Abstract
Aims
To determine the effect of diltiazem on intestinal CYP3A activity and protein and mRNA expression in vivo in healthy subjects.
Methods
Intestinal biopsies were obtained from ten healthy controls and from ten healthy subjects after receiving diltiazem 120 mg bid for 7 days. Intestinal CYP3A activity, CYP3A4 protein and mRNA concentrations were quantified in both groups. Intestinal CYP3A activity was determined by incubation of small bowel homogenate with midazolam (25 µM) and NADPH for 5 min and the rate of formation of 1’-hydroxymidazolam was quantified.
Results
All subjects in the treatment group had detectable diltiazem concentration in the serum. While there was no significant difference in CYP3A4 protein and mRNA expression between the control and treatment groups, the formation of 1’-hydroxymidazolam (446 pmol min−1 mg−1 6 (control) vs. 170 (CI 112, 228) pmol min−1 mg−1 95% confidence interval (CI 269, 623) (diltiazem group)) was significantly reduced (P < 0.05).
Conclusion
Diltiazem decreased small bowel CYP3A activity by 62% as a result of irreversible inhibition with no corresponding change in intestinal CYP3A4 mRNA or protein concentrations.
Keywords: Diltiazem, CYP3A, intestinal biopsy, midazolam, irreversible inhibition
Introduction
The CYP3A subfamily is responsible for the metabolism of a large proportion of the drugs in current clinical use, including calcium channel blockers, benzodiazepines, immunosuppressants, and anticonvulsants [1]. In addition to hepatic expression, CYP3A4 is also the most abundant form of cytochrome P450 (CYP) found in the small intestinal epithelia of the human adults [2]. Diltiazem is a calcium channel antagonist widely used for the treatment of hypertension, chronic stable angina pectoris, and supraventricular arrythmias [3]. In vivo and in vitro studies have demonstrated that diltiazem is an inhibitor of CYP3A-dependent metabolism of many drugs [4]. Thus, diltiazem increases the bioavailability of several CYP3A substrates such as triazolam [5], cisapride [6], midazolam [7], simvastatin [8], pravastatin and lovastatin [9]. The in vivo metabolism of diltiazem involves N-demethylation to N-desmethyldiltiazem (MA) [10], which is catalysed primarily by CYP3A with less significant contributions from CYP2C8 and CYP2C9 [11]. Diltiazem and its metabolite MA are competitive inhibitors of CYP3A in human liver microsomes, with competitive inhibition constants (Ki) approaching 60 µm for diltiazem and 2 µm for MA [11, 12]. The steady state plasma concentration of diltiazem in humans during chronic diltiazem treatment is approximately 0.3 µm[10] and therefore, significant competitive inhibition of CYP3A by diltiazem via inhibition is not expected. Similarly, the reported steady-state plasma concentration of MA is 0.15 µm[10], which again, is much lower than the reported Ki, and thus would not explain the inhibition of CYP3A by diltiazem or its metabolites through a reversible mechanism. Diltiazem forms a metabolic intermediate complex (MIC) in vitro and in vivo in dexametasone and phenobarbital induced rat liver microsomes [13]. When human liver microsomes were preincubated with diltiazem for 60 min, there was more than 80% inhibition of midazolam 1′-hydroxylation [14] This occurs through the formation of a MIC, which results in a catalytically inactive enzyme [10]. When lovastatin was administered after oral diltiazem (120 mg bid for 7 days), there was 3.6-fold increase in oral AUC of the former drug without a change in half-life, which was consistent with a first pass metabolism secondary to intestinal CYP3A inhibition [9]. In vivo the effect of diltiazem on intestinal CYP3A protein and mRNA expression is unknown. The aim of this study was to determine the effect of diltiazem on intestinal CYP3A catalytic activity, CYP3A4 protein and mRNA expression in vivo in healthy subjects.
