Abstract
Presynaptic inhibition mediated by G protein-coupled receptors (GPCRs) can develop and decay in a few seconds. This time course is too rapid to be accounted for by the intrinsic GTPase activity of Gα subunits alone. Here, we test the hypothesis that endogenous regulators of G protein signaling (RGS proteins) are required for rapid, brief presynaptic inhibition. Endogenous G protein α subunits were uncoupled from GPCRs by treating cultures with pertussis toxin (PTX). Adenoviral expression of mutant PTX-insensitive (PTX-i) Gαi1–3 or Gαo subunits rescued adenosine-induced presynaptic inhibition in cultured hippocampal neurons. Expression of double mutant Gαi1 or Gαo subunits that were both PTX-insensitive and unable to bind RGS proteins (PTX/RGS-i) also rescued presynaptic inhibition. Presynaptic inhibition mediated by PTX/RGS-i subunits decayed much more slowly after agonist removal than that mediated by PTX-i subunits or native G proteins. The onset of presynaptic inhibition mediated by PTX/RGS-i Gαo was also slower than that mediated by PTX-i Gαo. In contrast, the onset of presynaptic inhibition mediated by PTX/RGS-i Gαi1 was similar to that mediated by PTX-i Gαi1. These results suggest that endogenous RGS proteins regulate the time course of G protein signaling in mammalian central nervous system presynaptic terminals.
The release of neurotransmitter from most presynaptic terminals is regulated by activation of G protein-coupled receptors (GPCRs) (1, 2). These receptors may be autoreceptors for ligands released at the same synapse, or heteroreceptors for a wide variety of small molecules [e.g., γ-aminobutyric acid (GABA), glutamate, or adenosine] or peptides. The most common type of regulation is presynaptic inhibition mediated by heterotrimeric G proteins composed of pertussis toxin (PTX)-sensitive α subunits (Gαi and/or Gαo) and βγ dimers. Receptor activation promotes nucleotide exchange, which is followed by dissociation of α-GTP and βγ. Free βγ dimers directly alter the gating of the calcium channels that mediate neurotransmitter release in a manner that makes these channels reluctant to open (3). The resulting inhibition of presynaptic calcium influx can largely account for presynaptic inhibition mediated by GPCRs (4, 5), although mechanisms downstream from calcium influx also may be important in some cases (5). Presynaptic inhibition is terminated by heterotrimer reassociation after hydrolysis of α-GTP to α-GDP (6).
Presynaptic inhibition decays in a few seconds after agonist removal (7, 8). This agrees well with the time course of inhibition of somatic calcium channels (7, 9–11) and inhibition of presynaptic calcium influx (8). Activity-dependent release of endogenous agonists produces presynaptic inhibition with a similarly rapid time course (8, 12–14). The decay of presynaptic inhibition is far too rapid to be accounted for by the intrinsic GTPase activity of PTX-sensitive α subunits alone (15). In other systems regulators of G protein signaling (RGS proteins) act as GTPase-accelerating proteins, and thereby regulate the time course of G protein signals (16–21). For example, heterologous overexpression of RGS proteins can accelerate the activation and deactivation of G protein-activated inwardly rectifying potassium (GIRK) channels (22, 23), and accelerate recovery from calcium channel inhibition (24, 25). However, few studies have examined the function of endogenous RGS proteins. Many cells express several RGS proteins (26), thus precluding simple genetic deletion, and, at present, no method is available that selectively disrupts RGS function.
We have used adenovirus-mediated expression of mutant Gα subunits (Gαo and Gαi1–3) to assess the ability of endogenous RGS proteins to regulate presynaptic inhibition in hippocampal nerve terminals. Native G protein α subunits were uncoupled from receptors by treating cultured neurons with PTX, and presynaptic inhibition was rescued by expressing mutant α subunits that were either PTX-insensitive (PTX-i), or both PTX-insensitive and RGS-insensitive (PTX/RGS-i). We find that presynaptic inhibition mediated by PTX-i α subunits resembles that mediated by native G proteins, whereas that mediated by PTX/RGS-i G proteins decays much more slowly. Endogenous RGS proteins also appear to regulate the onset kinetics of presynaptic inhibition, although this regulation depends on the α subunit involved. These results show that RGS proteins are an integral part of the signaling machinery used by presynaptic GPCRs.
Methods
Adenoviral Vectors.
Adenoviruses coexpressing enhanced green fluorescent protein (EGFP) and PTX-i or PTX/RGS-i G protein α subunits were generated by using the AdEasy system (27). To generate PTX-i Gαo/i subunits, we used PCR to introduce a cysteine to isoleucine mutation in the site for PTX-catalyzed ADP ribosylation (the −4 position) in Gαo and Gαi1–3. Mutants were generated by incorporating altered bases into the reverse primers. Forward primers were designed such that the start codon was under the optimal context for translation initiation (28). Vectors encoding wild types of rat Gαo/i1–3 were provided by R. R. Reed (Johns Hopkins University, Baltimore, MD). To generate PTX/RGS-i mutants of Gαo and Gαi1, vectors encoding rat G184S αo and G183S αi1 (29) (provided by R. R. Neubig, University of Michigan, Ann Arbor) were used as templates. After amplification, the coding region of each subunit was subcloned into a shuttle vector. The shuttle vector used (pAdTrack-CMV) incorporates EGFP driven by a cytomegalovirus (CMV) promoter to allow visualization of virus production and identification of infected cells (27). The resultant plasmids were linearized with EcoRI and cotransformed with an adenoviral backbone vector (pAdEasy-1) into Escherichia coli BJ5183 cells. Recombination was confirmed by restriction analysis and sequencing. The shuttle and backbone vectors, BJ5183 cells, and the adenoviral vector pAdEasy1-GFP + β-galactosidase (β-gal) were provided by T.-C. He (Johns Hopkins University). High titer stocks were generated by transfecting PacI linearized recombinant plasmid into 293 cells. Adenoviruses were purified by CsCl banding and dialysis (30). The effective titer was determined by the frequency of EGFP-positive 293 cells 30 h after infection.
