Abstract
Aims
The aim of the study was to determine whether a correlation exists between MDR1 (ABCB1) gene polymorphisms at positions 3435 (C3435T) and 2677 (G2677T(A)) and the expression of human hepatic P-glycoprotein (P-gp).
Methods
P-gp protein expression in 26 human livers was assessed by Western blotting and ABCB1 mRNA expression was determined by real time RT-PCR. The C3435T and G2677T(A) polymorphisms were identified by RFLP and direct sequence analysis, respectively.
Results
The C and G allele frequencies for the C3435T and G2677T(A) polymorphisms were 0.48 and 0.79, respectively, and the genotypes were in Hardy–Weinberg equilibrium. There was a 200- and 20-fold variation in the expression of ABCB1 mRNA and Pgp protein expression, respectively. There were no differences in mRNA and protein expression identified amongst the different genotypes attributable to the C3435T and G2677T(A) polymorphisms in the ABCB1 gene. Exposure to a PXR ligand prior to death did not influence mRNA or protein expression.
Conclusions
There is substantial variability in the expression of Pgp in human liver, but this is not due to the presence of C3435T and G2677T(A) polymorphisms in the ABCB1 gene, although our study is limited by a small sample size.
Keywords: ABCB1, C3435T, expression, G2677T/A, MDR1, P-glycoprotein, SNP
Introduction
Although the physiological role of P-glycoprotein (P-gp) in man is not completely understood, its importance in drug absorption from the gastrointestinal tract and drug elimination via the bile and urine is clear. Variation in the expression of the ABCB1 (MDR1) gene is likely to be an important source of interindividual variability in pharmacokinetics and pharmacodynamics of many drugs [1, 2]. Additionally, given its role in the disposition of other chemicals, the ABCB1 gene may also act as a susceptibility factor for cancer [3].
Hoffmeyer et al. [4] identified 15 single nucleotide polymorphisms (SNPs) in the ABCB1 gene in Caucasians, of which six were in the coding-region. However, only the C3435T SNP correlated with Pgp expression and function such that TT homozygotes had significantly decreased duodenal ABCB1 expression and increased digoxin (a substrate of P-gp) plasma concentrations. A similar relationship has also been demonstrated in CD56 + natural killer cells [5] and in cell lines derived from kidney, mammary, ovarian and colorectal tumours [3, 6, 7]. The C3435T SNP does not lead to an amino acid substitution and is therefore unlikely to influence P-gp expression directly. However, it may be linked to other variants in the gene that control expression, or in sequences that are important for mRNA processing.
Not all studies have demonstrated that the C3435T SNP has a functional effect. For example, P-gp expression was shown to be unrelated to the C3435T genotype in both placenta [8] and CD34+ haematopoietic stem cells [9]. In the placenta, Pgp expression was correlated with a promoter SNP, T-129C, and the nonsynonomous SNP G2677T(A), which results in an Ala→Ser amino acid change at position 893 of the protein. G2677T(A) has been shown to be in linkage disequilibrium with C3435T [10, 11].
To date, functional effects of ABCB1 SNPs have been shown in gut, haemopoietic cells and placenta, but not in the liver. Accordingly, in this study we have utilized RFLP analysis, Western blot analysis and quantitative real-time PCR to determine whether human liver P-gp expression is influenced by the C3435T or G2677T(A) SNPs.
Methods
Materials
The QIAamp DNA mini kit was obtained from QIAGEN Ltd (West Sussex, UK) and ultraPURE TRIzol reagent and molecular biology grade agarose were purchased from Gibco BRL (Life technologies, Paisley, UK). Nucleon PCR®/oligo clean up kit was obtained from Tepnel Life Sciences (Manchester, UK). Xylene was purchased from Fisher Scientific (Loughborough, Leicestershire, UK) and ethanol was obtained from BDH (Poole, Dorset, UK). All synthetic oligonucleotides and primers as well as MboI restriction endonuclease were obtained from Invitrogen Ltd (Paisley, UK). PCR and real-time PCR reagents were from Applied biosystems (California, USA) and Roche Diagnostics Ltd (East Sussex, UK), respectively. BIO-RAD protein assay solution was obtained from BIO-RAD (California, USA). All other reagents were obtained from Sigma (Dorset, UK).