Materials and methods
Subjects
Twenty healthy subjects were randomly assigned to either the diltiazem or the control group. In the former group the subjects underwent intestinal biopsies after receiving diltiazem (120 mg bid for 7 days) and the controls had biopsies at the same time without receiving any medication. All subjects were 18 years or older, received no prescription or over-the-counter medications for 2 weeks before the study. Individuals with intolerance to diltiazem or benzodiazepines, significant medical history, and who were smokers or who drank alcohol were excluded from the study. One week prior to and during the study, subjects abstained from consuming grapefruit or juice, apple juice, citrus products, or vegetables from the mustard green family. Clarian and Indiana University Purdue University Indianapolis (IUPUI) Institutional Review Board approved this study. All the subjects provided written informed consent.
Experimental design
After overnight fast, four proximal small bowel mucosal biopsy specimens were obtained by upper intestinal endoscopy from each subject in the control group and after 7 days of treatment with diltiazem (120 mg bid) in the treatment group. Subjects received intravenous midazolam (Roche Pharmaceuticals, Nutley, NJ) for conscious sedation for endoscopy and the dose varied between subjects. A single random blood sample was drawn for the determination of the serum midazolam and concentration. In the diltiazem group a blood sample was obtained immediately before endoscopy on day 8 to measure serum concentrations of the drug and its metabolite MA. Subjects were questioned to monitor compliance with diet, alcohol and drug restrictions. All the biopsies were immediately snap frozen in liquid nitrogen and stored at −80 °C until analysis.
Preparation of duodenal homogenates
Duodenal biopsies (approximately 20 µg wet tissue weight) were homogenized with 20 strokes of a hand-held glass homogenizer in an ice jacket with 1 ml ice cold buffer consisting of 50 m m tris (hydroxymethyl-aminomethane), 2 m m ethylenediaminetetraacetic acid, 1 m m phenylmethylsulphonyl fluoride, 1 mm benzamidine, 50 µg apotinin, and 20% glycerol (pH 7.4). The resultant homogenate was then snap frozen in liquid nitrogen and stored at −80 °C until assayed. Homogenate protein concentration was determined by the micro Lowry assay using bovine serum albumin [15].
Determination of CYP3A activity using midazolam
An aliquot of 0.5 ml of duodenal homogenate was diluted with 0.5 ml 100 mm sodium phosphate buffer (pH = 7.4) and incubated at 37 °5C with 25 µm midazolam. The reaction was started by the addition of 1µmol NADPH, and was quenched after 10 min with 1 ml of ethyl acetate/hexane (50/50; v/v) and then 1 ml of 1N NaOH 1M glycine pH = 11.3 buffer on ice. N-desmethyldiazepam was added as an internal standard, and the mixture was extracted with an additional 3 ml ethyl acetate/hexane (50/50; v/v). The organic phase was dried under vacuum and the samples were reconstituted in 100 µl of mobile phase (20 m m ammonium acetate pH 7.4, acetonitrile, and methanol; 40/40/20: v/v/v). N-desmethyldiazapam, 1′-hydroxymidazolam and were analysed by HPLC/MS (Navigator, Finnigan, San Jose, CA). The separation was achieved with an isocratic flow of 1 ml min−1 to a Phenomenex Luna C-18 column (5 µm; 150 mm × 4.6 mm ID). The eluents were analysed by atmospheric pressure chemical ionization (APCI) in the positive mode with a cone voltage of 25 V and source and probe temperatures of 200 °C and 550 °C, respectively. N-desmethyldiazapam, 1′-hydroxymidazolam were detected with selected ion monitoring at m/z 271 and 342, respectively. The limit of quantification of the hydroxylated metabolite of midazolam was 0.2 ng and the precision at this concentration was 3.2%.
Preparation and analysis of CYP3A4 mRNA
Total RNA from the samples was prepared using Trizol® reagent (Invitrogen Corp., Carlsbad, CA) according the manufacturer's instructions. RNA yield was determined by spectrophotometry (Beckman DU® 640, Beckman Coulter, Inc., Fullerton, CA) and its quality was assessed by the ratio of its absorption at 260 nm : 280 nm. Following RNA isolation all samples were stored at −80 °C until cDNA preparation. In a 20-µl reaction containing random hexamers, 2 µg of total RNA was reverse transcribed into cDNA using the Promega Reverse Transcription system (Promega Corp., Madison WI) according to the manufacturer's instructions.