Cell Culture and Infection.
Hippocampal neurons were grown on collagen/polylysine microislands essentially as described (31, 32). Hippocampi were dissected from newborn rats and digested with papain (≈25 units ml−1). After dissociation, 5 × 104 neurons were plated in 35-mm dishes that had been coated with 0.15% agarose, then sprayed with a 1:4 (vol/vol) mixture of rat tail collagen (3.6 mg ml−1) and poly-d-lysine (0.5 mg ml−1). Neurons were fed weekly by adding 0.5 ml of conditioned growth medium containing 50 μM d(−)-2-amino-5-phosphonopentanoic acid (APV), and were used for experiments after 13–19 days in culture. Neurons were infected by adding 100 μM APV to minimize toxicity, then adding ≈1–3 × 107 infectious units per dish. As appropriate, neurons were incubated with 100 ng ml−1 PTX for 24–72 h before recording.
Electrophysiology and Data Analysis.
Whole-cell patch–clamp recordings were made from neurons expressing EGFP 40–72 h after infection. Neurons were held at −60 mV and depolarized with pairs (50-ms interval) of 2-ms square commands every 2 s. These commands evoked unclamped action currents and synaptic (autaptic) currents (32). Series resistance (after compensation) was monitored during recording, and cells were discarded if a significant increase occurred. Currents were digitized and recorded by using winwcp software (provided by J. Dempster, Strathclyde University, Glasgow, U.K.). Patch electrodes were filled with a solution containing 140 mM K-gluconate, 5 mM KCl, 0.2 mM EGTA, 10 mM Hepes, 3 mM MgATP, 0.3 mM Na2GTP (pH 7.2, ≈295 mOsm kg−1 H2O). The external solution contained 145 mM NaCl, 2.5 mM KCl, 10 mM Hepes, 10 mM glucose, 1.5 mM CaCl2, 2.5 mM MgCl2 (pH 7.2, ≈310 mOsm kg−1 H2O). Recordings were made at room temperature. Drugs were applied by means of an array of gravity-fed fused silica tubes (i.d. 200 μm). Switching between drug solutions was accomplished by moving the array with stepper motor. The solution exchange time was measured by switching between normal and diluted (50%) external solutions. The change in junction potential after a solution switch began after a delay of ≈30 ms, and could be fitted with a single exponential with a time constant of ≈30 ms, thus solution exchange (at the tip of the pipette) was complete within a few hundred milliseconds. Numerical values are expressed as mean ± SEM; statistical comparisons were made by using Student's t test or ANOVA.
Results
Rescue of Presynaptic Inhibition by Adenoviral Expression of PTX-i α Subunits.
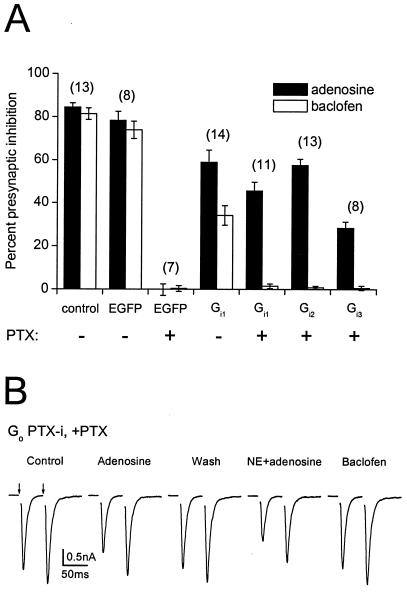
To study presynaptic G proteins, we made use of synapses (autapses) made by isolated neurons onto themselves in culture. Cultured hippocampal neurons are amenable to adenoviral infection yet retain many of the features of more intact preparations, including robust, PTX-sensitive presynaptic inhibition mediated by adenosine A1 receptors and GABAB receptors (33, 34). In uninfected control neurons, application of adenosine (20 μM) or baclofen (50 μM) produced a marked inhibition of excitatory postsynaptic currents (EPSCs; 84 ± 2% and 81 ± 3% inhibition, respectively, n = 13; Fig. 1A). Infection with an adenovirus expressing only EGFP and β-gal (pAdEasy1-GFP + β-gal) had no significant effect on presynaptic inhibition (adenosine: 78 ± 4%; baclofen: 74 ± 4%; n = 8, P > 0.05 compared with uninfected cells). PTX pretreatment (100 ng ml−1, >24 h) abolished adenosine- and baclofen-induced presynaptic inhibition, as reported (33, 34), in uninfected control neurons (data not shown; n = 18), and in pAdEasy1-GFP + β-gal-infected neurons (adenosine: 0 ± 3%; baclofen: 0 ± 1%; n =7; Fig. 1A).
Figure 1.
