysis of chlorzoxazone.
References
- 1.Nebert DW, Russell DW. Clinical importance of the cytochromes P450. Lancet. 2002;360:1155–62. doi: 10.1016/S0140-6736(02)11203-7. [DOI] [PubMed] [Google Scholar]
- 2.Ekins S, Erickson JA. A pharmacophore for human pregnane X receptor ligands. Drug Metab Disp. 2002;30:96–9. doi: 10.1124/dmd.30.1.96. [DOI] [PubMed] [Google Scholar]
- 3.Shimada T, Yamazaki H, Mimura M, Inui Y, Guengerich FP. Interindividual variations in human liver cytochrome P-450 enzymes involved in the oxidation of drugs, carcinogens and toxic chemicals. Studies with liver microsomes of 30 Japanese and 30 Caucasians. J Pharmacol Exp Ther. 1994;270:414–23. [PubMed] [Google Scholar]
- 4.Koukouritaki SB, Manro JR, Marsh SA, Stevens JC, Rettie AE, McCarver DG, Hines RN. Developmental Expression of Human Hepatic CYP2C9 and CYP2C19. J Pharmacol Exp Ther. 2004;308:965–74. doi: 10.1124/jpet.103.060137. [DOI] [PubMed] [Google Scholar]
- 5.Rodrigues AD, Rushmore TH. Cytochrome P450 pharmacogenetics in drug development: in vitro studies and clinical consequences. Curr Drug Metab. 2002;3:289–309. doi: 10.2174/1389200023337522. [DOI] [PubMed] [Google Scholar]
- 6.Sallee FR, DeVane CL, Ferrell RE. Fluoxetine-related death in a child with cytochrome P-450 2D6 genetic deficiency. J Child Adolesc Psychopharmacol. 2000;10:27–34. doi: 10.1089/cap.2000.10.27. [DOI] [PubMed] [Google Scholar]
- 7.Kidd RS, Gallagher SJ, Gallagher S, Edeki T, Blaisdell J, Goldstein JA. Identification of a null allele of CYP2C9 in an African-American exhibiting toxicity to phenytoin. Pharmacogenetics. 2001;11:803–8. doi: 10.1097/00008571-200112000-00008. [DOI] [PubMed] [Google Scholar]
- 8.Bertilsson L, Aberg-Wistedt A, Gustafsson LL, Nordin C. Extremely rapid hydroxylation of debrisoquine: a case report with implication for treatment with nortriptyline and other tricyclic antidepressants. Ther Drug Monit. 1985;7:478–80. [PubMed] [Google Scholar]
- 9.Kirchheiner J, Bauer S, Meineke I, Rohde W, Prang V, Meisel C, Roots I, Brockmoller J. Impact of CYP2C9 and CYP2C19 polymorphisms on tolbutamide kinetics and the insulin and glucose response in healthy volunteers. Pharmacogenetics. 2002;12:101–9. doi: 10.1097/00008571-200203000-00004. [DOI] [PubMed] [Google Scholar]
- 10.Stamer UM, Bayerer B, Wolf S, Hoeft A, Stuber F. Rapid and reliable method for cytochrome P450 2D6 genotyping. Clin Chem. 2002;48:1412–17. [PubMed] [Google Scholar]
- 11.Chen S, Chou WH, Blouin RA, Mao Z, Humphries LL, Meek QC, Neill JR, Martin WL, Hays LR, Wedlund PJ. The cytochrome P450 2D6 (CYP2D6) enzyme polymorphism: screening costs and influence on clinical outcomes in psychiatry. Clin Pharmacol Ther. 1996;60:522–34. doi: 10.1016/S0009-9236(96)90148-4. [DOI] [PubMed] [Google Scholar]
- 12.Marez D, Legrand M, Sabbagh N, Lo Guidice JM, Spire C, Lafitte JJ, Meyer UA, Broly F. Polymorphism of the cytochrome P450 CYP2D6 gene in a European population. Characterization of 48 mutations and 53 alleles, their frequencies and evolution. Pharmacogenetics. 1997;7:193–202. doi: 10.1097/00008571-199706000-00004. [DOI] [PubMed] [Google Scholar]
- 13.Kivisto KT, Kroemer HK. Use of probe drugs as predictors of drug metabolism in humans. J Clin Pharmacol. 1997;37:40S–48S. doi: 10.1177/009127009703700121. [DOI] [PubMed] [Google Scholar]
- 14.Frye RF, Matzke GR, Adedoyin A, Porter JA, Branch RA. Validation of the five-drug ‘Pittsburgh cocktail’ approach for assessment of selective regulation of drug-metabolizing enzymes. Clin Pharmacol Ther. 1997;62:365–76. doi: 10.1016/S0009-9236(97)90114-4. [DOI] [PubMed] [Google Scholar]