Human liver samples
Histologically normal livers (n = 26) were obtained from Caucasian transplant donors. The median age of the patients was 36 years (IQR 22–48 years) and 18 (69%) were males. The certified cause of death was in each case traumatic injury due to a road traffic accident. The patients were being treated with a number of drugs. Five were receiving drugs that are known PXR ligands [12–15] (one taking phenobarbitone and phenytoin, one phenytoin, one dexamethasone, one dexamethasone and phenytoin, and one hydrocortisone). The details of dose and duration of therapy were not available, although in many cases, these drugs had been started acutely prior to death. The liver samples were transferred to the laboratory within 30 min of death. They were portioned, frozen in liquid nitrogen, and stored at −80 °C. All the livers that were available at the time were used for this study. Approval was granted by the Liverpool local research ethics committee and prior consent was obtained from the donors' relatives.
Amplification of ABCB1 exon 26 by polymerase chain reaction
Total liver genomic DNA was isolated using QIAamp DNA mini kit and amplification of ABCB1 exon 26 was carried out with primers designed from the ABCB1 sequence (medline accession M29445). Forward and reverse primers were 5′-GATCTGTGAACTCTTGT TTCA-3′ and 3′-GAAGAGAGCTTACATTAGG-5′, respectively. The DNA (40 ng µl−1; 1µl) was combined with ABCB1 sense (4 µm) and antisense (4 µm) primers, 50 µm each of the four dNTPs, 1 U of amplitaq gold DNA polymerase, and 5 µl of PCR buffer, in a total volume of 50 µl. The annealing temperature was 59 °C.
RFLP analysis for the C3435T genotype
RFLP analysis with MboI restriction endonuclease was carried out on PCR products in order to determine C3435T genotype. PCR product (5 µl) was combined with 4 units of MboI, 2 µl of 10x SuRE/Cut Buffer A and 0.1 µl of acetylated bovine serum albumin (10 µg µl−1) in a total volume of 20 µl. Samples were digested for 4 h at 37 °C and products electrophoresed on a 2% agarose gel containing ethidium bromide (0.5 µg ml−1). Heterozygotes, wild-type homozygotes and mutant homozygotes appeared as four, three and two bands, respectively. Results were confirmed for two individuals for each genotype by direct sequence analysis.
Amplification of ABCB1 exon 21 by polymerase chain reaction
Amplification of ABCB1 exon 21 was carried out with primers designed from the ABCB1 sequence (medline accession M29440). The forward and reverse primers for the PCR were 5′-CTATGGTTGGCAACTAACA CTG-3′ and 3′-CATATTTAGTTTGACTCACCTTGC TAG-3′, respectively. The DNA (20 ng µl−1; 1 µl) was combined with ABCB1 sense (4 µm) and antisense (4 µm) primers, 50 µm each of the four dNTPs, 1 U of hotmaster taq DNA polymerase, and 5 µl of PCR buffer, in a total volume of 50 µl. The annealing temperature was 68 °C.
Sequencing of G2677T(A) polymorphism
Exon 21 amplicons were cleaned according to the manufacturer's instructions using the nucleon PCR®/oligo clean up kit. Automated DNA sequencing was performed on an ABI 377 sequencer by using BigDye Terminator Version 2 reactions (Perkin Elmer/Applied Biosystems, UK) and the forward primer used for amplification of exon 21 (5′-CTATGGTTGGCAACTAACACTG-3′).
Western blot analysis of P-glycoprotein
Sections of liver (∼100 mg) were homogenized with a glass-Teflon homogenizer and protein concentration determined, as described previously [16]. An aliquot containing 50 µg of total protein was then taken and added to an equal volume of loading buffer. Samples were then boiled at 95 °C for 3 min and proteins separated by electrophoresis on SDS-polyacrylamide gels using the discontinuous buffer system described by Laemmli [17]. Proteins were then transferred to nitrocellulose membranes at 100 V for 1 h at 4 °C. Membranes were blocked for 1 h in 10% milk and probed for 2 h with goat anti-Pgp IgG antibody (C219; 1 : 2000 (v/v). Following further 1 h incubation with peroxide-conjugated rabbit antigoat IgG antibody (1 : 5000 (v/v)), the protein-antibody conjugates were visualized using the ECL Western blotting detection system. All samples were quantified from the same gel/membrane.