A real-time PCR method was used to measure the amount of CYP3A4 mRNA in intestinal biopsy specimen using a previously described method [16] with minor modifications. Primers specific to CYP3A4 transcripts were purchased from Integrated DNA Technologies (Coralville, IA). The CYP3A4 forward primer sequence was 5′-CAT TCC TCA TCC CAA TTC TTG AAG T-3′ and the reverse primer sequence was 5′-CCA CTC GGT GCT TTT GTG TAT CT-3′. As a control, each sample was assayed for the expression of the housekeeping gene villin using the forward primer 5′-GAA GGT GAA GGT CGG AGT C-3′ and the reverse primer 5′-GAA GAT GGT GAT GGG ATT TC-3′. The cDNA samples were assayed for the expression of CYP3A4 and villin using the SYBR Green Core Reagents (Applied Biosystems, Foster City, CA) on a Bio-Rad iCycler Thermal Cycler. The running conditions include three steps: 95 °C for 10 min (activation of Taq); 95 °C for 15 s (denaturation); 58 °C for 1 min (annealing step and extension). Steps 2 and 3 were repeated for 40 cycles. Melt-curve analysis was used to ensure that the expected PCR products are being generated continuously and reproducibly in each real-time PCR reaction. The presence of a single inflection point on the melt-curve, at the previously established temperature, indicates that a single PCR product is generated. A dilution series containing 200 femtograms to 0.2 attograms of CYP3A4 and villin cDNA was used as positive controls and to generate calibrator standards. These controls are required to determine the PCR efficiencies and were included with every assay. The slope of the plot of the threshold (CT) values against original cDNA concentrations was used to calculate efficiency using the formula E = 10(1/slope)− 1. The relative expression of CYP3A4 gene, expressed as fold variation over control, were calculated after the determination of the difference between CTfor the CYP3A4 gene and that of the calibrator gene villin in the intestinal biopsy samples.
Immunoblotting
An aliquot (50 µg) of small bowel homogenate, CYP3A4, or villin protein was mixed with Laemmli sample buffer containing sodium lauryl sulphate (SDS) and 2-mercaptoethanol and heated to 95 °C for 4 min. The samples were electrophoresized in 10% polyacrylamide/0.1% SDS gels until the bromophenol blue dye front ran off the gel. The samples were transferred onto Hybond-P, hydrophobic polyvinylidene difluoride (PVDF) membranes (Amersham Pharmacia Biotech, Piscataway, NJ) with a 25 mm Tris and 192 mm glycine buffer and methanol (60/40%; v/v) electrophoresed at 100 V for 2 h. The blots were blocked for 1 h in Tris-buffered saline containing 0.1% Tween 20 and 5% nonfat dry milk. The membrane was then probed for 1 h with mouse polyclonal antibody to CYP3A4 (Gentest, Woburn, MA) and a mouse monoclonal antibody to chick villin that is known to cross-react with human villin (Chemicon, Temecula, CA). After extensive washing with Tris-buffered saline the membranes were probed with a fluoresein linked alkaline phosphatase conjugated anti mouse IgG, and developed with an ECF Western blotting chemifluoresence kit (Amersham Pharmacia Biotech, Piscataway, NJ). The membrane was exposed to fluorescent substrate for 20 min and band densities were measured with a STORM 860 imager (Molecular Dynamics, Sunnyvale, CA).
Correction for interbiopsy variation of enterocyte content
CYP3A4 protein is expressed exclusively in mature enterocytes. Enterocytes represent only a small portion of tissue obtained in the pinch biopsy of the intestine [18]. It is well known that there is significant interbiopsy variation in the content of enterocytes among biopsy obtained from single individual will alter the amount of immunoreative protein observed on a blot. Villin, an enterocyte specific protein is able to control for the variation in biopsy content of mature enterocytes [18]. Therefore biopsy levels of CYP3A4 were expressed as a ratio with the villin level in the same sample. These villin corrected values provide a relative measure of enterocyte CYP3A4 concentration.