Rescue of presynaptic inhibition by PTX-insensitive Gαi and Gαo subunits after PTX pretreatment. (A) Summary of experiments comparing presynaptic inhibition induced by baclofen (50 μM) or adenosine (20 μM) under control conditions, with or without infection with adenoviruses expressing various proteins, and with or without pretreatment with PTX (100 ng ml−1, >24 h). Percent presynaptic inhibition was calculated as the difference between peak EPSC amplitudes under control conditions and in the presence of the drug divided by the control EPSC amplitude, multiplied by 100. Bars represent mean ± SEM, with the number of experiments (n) in parentheses. The experimental conditions (virus and toxin treatment) are indicated below each bar. (B) Traces recorded from a neuron expressing PTX-insensitive Gαo after pretreatment with PTX. Adenosine (20 μM) or combined application of adenosine and norepinephrine (NE; 10 μM) depressed evoked EPSCs (with a shift toward paired-pulse facilitation), whereas baclofen (50 μM) was without effect. EPSCs were evoked by pairs of 2-ms depolarizing commands (50-ms interpulse interval) every 2 s (arrows). Each trace represents the average of five consecutive recordings; currents evoked by the depolarizing commands are blanked.
To study nonnative Gα subunits in presynaptic terminals, we adopted a strategy used by others (35–39, 49) wherein the site for PTX-catalyzed ADP ribosylation (the −4 cysteine residue) is changed to an isoleucine, rendering Gαo and Gαi1–3 subunits insensitive to PTX. We first tested the ability of PTX-i Gα subunits to rescue presynaptic inhibition after treatment with PTX. As is shown in Fig. 1A, after treatment with PTX, expression of PTX-i Gαi1, Gαi2, or Gαi3 partially rescued adenosine-induced presynaptic inhibition (46 ± 4%, 58 ± 3%, and 28 ± 3%; n = 11, 13, and 8, respectively). In contrast, expression of these Gα subunits did not significantly rescue baclofen-induced presynaptic inhibition in the same cells (1 ± 1%, 1 ± 1%, and 1 ± 1%, respectively). There are a number of possible reasons for differential rescue of adenosine- vs. baclofen-induced presynaptic inhibition, the most obvious being that the C/I mutation disrupts coupling to GABAB receptors (see Discussion). We also found that expression of PTX-i subunits selectively inhibited baclofen-induced presynaptic inhibition in cells that were not treated with PTX, as would be expected if native and exogenous G proteins compete in some way for access to presynaptic GPCRs. For example, in Gαi1-expressing neurons, adenosine-induced presynaptic inhibition was 59 ± 6%, whereas baclofen-induced presynaptic inhibition was 34 ± 5% (n = 14; Fig. 1A). The ratio of baclofen- to adenosine-induced inhibition was 0.94 ± 0.01 in EGFP-expressing neurons, and 0.60 ± 0.06 in PTX-i Gαi1-expressing neurons (P < 0.05). Similar results were obtained with other PTX-i subunits without PTX treatment (data not shown). This result is consistent with the idea that PTX-i subunits enter a pool of G proteins available to both receptors, but are unable to couple to GABAB receptors (43). It should be noted that adenosine-induced presynaptic inhibition mediated by any of the PTX-i Gαi subunits was significantly less than that mediated by native Gα subunits (see Discussion).
Rescue of adenosine-induced presynaptic inhibition by expression of PTX-i Gi subunits was very reliable. Expression of either Gαi1 or Gαi2 rescued adenosine-induced presynaptic inhibition in every neuron tested, whereas Gαi3 was effective in 10 of 16 neurons. In contrast, expression of PTX-i Gαo did not significantly rescue either adenosine- or baclofen-induced presynaptic inhibition in the large majority (≈90%) of infected neurons. In a small population of cells, there was modest rescue of presynaptic inhibition mediated by adenosine receptors (16 ± 2%, n = 9; Fig. 1B). Differential rescue by PTX-i Gαi vs. Gαo subunits could be due to differences in receptor coupling, as Gαo differs substantially from Gαi1–3 in the C-terminal region (44). The particular PTX-i Gαo subunit we used (C351I) is known to couple to α2A-adrenoreceptors (α2-ARs) (38). Therefore, in an attempt to maximize the number of active presynaptic receptors capable of coupling to PTX-i Gαo C351I, we coapplied norepinephrine (NE; 10 μM). NE induces PTX-sensitive presynaptic inhibition in cultured hippocampal neurons by activating α2-ARs (45). NE-induced presynaptic inhibition is less consistent than that produced by adenosine or baclofen, in that only about half of control neurons respond (45) (unpublished observation). Nonetheless, in five neurons (of 21 tested) expressing PTX-i Gαo, combined application of adenosine and NE inhibited EPSCs by 35 ± 6% (Fig. 1B). In the same cells, baclofen was without effect, and, in control cells, PTX completely blocked presynaptic inhibition produced by the combined application of adenosine and NE (n =16, data not shown) (45). Thus, it appears that all of the PTX-i Gα subunits tested (Gαi1–3 and Gαo) are capable of mediating presynaptic inhibition to varying degrees.
Rescue of Presynaptic Inhibition by Adenoviral Expression of PTX/RGS-i α Subunits.