Quantification of MDR1 mRNA by real-time reverse-transcription polymerase chain reaction
Total cellular RNA was extracted as described previously [18]. Complementary DNA (cDNA) was synthesized from total RNA using the Promega Reverse Transcription System. Quantification of MDR1 mRNA was achieved by real-time PCR using the Roche lightcycler. cDNA (100 ng) was combined with MgCl2 (4 µm), sense (AAGCGACTGAATGTTCAGTG), and antisense primers (AGAGCTGAGTTCCTTTGTCT; 0.4 µm each) and CYBR green (2 µl) in a final volume of 10 µl. Amplification was carried out for 40 cycles with an annealing temperature of 56 °C. The above primers were used to amplify the MDR1 cDNA and the amplicon was electropheresed in a 1% agarose gel containing ethidium bromide. The band was then excised and gel purified using Qiagen gel purification kit. The amplicon was then quantified by reading the absorbance at 260 nm and the number of copies calculated according to the following equation:
A standard curve was then constructed alongside the samples in the range of 103−109 copies of the ABCB1 amplicon per reaction. The precision of the analyses was calculated as:
Assays were performed using the same amplicon stock solutions. The accuracy of the method was assessed at all amplicon concentrations and determined as:
The integrity of total RNA in all samples relative to freshly isolated RNA was assessed prior to reverse transcription. RNA samples (1 µl) were added to RNA loading buffer [10% (w/v) ficoll; 0.25% (w/v) bromophenol blue; 0.25% (w/v) xylene cyanole made up in nuclease free water; 1 µl] and nuclease free water (8 µl). The diluted samples were then electrophoresed on 1% agarose gel containing ethidium bromide (0.5 µg ml−1) prepared using 0.5x TBE buffer made up with DEPC-treated water [1 : 100 (v/v)].
Statistical analysis
All results are presented as mean ± SD. Statistical analysis was performed using one way analysis of variance followed by Scheffe's test for multiple comparisons, accepting P < 0.05 as being significant. Prior to all analyses, Hardy–Weinberg equilibrium testing was performed for all genotypes using the chi-squared test.
Results
Exon 26 was amplified from the genomic DNA of 26 livers and the C3435T genotype determined by RFLP analysis using the MboI restriction endonuclease. Fifty-eight % of the samples were heterozygotes and 23% were CC homozygotes (Table 1), and the genotypes were in Hardy–Weinberg equilibrium (number of observed CC, CT and TT genotypes = 6, 15 and 5, respectively; number of predicted CC, CT and TT genotypes = 7, 13 and 6, respectively; χ2 = 0.31; P = 0.86).
Table 1.
Frequency and distribution of C3435T amd G2677T(A) genotypes in human livers (n = 26)
| Single nucleotide polymorphisms | |||||||
|---|---|---|---|---|---|---|---|
| C3435T | G2677T(A) | ||||||
| CC | CT | TT | GG | GT | TT | GA | |
| Genotype frequency | 5 (19.2%) | 15 (57.7%) | 6 (23.1%) | 18 (69.2%) | 4 (15.5%) | 3 (11.5%) | 1 (3.8%) |
| Allele frequency | C/T 0.48/0.52 | G/T/A 0.52/0.19/0.02 | |||||
The G2677T(A) polymorphism in exon 21 was determined by PCR amplification and direct sequencing. Fifteen point five % of the sample set were heterozygotes while 11.5% were TT homozygotes (Table 1), and the genotypes obeyed Hardy–Weinberg equilibrium (number of observed GG, GT and TT genotypes = 19, 4 and 3, respectively; number of predicted GG, GT and TT genotypes = 17, 8 and 1, respectively; χ2 = 2.44; P = 0.29).
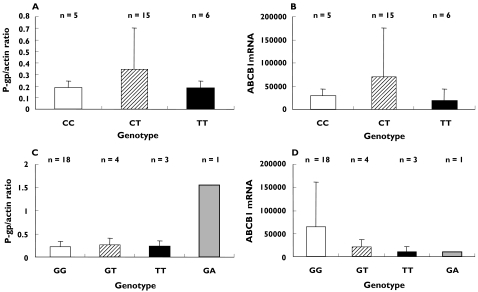
Hepatic P-gp content was assessed by Western blot analysis. Normalization of the optical density of the P-gp band against the optical density of the actin band gave an expression ratio of 0.27 (95% CI 0.16–0.39). The monoclonal antibody C219 has previously been shown to cross react with MDR3 P-glycoprotein [19]. However, no band was observed corresponding to the molecular weight of MDR3 (140 kDa). There was no relationship between either the C3435T genotypes (Figure 1A) or the G2677T(A) genotypes (Figure 1C) and P-gp expression.
Figure 1.