Drug and metabolite analysis
Serum midazolam and intestinal biopsy homogenate 1′hydroxymidazolam concentrations were measured by liquid chromatography-mass spectrometry [17]. Serum diltiazem concentrations were measured by a high performance liquid chromatography method that has been reported previously [9]. The intraday and interday coefficients of variation were 10% or less.
Statistical analysis
Data are reported as mean and standard deviation. The effect of diltiazem was determined by paired students t-test and significance was associated with P ≤ 0.05.
Results
Four healthy men and six women in each of the the control and diltiazem group completed the study without endoscopy or sedation related side-effects. All the subjects in the diltiazem group had detectable serum concentrations of diltiazem (105.6 ng ml−1 (CI 63.8, 147.4)) and MA metabolite (72.7 ng ml−1 (CI 33.8, 111.6)).
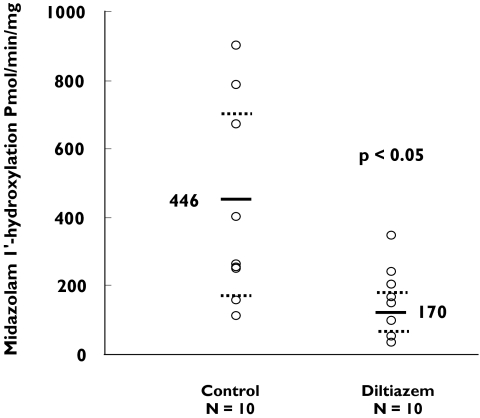
The rate of formation of 1′-hydroxymidazolam in intestinal biopsy homogenates after incubation with 25 µm midazolam for 10 min is shown in Figure 1. There was a significant (62%) decrease (P < 0.05) in the mean rate of formation of 1′-hydroxymidazolam following 7 days of treatment with diltiazem (170 (CI 112.228) pmol min−1 mg−1) compared with that of the control group [CI 269.623] (446 ± 286 pmol min−1 mg−1).
Figure 1.
The effect of diltiazem treatment (120 mg bid for 7 days) on intestinal CYP3A activity as determined by 1′-hydroxymidazolam formation from midazolam in intestinal biopsies from healthy subjects. The solid lines and the dotted lines represent the mean and standard deviation rates of 1′-hydroxymidazolam formation, respectively
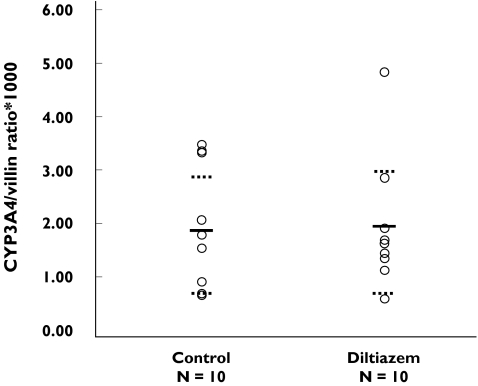
There was no significant change in intestinal CYP3A4 protein concentration in the diltiazem group (1.92 CI 1.26, 2.58) when compared with the control group (1.83 CI 1.09, 2.57) (Figure 2).
Figure 2.
Intestinal CYP3A protein concentration in controls and in the group that received diltiazem (120 mg bid for 7 days)
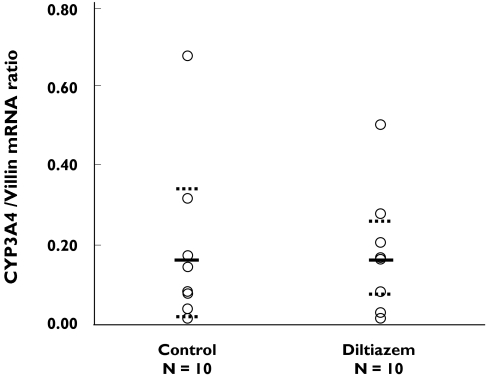
The mean enterocyte CYP3A4 mRNA concentration expressed as the ratio of CYP3A4/villin mRNA was 0.18 CI and 0.07, 0.30 in the diltiazem group and 0.18 CI 0.06, 0.31 in the control group (Figure 3).