- 15.Lennard MS, Silas JH, Freestone S, Ramsay LE, Tucker GT, Woods HF. Oxidation phenotype – A major determinant of metoprolol metabolism and response. N Engl J Med. 1982;307:1558–60. doi: 10.1056/NEJM198212163072505. [DOI] [PubMed] [Google Scholar]
- 16.Miners J. CYP2C9 polymorphism. Impact on tolbutamide pharmacokinetics and response. Pharmacogenetics. 2002;12:91–2. doi: 10.1097/00008571-200203000-00002. [DOI] [PubMed] [Google Scholar]
- 17.Streetman DS, Bertino JSJ, Nafziger AN. Phenotyping of drug-metabolizing enzymes in adults. A review of in-vivo cytochrome P450 phenotyping probes. Pharmacogenetics. 2000;10:187–216. doi: 10.1097/00008571-200004000-00001. [DOI] [PubMed] [Google Scholar]
- 18.Berthou F, Goasduff T, Lucas D, Dréano Y, Le Bot MH, Ménez JF. Interaction between two probes used for phenotyping cytochromes P4501A2 (caffeine) and P4502E1 (chlorzoxazone) in humans. Pharmacogenetics. 1995;5:72–9. doi: 10.1097/00008571-199504000-00003. [DOI] [PubMed] [Google Scholar]
- 19.Streetman DS, Bleakley JF, Kim JS, Nafziger AN, Leeder JS, Gaedigk A, Gotschall R, Kearns GL, Bertino JSJ. Combined phenotypic assessment of CYP1A2, CYP2C19, CYP2D6, CYP3A, N-acetyltransferase-2, and xanthine oxidase with the ‘Cooperstown cocktail’. Clin Pharmacol Ther. 2000;68:375–83. doi: 10.1067/mcp.2000.109519. [DOI] [PubMed] [Google Scholar]
- 20.Zhu B, Ou-Yang DS, Chen XP, Huang SL, Tan ZR, He N, Zhou HH. Assessment of cytochrome P450 activity by a five-drug cocktail approach. Clin Pharmacol Ther. 2001;70:455–61. doi: 10.1067/mcp.2001.119813. [DOI] [PubMed] [Google Scholar]
- 21.Schellens JHM, Van Der Wart JHF, Brugman M, Breimer DD. Influence of enzyme induction and inhibition on the oxidation of nifedipine, sparteine, mephenytoin and antipyrine in humans as assessed by a ‘cocktail’ study design. J Pharmacol Exp Ther. 1989;249:638–45. [PubMed] [Google Scholar]
- 22.Wandel C, Bocker RH, Bohrer H, DeVries JX, Hofmann W, Walter K, Kleingeist B, Neff S, Ding R, Walter-Sack I, Martin E. Relationship between hepatic cytochrome P450 3A content and activity and the disposition of midazolam administered orally. Drug Metab Disp. 1998;26:110–14. [PubMed] [Google Scholar]
- 23.Kalow W. The genetic defect of mephenytoin hydroxylation. Xenobiotica. 1986;16:379–89. doi: 10.3109/00498258609050246. [DOI] [PubMed] [Google Scholar]
- 24.Streetman DS, Kashuba AD, Bertino JSJ, Kulawy R, Rocci MLJ, Nafziger AN. Use of midazolam urinary metabolic ratios for cytochrome P450, 3A (CYP3A) phenotyping. Pharmacogenetics. 2001;11:349–55. doi: 10.1097/00008571-200106000-00010. [DOI] [PubMed] [Google Scholar]
- 25.Tamminga WJ, Wemer J, Oosterhuis B, Wieling J, Touw DJ, De Zeeuw RA, deLeij LFJJH. Mephenytoin as a probe for CYP2C19 phenotyping: effect of sample storage, intra-individual reproducibility and occurrence of adverse events. Br J Clin Pharmacol. 2001;51:471–4. doi: 10.1046/j.1365-2125.2001.01331.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.May DG, Porter J, Wilkinson GR, Branch RA. Frequency distribution of dapsone N-hydroxylase, a putative probe for P4503A4 activity, in a white population. Clin Pharmacol Ther. 1994;55:492–500. doi: 10.1038/clpt.1994.62. [DOI] [PubMed] [Google Scholar]
- 27.May DG, Porter JA, Uetrecht JP, Wilkinson GR, Branch RA. The contribution of N-hydroxylation and acetylation to dapsone pharmacokinetics in normal subjects. Clin Pharmacol Ther. 1990;48:619–27. doi: 10.1038/clpt.1990.204. [DOI] [PubMed] [Google Scholar]