Taken together, the above results suggest that exogenously expressed Gαi and Gαo subunits form heterotrimers with native βγ dimers, are transported to synaptic terminals, and enter the pool of heterotrimers available to presynaptic receptors and effectors. To test whether endogenous RGS proteins regulate presynaptic inhibition, we constructed doubly mutated (PTX/RGS-i) Gα subunits, which possessed the C-terminal C/I mutation used above together with a glycine (G) to serine (S) mutation at position 183/184 (in Gαi1 and Gαo, respectively) (29, 46). This mutation has been previously shown to greatly decrease the affinity of Gα subunits for RGS proteins (29, 46).
Expression of either PTX/RGS-i Gαi1 or Gαo reliably rescued adenosine-induced presynaptic inhibition after PTX pretreatment. Steady-state inhibition in the presence of adenosine was 47 ± 3% (n = 18) for PTX/RGS-i Gαi1, which was not significantly different from that mediated by PTX-i Gαi1 (P > 0.05). Steady-state inhibition in the presence of adenosine was 61 ± 5% (n = 12) for PTX/RGS-i Gαo, which was significantly greater than that mediated by PTX-i Gαo (P < 0.05). Adenosine-induced presynaptic inhibition mediated by either PTX/RGS-i subunit was significantly smaller than that in control uninfected or EGFP-expressing neurons (P < 0.05). Neither PTX/RGS-i protein rescued baclofen-induced presynaptic inhibition (PTX/RGS-i Gαi1: 1 ± 1%, n = 15; PTX/RGS-i Gαo: 0 ± 2%, n = 7).
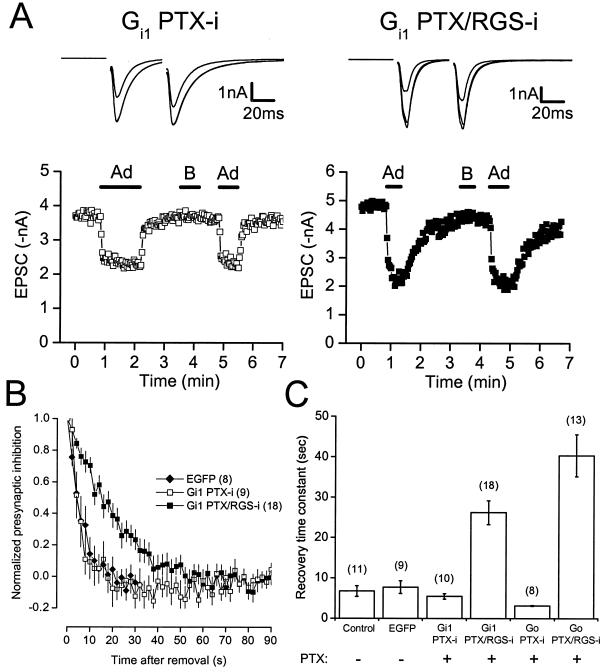
Consistent with the idea that endogenous RGS proteins accelerate GTP hydrolysis in presynaptic terminals, recovery from presynaptic inhibition mediated by PTX/RGS-i Gα subunits was much slower after agonist removal than recovery from presynaptic inhibition mediated by native or PTX-i subunits (Fig. 2). The recovery time course of EPSC amplitudes back to control after agonist removal could be fitted reasonably well with a single exponential in most experiments as reported (7). In control uninfected and EGFP-expressing neurons, the single exponential recovery time constants of adenosine-induced presynaptic inhibition were 6.7 ± 1.3 s (n = 11) and 7.7 ± 1.5 s (n = 9), respectively. These values agree well with previous studies of presynaptic inhibition (7, 8). In PTX-treated neurons expressing PTX-i or PTX/RGS-i Gαi1, presynaptic inhibition recovered with time constants of 5.5 ± 0.7 s (n =10) and 26.2 ± 3.0 s (n = 18; P < 0.05; Fig. 2B). In neurons expressing PTX-i or PTX/RGS-i Gαo, presynaptic inhibition recovered with time constants of 3.1 ± 0.1 s (n =8) and 40.4 ± 5.2 s (n = 13; P < 0.05; Fig. 2C). Recovery time constants in uninfected neurons and in neurons expressing EGFP, PTX-i Gαi1, or PTX-i Gαo were not significantly different (P = 0.053, F = 2.83; ANOVA). These results suggest that endogenous RGS proteins are required for rapid recovery from presynaptic inhibition.
Figure 2.
The recovery time course of adenosine-induced presynaptic inhibition mediated by PTX-insensitive and PTX/RGS-insensitive Gα subunits. (A) The time course of presynaptic inhibition is plotted for neurons pretreated with PTX (100 ng ml−1, >24 h) and expressing PTX-insensitive or PTX/RGS-insensitive Gαi1 subunits. Adenosine (20 μM) and baclofen (50 μM) were applied where indicated by the horizontal bars. Note the slow recovery from presynaptic inhibition mediated by PTX/RGS-insensitive Gαi1. Above each plot are superimposed traces (average of five consecutive recordings each) recorded under control conditions, in the presence of adenosine, and in the presence of baclofen. (B) Recovery from presynaptic inhibition (normalized to steady state) is plotted vs. time for neurons expressing EGFP (first 30 s only), PTX-insensitive Gαi1, and PTX/RGS-insensitive Gαi1. Points represent the mean ± SEM; the number of experiments (n) is given in parentheses. (C) Summary of recovery from presynaptic inhibition. Bars represent the mean ± SEM of recovery time constants derived from single exponential fits to plots of EPSC amplitude vs. time after drug removal. The experimental conditions (virus and toxin treatment) are indicated below each bar.