Figure 3.
Relative concentration of intestinal CYP3A mRNA in the group that received diltiazem (120 mg bid for 7 days) and in controls. The solid lines and the dotted lines represent the mean and standard deviation rates of CYP3A4/villin in mRNA ratio, respectively
Discussion
The CYP3A enzymes are responsible for the metabolism of approximately 50% of all drugs in current clinical use that undergo biotransformation, including calcium channel blockers, macrolide antibiotics, benzodiazepines, immunosuppressants, and anticonvulsants [1]. In humans, CYP3A enzymes are the most abundantly expressed CYPs in both liver and the enterocytes of the small intestine [2]. Intestinal wall metabolism makes a substantial contribution to the low bioavailibility of many CYP3A substrates, such as midazolam and cyclosporin, which have intestinal extraction ratios of 25–51% and 14–59%, respectively [1]. Similarly, intestinal CYP3A is a quantitatively important locus for a number of clinically significant oral drug–drug interactions including midazolam–saquinavir [19], midazolam–clarithromycin [20], midazolam–erythromycin [21], and terfenadine–ketoconazole [22].
The mechanism by which inhibitors decrease CYP3A catalytic activity is usually assumed to be both reversible and competitive, which can explain interactions such as ketoconazole–terfenadine [22] and saquinavir–midazolam [19]. However, more recently the potential importance of irreversible inhibition has been recognized. Irreversible inhibition of CYP3A enzymes results in delayed onset and offset of the inhibitory effect. As a result, the duration of the inhibitory effect is longer than predicted. When CYP3A is responsible for the elimination of the inhibitor, then auto-inhibition occurs resulting in nonlinear disposition. For example, the half-life of diltiazem is 3.7 h following single 20 mg intravenous dose, but the half-life is increased to 6.34 h after administration of oral diltiazem 120 mg bid for 14 days [23]. Diltiazem also forms a MIC with CYP4A in vitro and the rate of formation correlates with the loss of enzyme activity [14, 12]. However, in humans it is not known whether diltiazem forms a MIC with CYP3A in the liver and intestine in vivo.
In rat liver, diltiazem did not form a MIC with CYP3A in vivo when diltiazem was given alone, however, MIC formation was observed when diltiazem was administered to rats pretreated with dexamethasone [13]. In humans, CYP3A inhibitor troleandomycin 4.25 g orally over three days formed a MIC with hepatic CYP3A in vivo but the amount of CYP3A protein in the liver was increased by 5-fold and erythromycin demethylation activity was reduced compared with controls [24]. Similarly, erythromycin 500 mg tid for 7 days formed MIC with hepatic CYP3A in vivo and increased CYP3A protein by 2-fold in the livers of patients undergoing elective surgery [25]. The effect of diltiazem on expression of intestinal CYP3A protein or mRNA has not been studied in vivo. The aim of this study was to determine the effect of diltiazem on intestinal CYP3A catalytic activity, CYP3A protein and mRNA expression in vivo in healthy subjects.
In rodents treatment with troleandomycin for 5 days increased the expression of hepatic CYP3A mRNA [26]. In contrast, our study did not detect a significant change in intestinal CYP3A4 mRNA expression in the diltiazem group compared with controls. This discrepancy may reflect differences in the way CYP3A4 mRNA is regulated in humans and rodents, as well as differences in the effects of CYP3A4 inhibitors on mRNA expression within a given species. Additionally, there is evidence to suggest that intestinal and hepatic CYP3A4 are not co-regulated although transcriptional activation via PXR is common to both.