- 28.Lorenzo B, Reidenberg MM. Potential artifacts in the use of caffeine to determine acetylation phenotype. Br J Clin Pharmacol. 1989;28:207–8. doi: 10.1111/j.1365-2125.1989.tb05420.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hamelin BA, Xu K, Vallée F, Manseau L, Richer M, LeBel M. Caffeine metabolism in cystic fibrosis: enhanced xanthine oxidase activity. Clin Pharmacol Ther. 1994;56:521–9. doi: 10.1038/clpt.1994.173. [DOI] [PubMed] [Google Scholar]
- 30.Hamelin BA, Bouayad A, Méthot J, Jobin J, Desgagné P, Poirier P, Allaire J, Dumesnil J, Turgeon J. Significant interaction between the nonprescription antihistamine diphenhydramine and the CYP2D6 substrate metoprolol in healthy men with high or low CYP2D6 activity. Clin Pharmacol Ther. 2000;67:466–77. doi: 10.1067/mcp.2000.106464. [DOI] [PubMed] [Google Scholar]
- 31.Veronese ME, Miners JO, Randles D, Gregov D, Birkett DJ. Validation of the tolbutamide metabolic ratio for population screening with use of sulfaphenazole to produce model phenotypic poor metabolizers. Clin Pharmacol Ther. 1990;47:403–11. doi: 10.1038/clpt.1990.46. [DOI] [PubMed] [Google Scholar]
- 32.St-Hilaire S, Bélanger PM. Simultaneous determinations of tolbutamide and its hydroxy and carboxy metabolites in serum and urine: application to pharmacokinetic studies of tolbutamide in the rat. J Pharm Sci. 1989;78:863–6. doi: 10.1002/jps.2600781017. [DOI] [PubMed] [Google Scholar]
- 33.Frye RF, Stiff DD. Determination of chlorzoxazone and 6-hydroxychlorzoxazone in human plasma and urine by high-performance liquid chromatography. Chromatogr B Biomed Appl. 1996;686:291–6. doi: 10.1016/s0378-4347(96)00227-7. [DOI] [PubMed] [Google Scholar]
- 34.Dreisbach AW, Ferencz N, Hopkins NE, Fuentes MG, Rege AB, George WJ, Lertora JJL. Urinary excretion of 6-hydroxychlorzoxazone as an index of CYP2E1 activity. Clin Pharmacol Ther. 1995;58:498–505. doi: 10.1016/0009-9236(95)90169-8. [DOI] [PubMed] [Google Scholar]
- 35.Frye RF, Adedoyin A, Mauro K, Matzke GR, Branch RA. Use of chlorzoxazone as an in vivo probe of cytochrome P450 2E1: choice of dose and phenotypic trait measure. J Clin Pharmacol. 1998;38:82–9. doi: 10.1002/j.1552-4604.1998.tb04381.x. [DOI] [PubMed] [Google Scholar]
- 36.Scott RJ, Palmer J, Lewis IA, Pleasance S. Determination of a ‘GW cocktail’ of cytochrome P450 probe substrates and their metabolites in plasma and urine using automated solid phase extraction and fast gradient liquid chromatography tandem mass spectrometry. Rapid Commun Mass Spectrom. 1999;13:2305–19. doi: 10.1002/(SICI)1097-0231(19991215)13:23<2305::AID-RCM790>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 37.Fleming CM, Branch RA, Wilkinson GR, Guengerich FP. Human liver microsomal N-hydroxylation of dapsone by cytochrome P-4503A4. Mol Pharmacol. 1992;41:975–80. [PubMed] [Google Scholar]
- 38.Gill HJ, Tingle MD, Park BK. N-hydroxylation of dapsone by multiple enzymes of cytochrome P450: implications for inhibition of haemotoxicity. Br J Clin Pharmacol. 1995;40:531–8. doi: 10.1111/j.1365-2125.1995.tb05797.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Winter HR, Wang Y, Unadkat JD. CYP2C8/9 mediate dapsone N-hydroxylation at clinical concentrations of dapsone. Drug Metab Disp. 2000;28:865–8. [PubMed] [Google Scholar]