Relationship between the C3435T and G2677T(A) genotypes and hepatic P-gp and ABCB1 mRNA expression. (A) P-gp expression and C3435T genotype expressed as optical density of the P-gp band normalized against that of the β-actin band, (B) ABCB1 mRNA expression and C3435T genotype expressed as copies per 50 ng of cDNA, (C) P-gp expression and G2677T(A) genotype and (D) ABCB1 mRNA expression and G2677T(A) genotype
RNA integrity/degradation was assessed by electrophoresis of RNA in a 1% agarose gel. Hepatic ABCB1 mRNA expression was read from a parallel standard curve and the precision was calculated as 99%, 98%, 95.3%, 95.7% and 98.1% for 104, 105, 106, 107 and 108 copies, respectively. Similarly, the accuracy of the assay was calculated as 101 ± 0.3%, 97.1 ± 2.3%, 101.4 ± 2.1%, 101.7 ± 0.5% and 98.9 ± 0.8% for 104, 105, 106, 107 and 108 copies, respectively. ABCB1 mRNA expression did not differ between the C3435T genotypes (Figure 1B) and the G2677T(A) genotypes (Figure 1D).
Comparison of the five livers exposed to PXR ligands with the remaining 21 livers showed that there was no difference in either ABCB1 mRNA expression (mean different 2574 arbitrary units, 95% CI for the difference −84,870, 90,019, P = NS) or protein expression (mean difference in Pgp/actin ratio 0.13 arbitrary units, 95% CI −0.16, 0.43, P = NS) between the two groups.
There was 200- and 20-fold variability in expression of the mRNA and protein, respectively, between the different livers. However, there was no correlation between ABCB1 mRNA expression and the amount of P-gp protein as determined by Spearman rank correlation analysis (r = −0.11; 95% CI −0.48, 0.29).
Discussion
The aim of this study was to determine whether hepatic ABCB1 mRNA or P-gp expression varied with the C3435T or the G2677T(A) SNPs. In our sample, the frequency of the C and T alleles at position 3435 was not significantly different from previous studies [4, 8]. At position 2677, the allele frequencies in our study (78.8%, 19.2% and 2% for G, T and A alleles, respectively) also did not differ statistically from those reported recently in a Caucasian population (66% and 34% for G and T alleles, respectively [20]). However, the frequency of the 2677GG genotype in our study (69%) is higher than that reported previously (31%[11, 21]), despite the fact that the genotypes were in Hardy–Weinberg equilibrium. This is not due to sequencing error, since we have confirmed the genotypes using Taqman assays (data not shown), but may be a chance finding reflecting the small sample size.
Our data show that the substantial interindividual variability in ABCB1 mRNA and protein expression in livers could not be accounted for by either the C3435T or the G2677T(A) SNPs. This observation is unlikely to be due to degradation of RNA, since integrity was shown to be comparable with that of RNA from freshly prepared lymphocytes. Protein degradation within these samples is also therefore unlikely to have occurred given the stability of protein relative to mRNA, and no degradation has been detected in previous studies [25].
These data contradict reports present in the literature that have shown the TT genotype at position 3435 is associated with decreased protein expression in the duodenum [4] and decreased activity in CD56 + cells [5]. By contrast, other studies using digoxin pharmacokinetics as a marker of Pgp function have shown that the TT genotype is associated with increased activity [22, 23]. Our data are supported by other studies that have shown no effect of the C3435T genotype [24]. There are similar discrepancies in the data for the exon 21 G2677T SNP, with some studies showing that the TT genotype is associated with higher expression of Pgp [11], while others suggest it is associated with lower expression [8, 26, 27]. Our data showing that the G2677T SNP has no effect on expression are also supported by other reports [28–30].
A limitation of our study was the small sample size since only 26 livers were available for this work. Clearly, a larger cohort would be needed in order to examine whether these SNPs and the corresponding haplotypes have a minor role in determining variability of Pgp expression.
In summary, no correlation was seen between the C3435T or G2677T(A) polymorphisms of the ABCB1 gene and hepatic P-gp expression. Our data add to the growing literature that suggests the relationship between genetic variation in the ABCB1 (MDRI) gene and the function of the Pgp protein is complex and incompletely understood, and will require larger and more detailed studies of haplotypes.