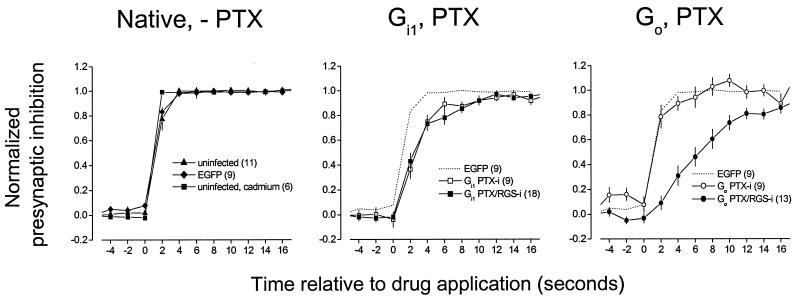
Heterologous overexpression of RGS proteins can accelerate both the onset and decay of G protein-mediated activation of GIRK channels (22, 23). We therefore examined the onset kinetics of adenosine-induced presynaptic inhibition mediated by PTX-i and PTX/RGS-i Gα subunits. In uninfected control neurons, adenosine-induced presynaptic inhibition was substantially complete 2 s after drug application, having reached 77 ± 9% (n = 11) of the eventual steady-state inhibition. In a series of control experiments, we determined that this onset time course was not limited by slow solution exchange or a slow binding of a suboptimal concentration of adenosine. Presynaptic inhibition produced by cadmium (100 μM), which directly blocks presynaptic calcium channels, was 99 ± 1% complete by 2 s (P < 0.05 vs. control), and presynaptic inhibition produced by 100 μM adenosine was not significantly faster than that produced by 20 μM adenosine (75 ± 9% and 81 ± 8% at 2 s, respectively; n = 6; P > 0.05, paired t test). This onset time course is consistent with previous studies of the kinetics of GPCR-mediated presynaptic inhibition (7, 8). In neurons expressing EGFP, adenosine-induced presynaptic inhibition was 83 ± 4% (n = 9) of steady-state by 2 s, which was not significantly different from the control value (P > 0.05; Fig. 3). The onset of adenosine-induced presynaptic inhibition was slower when mediated by either PTX-i or PTX/RGS-i Gαi1 subunits, having reached only 37 ± 8% (n = 9) and 43 ± 6% (n = 18) of steady-state by 2 s, respectively (Fig. 3). These values were not significantly different from each other (P > 0.05), but both were significantly different from control values (P < 0.05). In contrast, the onset of presynaptic inhibition mediated by PTX-i Gαo subunits (after application of either adenosine or adenosine and NE combined) resembled the control situation (79 ± 9% of steady state by 2 s, n = 9), whereas that mediated by PTX/RGS-i Gαo subunits was much slower (9 ± 6%, n = 13; Fig. 3). The value for PTX-i Gαo was not significantly different from either control value (P > 0.05), whereas the value for PTX/RGS-i Gαo was significantly different from both (P < 0.05). These results suggest that RGS proteins are important for the rapid onset kinetics of presynaptic inhibition, and that this effect differs depending on the Gα subunit involved.
Figure 3.
The onset time course of presynaptic inhibition mediated by PTX-insensitive and PTX/RGS-insensitive Gα subunits. Superimposed plots of normalized presynaptic inhibition vs. time, with drug application at 0 s. Each point represents the mean ± SEM, with the number of experiments (n) given in parentheses in the legend. Presynaptic inhibition was normalized by using data points 6 s before and at least 20 s after drug application (20 μM adenosine, 50 μM baclofen). In all experiments where Gα subunits were expressed, neurons were pretreated with PTX.
Discussion
G proteins transduce signals on time scales that range from milliseconds to hours. It is now apparent that RGS proteins can accelerate G protein-mediated signal onset and/or decay (21). This implies that these proteins may be essential for physiological processes involving rapid, brief G protein-mediated signals. For example, it was recently shown that RGS9–1 is required for rapid (subsecond) termination of transducin (Gαt)-mediated photoresponses in rod outer segments (47). Like phototransduction, presynaptic inhibition mediated by PTX-sensitive G proteins in the mammalian central nervous system is a fairly rapid process—often rising to a peak in a few hundred ms, and decaying in less than 1 s (7, 8, 12–14). The purpose of the experiments described here was to determine whether RGS proteins are required for rapid signaling by PTX-sensitive G proteins in presynaptic terminals.
These experiments made use of a strategy devised by others to replace native PTX-sensitive G proteins with PTX-insensitive mutants (35, 36) and PTX- and RGS-insensitive double mutants (29, 46, 49). To date, more than 20 RGS proteins have been identified (16), at least seven of which could be expressed in hippocampal neurons (26). Given the likelihood that some of these proteins are functionally redundant, a genetic deletion approach to the question of RGS function in presynaptic terminals seems unlikely to succeed. We chose instead to make use of a mutation first recognized in yeast, and later applied to mammalian cells, which greatly reduces the affinity of Gα subunits for RGS proteins (29, 46). This mutation does not appear to change other aspects of Gα function, such as the rates of nucleotide exchange and intrinsic GTP hydrolysis (29), and thus is a useful tool for studying endogenous RGS proteins. We have shown that this combined strategy can be applied effectively to study G proteins in presynaptic terminals. Expression of Gα subunits that were both PTX-i and defective with respect to RGS protein binding revealed a role for the latter proteins in regulating the kinetics of presynaptic inhibition. Presynaptic inhibition mediated by PTX/RGS-i Gα subunits decayed much more slowly than that mediated by native subunits or PTX-i subunits. In addition, the onset of presynaptic inhibition mediated by Gαo appeared to be accelerated by an interaction with endogenous RGS proteins, whereas the onset of presynaptic inhibition mediated by Gαi1 was the same with or without RGS binding.