- 40.Mitra AK, Thummel KE, Kalhorn TF, Kharasch ED, Unadkat JD, Slattery JT. Metabolism of dapsone to its hydroxylamine by CYP2E1 in vitro and in vivo. Clin Pharmacol Ther. 1995;58:556–66. doi: 10.1016/0009-9236(95)90176-0. [DOI] [PubMed] [Google Scholar]
- 41.Zuidema J, Hilbers-Modderman ES, Merkus FW. Clinical pharmacokinetics of dapsone. Clin Pharmacokinet. 1986;11:299–315. doi: 10.2165/00003088-198611040-00003. [DOI] [PubMed] [Google Scholar]
- 42.Soper JW, Chaturvedi AK, Canfield DV. Prevalence of chlorpheniramine in aviation accident pilot fatalities, 1991–96. Aviat Space Environ Med. 2000;71:1206–9. [PubMed] [Google Scholar]
- 43.Anderson DT, Fritz KL, Muto T. Distribution of mirtazapine (Remeron) in thirteen postmortem cases. J Anal Toxicol. 1999;23:544–8. doi: 10.1093/jat/23.6.544. [DOI] [PubMed] [Google Scholar]
- 44.Tingle MD, Mahmud R, Maggs JL, Pirmohamed M, Park BK. Comparison of the metabolism and toxicity of dapsone in rat, mouse and man. J Pharmacol Exp Ther. 1997;283:817–23. [PubMed] [Google Scholar]
- 45.Israili ZH, Cucineil SA, Vaught J, Davis E, Lesser JM, Dayton PG. Studies of the metabolism of dapsone in man and experimental animals. formation of N-hydroxy metabolites. J Pharmacol Exp Ther. 1973;187:138–51. [PubMed] [Google Scholar]
- 46.Labbé L, Sirois C, Pilote S, Arseneault M, Robitaille NM, Turgeon J, Hamelin BA. Effect of gender, sex hormones, time variables and physiological urinary pH on apparent CYP2D6 activity as assessed by metabolic ratios of marker substrates. Pharmacogenetics. 2000;10:425–38. doi: 10.1097/00008571-200007000-00006. [DOI] [PubMed] [Google Scholar]
- 47.Lennard MS, Tucker GT, Silas JH, Woods HF. Debrisoquine polymorphism and the metabolism and action of metoprolol, timolol, propranolol and atenolol. Xenobiotica. 1986;16:435–47. doi: 10.3109/00498258609050250. [DOI] [PubMed] [Google Scholar]
- 48.Miners JO, Birkett DJ. Use of tolbutamide as a substrate probe for human hepatic cytochrome P450 2C9. Meth Enzymol. 1996;272:139–45. doi: 10.1016/s0076-6879(96)72017-7. [DOI] [PubMed] [Google Scholar]
- 49.Shon JH, Yoon YR, Kim KA, Lim YC, Lee KJ, Park JY, Cha IJ, Flockhart DA. Effects of CYP2C19 and CYP2C9 genetic polymorphisms on the disposition of and blood glucose lowering response to tolbutamide in humans. Pharmacogenetics. 2002;12:111–19. doi: 10.1097/00008571-200203000-00005. [DOI] [PubMed] [Google Scholar]
- 50.Veronese ME, Miners JO, Rees DLP, Birkett DJ. Tolbutamide hydroxylation in humans: lack of bimodality in 106 healthy subjects. Pharmacogenetics. 1993;3:86–93. doi: 10.1097/00008571-199304000-00004. [DOI] [PubMed] [Google Scholar]
- 51.Peter R, Bocker R, Beaune PH, Iwasaki M, Guengerich FP, Yang CS. Hydroxylation of chlorzoxazone as a specific probe for human liver cytochrome P-450IIE1. Chem Res Toxicol. 1990;3:566. doi: 10.1021/tx00018a012. [DOI] [PubMed] [Google Scholar]
- 52.Adedoyin A, Frye RF, Mauro K, Branch RA. Chloroquine modulation of specific metabolizing enzymes activities: investigation with selective five drug cocktail. Br J Clin Pharmacol. 1998;46:215–19. doi: 10.1046/j.1365-2125.1998.00765.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chainuvati S, Nafziger AN, Leeder JS, Gaedigk A, Kearns GL, Sellers E, Zhang Y, Kashuba ADM, Rowland E, Bertino JSJ. Combined phenotypic assessment of cytochrome P450 1A2, 2C9, 2C19, 2D6, and 3A, N-acetyltransferase-2, and xanthine oxidase activities with the ‘Cooperstown 5 + 1 cocktail’. Clin Pharmacol Ther. 2003;74:437–47. doi: 10.1016/S0009-9236(03)00229-7. [DOI] [PubMed] [Google Scholar]