References
- 1.Singh D, Alexander J, Owen A, et al. Whole-blood cultures from renal-transplant patients stimulated ex vivo show that the effects of cyclosporine on lymphocyte proliferation are related to P-glycoprotein expression. Transplantation. 2004;77:557–61. doi: 10.1097/01.tp.0000114594.21317.a5. [DOI] [PubMed] [Google Scholar]
- 2.Brinkmann U. Functional polymorphisms of the human multidrug resistance (MDR1) gene: correlation with P glycoprotein expression and activity in vivo. Novartis Found Symp. 2002;243:207–10. doi: 10.1002/0470846356.ch15. discussion•(210–2): 231–5. [DOI] [PubMed] [Google Scholar]
- 3.Siegsmund M, Brinkmann U, Schaffeler E, et al. Association of the P-glycoprotein transporter MDR1 (C3435T) polymorphism with the susceptibility to renal epithelial tumors. J Am Soc Nephrol. 2002;13:1847–54. doi: 10.1097/01.asn.0000019412.87412.bc. [DOI] [PubMed] [Google Scholar]
- 4.Hoffmeyer S, Burk O, von Richter O, et al. Functional polymorphisms of the human multidrug-resistance gene: multiple sequence variations and correlation of one allele with P-glycoprotein expression and activity in vivo. Proc Natl Acad Sci U S A. 2000;97:3473–8. doi: 10.1073/pnas.050585397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hitzl M, Drescher S, van der Kuip H, et al. The C3435T mutation in the human MDR1 gene is associated with altered efflux of the P-glycoprotein substrate rhodamine 123 from CD56+ natural killer cells. Pharmacogenetics. 2001;11:293–8. doi: 10.1097/00008571-200106000-00003. [DOI] [PubMed] [Google Scholar]
- 6.Potocnik U, Ravnik-Glavac M, Golouh R, Glavac D. Naturally occurring mutations and functional polymorphisms in multidrug resistance 1 gene. Correlation with microsatellite instability and lymphoid infiltration in colorectal cancers. J Med Genet. 2002;39:340–6. doi: 10.1136/jmg.39.5.340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sauer G, Kafka A, Grundmann R, et al. Basal expression of the multidrug resistance gene 1 (MDR-1) is associated with the TT genotype at the polymorphic site C3435T in mammary and ovarian carcinoma cell lines. Cancer Lett. 2002;185:79–85. doi: 10.1016/s0304-3835(02)00232-x. [DOI] [PubMed] [Google Scholar]
- 8.Tanabe M, Ieiri I, Nagata N, et al. Expression of P-glycoprotein in human placenta: relation to genetic polymorphism of the multidrug resistance (MDR)-1 gene. J Pharmacol Exp Ther. 2001;297:1137–43. [PubMed] [Google Scholar]
- 9.Calado RT, Falcao RP, Garcia AB, et al. Influence of functional MDR1 gene polymorphisms on P-glycoprotein activity in CD34+ hematopoietic stem cells. Haematologica. 2002;87:564–8. [PubMed] [Google Scholar]
- 10.Furuno T, Landi MT, Ceroni M, et al. Expression polymorphism of the blood–brain barrier component P-glycoprotein (MDR1) in relation to Parkinson's disease. Pharmacogenetics. 2002;12:529–34. doi: 10.1097/00008571-200210000-00004. [DOI] [PubMed] [Google Scholar]
- 11.Kim RB, Leake BF, Choo EF, et al. Identification of functionally variant MDR1 alleles among European Americans and African Americans. Clin Pharmacol Ther. 2001;70:189–99. doi: 10.1067/mcp.2001.117412. [DOI] [PubMed] [Google Scholar]
- 12.El-Sankary W, Plant NJ, Gibson GG, Moore DJ. Regulation of the CYP3A4 gene by hydrocortisone and xenobiotics. Role of the glucocorticoid and pregnane X receptors. Drug Metab Dispos. 2000;28:493–6. [PubMed] [Google Scholar]
- 13.Pascussi JM, Drocourt L, Gerbal-Chaloin S, et al. Dual effect of dexamethasone on CYP3A4 gene expression in human hepatocytes. Sequential role of glucocorticoid receptor and pregnane X receptor. Eur J Biochem. 2001;268:6346–58. doi: 10.1046/j.0014-2956.2001.02540.x. [DOI] [PubMed] [Google Scholar]
- 14.Sueyoshi T, Negishi M. Phenobarbital response elements of cytochrome P450 genes and nuclear receptors. Annu Rev Pharmacol Toxicol. 2001;41:123–43. doi: 10.1146/annurev.pharmtox.41.1.123. [DOI] [PubMed] [Google Scholar]