In addition to lack of change in intestinal CYP3A4 mRNA expression, diltiazem did not have any effect on intestinal CYP3A4 protein expression. Several studies have demonstrated that administration of CYP3A inhibitors such as erythromycin and troleandomycin results in increases in hepatic CYP3A content in both humans and rodents [25, 27]. In rodents clarithromycin administration for 7 days increased hepatic CYP3A protein content by 3-fold compared with controls [28]. The effect of diltiazem on intestinal CYP3A protein expression has not been studied in humans or animals. Administration of grapefruit juice (240 ml tid for 6 days), to healthy human subjects, decreased CYP3A4 concentration by 62% in the intestinal homogenate and the authors speculated that this occurred through suicide inhibition resulting in accelerated degradation of the enzyme [29]. In the present study we have shown that diltiazem does not change in the intestinal concentration of CYP3A4. Thus, it is likely that MIC formation does not result in stabilization or accelerated degradation of CYP3A4 protein.
In contrast to lack of change in intestinal CYP3A4 protein and mRNA expression after treatment with diltiazem, we found that there was significant 62% decrease in intestinal CYP3A activity in the diltiazem group compared with controls. We have previously demonstrated that when liver microsomes are preincubated with diltiazem, the formation of a MIC corresponds to loss of CYP3A activity in vitro [12]. Although in the current study, we did not measure diltiazem in the intestinal homogenate, the most likely mechanism of CYP3A inhibition by clarithromycin is irreversible inhibition. During the process of homogenate preparation, the intestinal biopsy tissue is diluted 50-fold and therefore it is unlikely that there is any free diltiazem in the incubation. In the present study, the observed average steady-state serum diltiazem concentration was 0.3 µm. Previous studies indicate that diltiazem is also a competitive inhibitor of CYP3A in vitro with a Ki value of 50 µm[30, 4]. Thus, the steady-state concentration is much lower than the reported K for reversible competitive inhibition of CYP3A4 by diltiazem in vitro. The negligible concentration of diltiazem in the intestinal homogenate and the known potency of diltiazem as an irreversible inhibitor strongly suggests that the mechanism of CYP3A inhibition by diltiazem is primarily by MIC formation.
In conclusion, administration of diltiazem 120 mg bid for 7 days irreversibly inhibits intestinal CYP3A activity with no corresponding change in intestinal CYP3A4 protein or mRNA expression in healthy adults. Unlike other CYP3A inhibitors such as erythromycin and troleandomycin, diltiazem did not increase CYP3A protein concentration following chronic administration.
Acknowledgments
This study was supported by National Institutes of Health grants T32G m08425, AG 13718, and MO1-RR00750; Food and Drug Administration Cooperative Agreement FDT-001756.
Competing interests: None to declare.
References
- 1.Thummel KE, Wilkinson GR. In vitro and in vivo drug interactions involving human CYP3A. Annu Rev Pharmacol Toxicol. 1998;38:389–430. doi: 10.1146/annurev.pharmtox.38.1.389. [DOI] [PubMed] [Google Scholar]
- 2.Watkins PB. Drug metabolism by cytochromes P450 in the liver and small bowel. Gastroenterol Clin North Am. 1992;21:511–26. [PubMed] [Google Scholar]