As is the case with any such study, the possibility exists that the mutations we introduced had unintended effects on G protein function. For example, it is possible that receptor coupling efficiency is changed, because the −4 residue is critical for receptor coupling (40). We chose C/I mutants because previous studies indicated that at least adenosine A1 receptors would couple to these proteins (39, 41, 42). However, biochemical evidence suggests that the PTX-i Gαi1 used here does have a lowered affinity for A1 adenosine receptors (ref. 41, but see ref. 39). Similarly, Franek et al. (43) have shown that an isoleucine in the −4 position prevents coupling of Gαqo fusion proteins to recombinant GABAB receptors. One likely manifestation of this problem in the present study was our inability to rescue GABAB receptor-mediated presynaptic inhibition with any of the PTX-i or PTX/RGS-i Gα subunits, although alternative explanations for this failure cannot be eliminated. A change in coupling efficacy also could account for our inability to completely rescue adenosine-induced presynaptic inhibition with PTX-i Gα subunits. It is possible that delivery of a greater number of PTX-i Gα subunits to presynaptic terminals would have resulted in more complete rescue of adenosine-induced presynaptic inhibition, and some rescue of baclofen-induced presynaptic inhibition. Thus, without precise information about coupling efficacy and G protein expression levels, differences in coupling specificity observed with exogenous G proteins should be interpreted with extreme caution. For example, the less effective rescue of adenosine-induced presynaptic inhibition with PTX-i Gαo vs. PTX-i Gαi that we observed should not be interpreted as evidence that A1 receptors prefer native Gαi over Gαo.
The same caution should be applied to interpretation of differences in onset kinetics between PTX-i Gα subunits. We found that the onset of adenosine-induced presynaptic inhibition was slower with Gαi subunits than Gαo subunits or native Gα subunits. At first glance, this result may seem to indicate that the native G proteins mediating presynaptic inhibition are not Gαi subunits. Although this is certainly possible, onset kinetics may be limited by receptor-G protein coupling, which may be differentially impaired in the PTX-i mutants used here. Our results indicate that all of the Gα subunits studied here can mediate presynaptic inhibition; however, the identity of the native Gα subunit(s) that participate in this response is still unknown.
One discrepancy arising from the present study is the much greater rescue of adenosine-induced presynaptic inhibition by PTX/RGS-i Gαo compared with PTX-i Gαo. If the inability of PTX-i Gαo to support adenosine-induced presynaptic inhibition were due entirely to poor coupling with A1 receptors (see above), we would expect coupling to PTX/RGS-i Gαo to be poor as well. It is possible that this is indeed the case, but that the lifetime of active PTX/RGS-i Gαo subunits is long enough to support robust presynaptic inhibition nonetheless. Another possibility is that Gαo subunits are better substrates for endogenous presynaptic RGS proteins than Gαi subunits. This would make it more difficult to rescue presynaptic inhibition with these G proteins, and would magnify the effect of disabling RGS binding. This explanation is plausible, as there is evidence that RGS proteins can be selective for specific signaling pathways (16, 48).
Our results are consistent with those of Jeong and Ikeda (49), who recently have shown that endogenous RGS proteins regulate the kinetics of calcium channel modulation in rat sympathetic neurons. Previous studies had indicated that exogenous RGS proteins can regulate calcium channel modulation (24, 50). A primary function of calcium channel modulation in neurons is to control neurotransmitter release, and presynaptic inhibition can largely be accounted for by calcium channel modulation (4, 5). Therefore, our results extend these observations by demonstrating the role of endogenous RGS proteins in the regulation of neurotransmitter release.
Jeong and Ikeda (49) found that endogenous RGS proteins contribute to the rapid onset of calcium channel modulation, and previous studies had indicated that RGS proteins can accelerate the onset of GIRK channel activation (22, 23). Similarly, we found that endogenous RGS proteins contribute to the rapid onset of presynaptic inhibition. Interestingly, this acceleration was apparent only for Gαo subunits. The slowed recovery of presynaptic inhibition mediated by Gαi1 should have led to a slower onset time to steady state as well (21). The fact that this was not observed (Fig. 3) suggests that RGS proteins also may decelerate signaling by these subunits. Thus, the precise effect of RGS proteins on signaling kinetics may depend on the particular Gα subunit involved, and effects on onset and recovery kinetics are at least partially dissociable. In this regard, it is interesting to note that RGS9–1 deletion prolonged the decay of photoresponses without substantially altering their onset rate (47). The mechanism by which RGS proteins accelerate G protein signal onset is not known, although it appears that GTPase-accelerating protein activity alone cannot account for these effects (21).
As pointed out by Zerangue and Jan (21), RGS proteins cannot be thought of simply as negative regulators of G protein signaling. Because these proteins can accelerate the onset of G protein-mediated responses, they may be essential for robust transduction of transient signals. Presynaptic GPCRs often are exposed to endogenous ligands transiently, for periods of tens or hundreds of milliseconds (7, 8, 12–14). The results of the present study indicate that some presynaptic G proteins may require RGS proteins to transduce this type of signal.