- 15.Raucy JL. Regulation of CYP3A4 expression in human hepatocytes by pharmaceuticals and natural products. Drug Metab Dispos. 2003;31:533–9. doi: 10.1124/dmd.31.5.533. [DOI] [PubMed] [Google Scholar]
- 16.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–54. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- 17.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–5. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 18.Kitteringham NR, Powell H, Clement YN, et al. Hepatocellular response to chemical stress in CD-1 mice: induction of early genes and gamma-glutamylcysteine synthetase. Hepatology. 2000;32:321–33. doi: 10.1053/jhep.2000.9602. [DOI] [PubMed] [Google Scholar]
- 19.Schinkel AH, Roelofs EM, Borst P. Characterization of the human MDR3 P-glycoprotein and its recognition by P-glycoprotein-specific monoclonal antibodies. Cancer Res. 1991;51:2628–35. [PubMed] [Google Scholar]
- 20.Schuetz EG, Furuya KN, Schuetz JD. Interindividual variation in expression of P-glycoprotein in normal human liver and secondary hepatic neoplasms. J Pharmacol Exp Ther. 1995;275:1011–18. [PubMed] [Google Scholar]
- 21.Drescher S, Schaeffeler E, Hitzl M, et al. MDR1 gene polymorphisms and disposition of the P-glycoprotein substrate fexofenadine. Br J Clin Pharmacol. 2002;53:526–34. doi: 10.1046/j.1365-2125.2002.01591.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sakaeda T, Nakamura T, Horinouchi M, et al. MDR1 genotype-related pharmacokinetics of digoxin after single oral administration in healthy Japanese subjects. Pharm Res. 2001;18:1400–4. doi: 10.1023/a:1012244520615. [DOI] [PubMed] [Google Scholar]
- 23.Oselin K, Nowakowski-Gashaw I, Mrozikiewicz PM, et al. Quantitative determination of MDR1 mRNA expression in peripheral blood lymphocytes: a possible role of genetic polymorphisms in the MDR1 gene. Eur J Clin Invest. 2003;33:261–7. doi: 10.1046/j.1365-2362.2003.01133.x. [DOI] [PubMed] [Google Scholar]
- 24.Cascorbi I, Gerloff T, Johne A, et al. Frequency of single nucleotide polymorphisms in the P-glycoprotein drug transporter MDR1 gene in white subjects. Clin Pharmacol Ther. 2001;69:169–74. doi: 10.1067/mcp.2001.114164. [DOI] [PubMed] [Google Scholar]
- 25.Powell H, Kitteringham NR, Pirmohamed M, Smith DA, Park BK. Expression of cytochrome P4502E1 in human liver: assessment by mRNA, genotype and phenotype. Pharmacogenetics. 1998;8:411–21. doi: 10.1097/00008571-199810000-00006. [DOI] [PubMed] [Google Scholar]
- 26.Moritra Y, Sakaeda T, Horinouchi M, et al. MDR1 genotype-related duodenal absorption rate of digoxin in healthy Japanese subjects. Pharm Res. 2003;20:552–6. doi: 10.1023/a:1023282312757. [DOI] [PubMed] [Google Scholar]
- 27.Verstuyft C, Schwab M, Schaeffeler E, et al. Digoxin pharmacokinetics and MDR1 genetic polymorphisms. Eur J Clin Pharmacol. 2003;58:809–12. doi: 10.1007/s00228-003-0567-5. [DOI] [PubMed] [Google Scholar]
- 28.Zheng H, Webber S, Zeevi A, et al. Tacrolimus dosing in pediatric heart transplant patients is related to CYP3A5 and MDR1 gene polymorphisms. Am J Transplant. 2003;3:477–83. doi: 10.1034/j.1600-6143.2003.00077.x. [DOI] [PubMed] [Google Scholar]
- 29.Oselin K, Gerloff T, Mrozikiewicz PM, Pahkla R, Roots I. MDR1 polymorphisms G2677T in exon 21 and C3435T in exon 26 fail to affect rhodamine 123 efflux in peripheral blood lymphocytes. Fundam Clin Pharmacol. 2003;17:463–9. doi: 10.1046/j.1472-8206.2003.00163.x. [DOI] [PubMed] [Google Scholar]
- 30.Siegmund W, Ludwig K, Giessmann T, et al. The effects of the human MDR1 genotype on the expression of duodenal P-glycoprotein and disposition of the probe drug talinolol. Clin Pharmacol Ther. 2002;72:572–83. doi: 10.1067/mcp.2002.127739. [DOI] [PubMed] [Google Scholar]