- 3.Chaffman M, Brogden RN. Diltiazem. A review of its pharmacological properties and therapeutic efficacy. Drugs. 1985;29:387–454. doi: 10.2165/00003495-198529050-00001. [DOI] [PubMed] [Google Scholar]
- 4.Renton KW. Inhibition of hepatic microsomal drug metabolism by the calcium channel blockers diltiazem and verapamil. Biochem Pharmacol. 1985;34:2549–53. doi: 10.1016/0006-2952(85)90541-6. [DOI] [PubMed] [Google Scholar]
- 5.Varhe A, Olkkola KT, Neuvonen PJ. Diltiazem enhances the effects of triazolam by inhibiting its metabolism. Clin Pharmacol Ther. 1996;59:369–75. doi: 10.1016/S0009-9236(96)90103-4. [DOI] [PubMed] [Google Scholar]
- 6.Thomas AR, Chan LN, Bauman JL, Olopade CO. Prolongation of the QT interval related to cisapride–diltiazem interaction. Pharmacotherapy. 1998;18:381–5. [PubMed] [Google Scholar]
- 7.Ahonen J, Olkkola KT, Salmenpera M, Hynynen M, Neuvonen PJ. Effect of diltiazem on midazolam and alfentanil disposition in patients undergoing coronary artery bypass grafting. Anesthesiology. 1996;85:1246–52. doi: 10.1097/00000542-199612000-00004. [DOI] [PubMed] [Google Scholar]
- 8.Mousa O, Brater DC, Sunblad KJ, Hall SD. The interaction of diltiazem with simvastatin. Clin Pharmacol Ther. 2000;67:267–74. doi: 10.1067/mcp.2000.104609. [DOI] [PubMed] [Google Scholar]
- 9.Azie NE, Brater DC, Becker PA, Jones DR, Hall SD. The interaction of diltiazem with lovastatin and pravastatin. Clin Pharmacol Ther. 1998;64:369–77. doi: 10.1016/S0009-9236(98)90067-4. [DOI] [PubMed] [Google Scholar]
- 10.Yeung PK, Buckley SJ, Hung OR, Pollak PT, Barclay KD, Feng JD, et al. Steady-state plasma concentrations of diltiazem and its metabolites in patients and healthy subjects. Ther Drug Monit. 1996;18:40–5. doi: 10.1097/00007691-199602000-00007. [DOI] [PubMed] [Google Scholar]
- 11.Sutton D, Butler AM, Nadin L, Murray M. Role of CYP3A4 in human hepatic diltiazem N-demethylation: inhibition of CYP3A4 activity by oxidized diltiazem metabolites. J Pharmacol Exp Ther. 1997;282:294–300. [PubMed] [Google Scholar]
- 12.Mayhew BS, Jones DR, Hall SD. An in vitro model for predicting in vivo inhibition of cytochrome P450 3A4 by metabolic intermediate complex formation. Drug Metab Dispos. 2000;28:1031–7. [PubMed] [Google Scholar]
- 13.Bensoussan C, Delaforge M, Mansuy D. Particular ability of cytochromes P450, 3A to form inhibitory P450–iron–metabolite complexes upon metabolic oxidation of aminodrugs. Biochem Pharmacol. 1995;49:591–602. doi: 10.1016/0006-2952(94)00477-4. [DOI] [PubMed] [Google Scholar]
- 14.Jones DR, Gorski JC, Hamman MA, Mayhew BS, Rider S, Hall SD. Diltiazem inhibition of cytochrome P-450 3A activity is due to metabolite intermediate complex formation. J Pharmacol Exp Ther. 1999;290:1116–25. [PubMed] [Google Scholar]
- 15.Lowry OH, Rosenbrough NH, Farr AL, Randall FJ. Protein measurement with Folin phenol reagent. J Biol Chem. 1951;193:265–75. [PubMed] [Google Scholar]
- 16.Westlind A, Malmebo S, Johansson I, Otter C, Andersson TB, Ingelman-Sundberg M, et al. Cloning and tissue distribution of a novel human cytochrome p450 of the CYP3A subfamily, CYP3A43. Biochem Biophys Res Commun. 2001;281:1349–55. doi: 10.1006/bbrc.2001.4505. [DOI] [PubMed] [Google Scholar]