Acknowledgments
We thank Dr. R. R. Reed (Johns Hopkins University) and Dr. R. R. Neubig (University of Michigan) for providing cDNA plasmids used in this study, Dr. T.-C. He (Johns Hopkins University) for making the AdEasy system available and providing materials, and Dr. J. Dempster (Strathclyde University) for providing data acquisition software. We thank Dr. Stephen R. Ikeda (Guthrie Research Institute) for introducing us to the double mutant strategy, for sharing data before publication, and for many helpful discussions. This study was supported by National Institutes of Health Grant NS 36455 and a Veterans Administration Merit Award.
Abbreviations
- GPCR
G protein-coupled receptor
- GABA
γ-aminobutyric acid
- PTX
pertussis toxin
- RGS
regulators of G protein signaling
- GIRK
G protein-activated inwardly rectifying potassium
- PTX-i
pertussis toxin-insensitive
- PTX/RGS-i
pertussis toxin and RGS-insensitive
- EGFP
enhanced green fluorescent protein
- β-gal
β-galactosidase
- EPSC
excitatory postsynaptic current
- NE
norepinephrine
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.230260397.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.230260397
References
- 1.Wu L G, Saggau P. Trends Neurosci. 1997;20:204–212. doi: 10.1016/s0166-2236(96)01015-6. [DOI] [PubMed] [Google Scholar]
- 2.Thompson S M, Capogna M, Scanziani M. Trends Neurosci. 1993;16:222–227. doi: 10.1016/0166-2236(93)90160-n. [DOI] [PubMed] [Google Scholar]
- 3.Ikeda S R, Dunlap K. Adv Second Messenger Phosphoprotein Res. 1999;33:131–151. doi: 10.1016/s1040-7952(99)80008-1. [DOI] [PubMed] [Google Scholar]
- 4.Wu L G, Saggau P. Neuron. 1994;12:1139–1148. doi: 10.1016/0896-6273(94)90321-2. [DOI] [PubMed] [Google Scholar]
- 5.Dittman J S, Regehr W G. J Neurosci. 1996;16:1623–1633. doi: 10.1523/JNEUROSCI.16-05-01623.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hepler J R, Gilman A G. Trends Biochem Sci. 1992;17:383–387. doi: 10.1016/0968-0004(92)90005-t. [DOI] [PubMed] [Google Scholar]
- 7.Pfrieger F W, Gottmann K, Lux H D. Neuron. 1994;12:97–107. doi: 10.1016/0896-6273(94)90155-4. [DOI] [PubMed] [Google Scholar]
- 8.Dittman J S, Regehr W G. J Neurosci. 1997;17:9048–9059. doi: 10.1523/JNEUROSCI.17-23-09048.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhou J, Shapiro M S, Hille B. J Neurophysiol. 1997;77:2040–2048. doi: 10.1152/jn.1997.77.4.2040. [DOI] [PubMed] [Google Scholar]
- 10.Sahara Y, Westbrook G L. J Neurosci. 1993;13:3041–3050. doi: 10.1523/JNEUROSCI.13-07-03041.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jones S W. Biophys J. 1991;60:502–507. doi: 10.1016/S0006-3495(91)82077-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mitchell J B, Lupica C R, Dunwiddie T V. J Neurosci. 1993;13:3439–3447. doi: 10.1523/JNEUROSCI.13-08-03439.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lambert N A, Wilson W A. J Neurophysiol. 1994;72:121–130. doi: 10.1152/jn.1994.72.1.121. [DOI] [PubMed] [Google Scholar]
- 14.Isaacson J S, Solis J M, Nicoll R A. Neuron. 1993;10:165–175. doi: 10.1016/0896-6273(93)90308-e. [DOI] [PubMed] [Google Scholar]
- 15.Linder M E, Ewald D A, Miller R J, Gilman A G. J Biol Chem. 1990;265:8243–8251. [PubMed] [Google Scholar]
- 16.De Vries L, Zheng B, Fischer T, Elenko E, Farquhar M G. Annu Rev Pharmacol Toxicol. 2000;40:235–271. doi: 10.1146/annurev.pharmtox.40.1.235. [DOI] [PubMed] [Google Scholar]
- 17.Watson N, Linder M E, Druey K M, Kehrl J H, Blumer K J. Nature (London) 1996;383:172–175. doi: 10.1038/383172a0. [DOI] [PubMed] [Google Scholar]
- 18.Hepler J R. Trends Pharmacol Sci. 1999;20:376–382. doi: 10.1016/s0165-6147(99)01369-3. [DOI] [PubMed] [Google Scholar]
- 19.Wieland T, Chen C K. Naunyn Schmiedebergs Arch Pharmacol. 1999;360:14–26. doi: 10.1007/s002109900031. [DOI] [PubMed] [Google Scholar]
- 20.Dohlman H G, Thorner J. J Biol Chem. 1997;272:3871–3874. doi: 10.1074/jbc.272.7.3871. [DOI] [PubMed] [Google Scholar]
- 21.Zerangue N, Jan L Y. Curr Biol. 1998;8:R313–R316. doi: 10.1016/s0960-9822(98)70196-4. [DOI] [PubMed] [Google Scholar]
- 22.Saitoh O, Kubo Y, Miyatani Y, Asano T, Nakata H. Nature (London) 1997;390:525–529. doi: 10.1038/37385. [DOI] [PubMed] [Google Scholar]