- 17.Belle DJ, Callaghan JT, Gorski JC, Maya JF, Mousa O, Wrighton SA, et al. The effects of an oral contraceptive containing ethinyloestradiol and norgestrel on CYP3A activity. Br J Clin Pharmacol. 2002;53:67–74. doi: 10.1046/j.0306-5251.2001.01521.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lown KS, Kolars JC, Thummel KE, Barnett JL, Kunze KL, Wrighton SA, et al. Interpatient heterogeneity in expression of CYP3A4 and CYP3A5 in small bowel. Lack of prediction by the erythromycin breath test. Drug Metab Dispos. 1994;22:947–55. [PubMed] [Google Scholar]
- 19.Palkama VJ, Ahonen J, Neuvonen PJ, Olkkola KT. Effect of saquinavir on the pharmacokinetics and pharmacodynamics of oral and intravenous midazolam. Clin Pharmacol Ther. 1999;66:33–9. doi: 10.1016/S0009-9236(99)70051-2. [DOI] [PubMed] [Google Scholar]
- 20.Gorski JC, Jones DR, Haehner-Daniels BD, Hamman MA, O'Mara EM, Jr, Hall SD. The contribution of intestinal and hepatic CYP3A to the interaction between midazolam and clarithromycin. Clin Pharmacol Ther. 1998;64:133–43. doi: 10.1016/S0009-9236(98)90146-1. [DOI] [PubMed] [Google Scholar]
- 21.Olkkola KT, Aranko K, Luurila H, Hiller A, Saarnivaara L, Himberg JJ, et al. A potentially hazardous interaction between erythromycin and midazolam. Clin Pharmacol Ther. 1993;53:298–305. doi: 10.1038/clpt.1993.25. [DOI] [PubMed] [Google Scholar]
- 22.Honig PK, Wortham DC, Zamani K, Conner DP, Mullin JC, Cantilena LR. Terfenadine–ketoconazole interaction. Pharmacokinetic and electrocardiographic consequences. JAMA. 1993;269:1513–8. [PubMed] [Google Scholar]
- 23.Abernethy DR, Montamat SC. Acute and chronic studies of diltiazem in elderly versus young hypertensive patients. Am J Cardiol. 1987;60:116I–20I. doi: 10.1016/0002-9149(87)90471-1. [DOI] [PubMed] [Google Scholar]
- 24.Watkins PB, Wrighton SA, Maurel P, Schuetz EG, Mendez-Picon G, Parker GA, et al. Identification of an inducible form of cytochrome P-450 in human liver. Proc Natl Acad Sci USA. 1985;82:6310–4. doi: 10.1073/pnas.82.18.6310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Larrey D, Funck-Brentano C, Breil P, Vitaux J, Theodore C, Babany G, et al. Effects of erythromycin on hepatic drug-metabolizing enzymes in humans. Biochem Pharmacol. 1983;32:1063–8. doi: 10.1016/0006-2952(83)90626-3. [DOI] [PubMed] [Google Scholar]
- 26.Hostetler KA, Wrighton SA, Molowa DT, Thomas PE, Levin W, Guzelian PS. Coinduction of multiple hepatic cytochrome P-450 proteins and their mRNAs in rats treated with imidazole antimycotic agents. Mol Pharmacol. 1989;35:279–85. [PubMed] [Google Scholar]
- 27.Wrighton SA, Campanile C, Thomas PE, Maines SL, Watkins PB, Parker G, et al. Identification of a human liver cytochrome P-450 homologous to the major isosafrole-inducible cytochrome P-450 in the rat. Mol Pharmacol. 1986;29:405–10. [PubMed] [Google Scholar]
- 28.Tinel M, Descatoire V, Larrey D, Loeper J, Labbe G, Letteron P, et al. Effects of clarithromycin on cytochrome P-450. Comparison with other macrolides. J Pharmacol Exp Ther. 1989;250:746–51. [PubMed] [Google Scholar]
- 29.Lown KS, Bailey DG, Fontana RJ, Janardan SK, Adair CH, Fortlage LA, et al. Grapefruit juice increases felodipine oral availability in humans by decreasing intestinal CYP3A protein expression. J Clin Invest. 1997. pp. 2545–53. [DOI] [PMC free article] [PubMed]
- 30.Pichard L, Fabre I, Fabre G, Domergue J, Saint AB, Mourad G, et al. Cyclosporin A drug interactions. Screening for inducers and inhibitors of cytochrome P-450 (cyclosporin A oxidase) in primary cultures of human hepatocytes and in liver microsomes. Drug Metab Dispos. 1990;18:595–606. [PubMed] [Google Scholar]