- 23.Doupnik C A, Davidson N, Lester H A, Kofuji P. Proc Natl Acad Sci USA. 1997;94:10461–10466. doi: 10.1073/pnas.94.19.10461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Melliti K, Meza U, Fisher R, Adams B. J Gen Physiol. 1999;113:97–110. doi: 10.1085/jgp.113.1.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jeong S W, Ikeda S R. Neuron. 1998;21:1201–1212. doi: 10.1016/s0896-6273(00)80636-4. [DOI] [PubMed] [Google Scholar]
- 26.Gold S J, Ni Y G, Dohlman H G, Nestler E J. J Neurosci. 1997;17:8024–8037. doi: 10.1523/JNEUROSCI.17-20-08024.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.He T C, Zhou S, da Costa L T, Yu J, Kinzler K W, Vogelstein B. Proc Natl Acad Sci USA. 1998;95:2509–2514. doi: 10.1073/pnas.95.5.2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kozak M. Gene. 1999;234:187–208. doi: 10.1016/s0378-1119(99)00210-3. [DOI] [PubMed] [Google Scholar]
- 29.Lan K L, Sarvazyan N A, Taussig R, Mackenzie R G, DiBello P R, Dohlman H G, Neubig R R. J Biol Chem. 1998;273:12794–12797. doi: 10.1074/jbc.273.21.12794. [DOI] [PubMed] [Google Scholar]
- 30.Giger R J, Ziegler U, Hermens W T, Kunz B, Kunz S, Sonderegger P. J Neurosci Methods. 1997;71:99–111. doi: 10.1016/s0165-0270(96)00130-6. [DOI] [PubMed] [Google Scholar]
- 31.Segal M M, Furshpan E J. J Neurophysiol. 1990;64:1390–1399. doi: 10.1152/jn.1990.64.5.1390. [DOI] [PubMed] [Google Scholar]
- 32.Bekkers J M, Stevens C F. Proc Natl Acad Sci USA. 1991;88:7834–7838. doi: 10.1073/pnas.88.17.7834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Scholz K P, Miller R J. J Physiol (London) 1991;444:669–686. doi: 10.1113/jphysiol.1991.sp018900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Scholz K P, Miller R J. J Physiol (London) 1991;435:373–393. doi: 10.1113/jphysiol.1991.sp018515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Taussig R, Sanchez S, Rifo M, Gilman A G, Belardetti F. Neuron. 1992;8:799–809. doi: 10.1016/0896-6273(92)90100-r. [DOI] [PubMed] [Google Scholar]
- 36.Wise A, Watson-Koken M A, Rees S, Lee M, Milligan G. Biochem J. 1997;321:721–728. doi: 10.1042/bj3210721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Delmas P, Abogadie F C, Milligan G, Buckley N J, Brown D A. J Physiol (London) 1999;518:23–36. doi: 10.1111/j.1469-7793.1999.0023r.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jeong S W, Ikeda S R. Proc Natl Acad Sci USA. 2000;97:907–912. doi: 10.1073/pnas.97.2.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Leaney J L, Tinker A. Proc Natl Acad Sci USA. 2000;97:5651–5656. doi: 10.1073/pnas.080572297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Conklin B R, Farfel Z, Lustig K D, Julius D, Bourne H R. Nature (London) 1993;363:274–276. doi: 10.1038/363274a0. [DOI] [PubMed] [Google Scholar]
- 41.Waldhoer M, Wise A, Milligan G, Freissmuth M, Nanoff C. J Biol Chem. 1999;274:30571–30579. doi: 10.1074/jbc.274.43.30571. [DOI] [PubMed] [Google Scholar]
- 42.Wise A, Sheehan M, Rees S, Lee M, Milligan G. Biochemistry. 1999;38:2272–2278. doi: 10.1021/bi982054f. [DOI] [PubMed] [Google Scholar]
- 43.Franek M, Pagano A, Kaupmann K, Bettler B, Pin J P, Blahos J. Neuropharmacology. 1999;38:1657–1666. doi: 10.1016/s0028-3908(99)00135-5. [DOI] [PubMed] [Google Scholar]
- 44.Milligan G. Biochem J. 1988;255:1–13. doi: 10.1042/bj2550001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Boehm S. J Physiol (London) 1999;519:439–449. doi: 10.1111/j.1469-7793.1999.0439m.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.DiBello P R, Garrison T R, Apanovitch D M, Hoffman G, Shuey D J, Mason K, Cockett M I, Dohlman H G. J Biol Chem. 1998;273:5780–5784. doi: 10.1074/jbc.273.10.5780. [DOI] [PubMed] [Google Scholar]
- 47.Chen C K, Burns M E, He W, Wensel T G, Baylor D A, Simon M I. Nature (London) 2000;403:557–560. doi: 10.1038/35000601. [DOI] [PubMed] [Google Scholar]
- 48.Cavalli A, Druey K M, Milligan G. J Biol Chem. 2000;275:23693–23699. doi: 10.1074/jbc.M910395199. [DOI] [PubMed] [Google Scholar]
- 49.Jeong S W, Ikeda S R. J Neurosci. 2000;20:4489–4496. doi: 10.1523/JNEUROSCI.20-12-04489.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Diverse-Pierluissi M A, Fischer T, Jordan J D, Schiff M, Ortiz D F, Farquhar M G, De Vries L. J Biol Chem. 1999;274:14490–14494. doi: 10.1074/jbc.274.20.14490. [DOI] [PubMed] [Google Scholar]