Abstract
Aims
The aim of this exploratory study was to investigate associations between irinotecan pharmacokinetic parameters and allelic variants in genes encoding for drug transporters and drug metabolizing enzymes that are involved in irinotecan disposition in Asian patients with cancer.
Methods
Irinotecan was administered at 100 mg m−2 over 90 min on a weekly schedule to 29 nasopharyngeal carcinoma patients and pharmacokinetic analysis was performed during the first cycle. All patients were genotyped for allelic variants in genes encoding drug metabolizing enzymes (CYP3A4, CYP3A5, UGT1A1) and drug transporters (ABCB1, ABCC2 and ABCG2) that are involved in irinotecan disposition.
Results
Of the six candidate genes that were analyzed, 11 genetic variants were found. Significant genotypic–phenotypic associations were apparent only for transporter genes. The Cmax of irinotecan was significantly lower in patients carrying the CC genotype at exon 26 of the ABCB1 gene compared with those harbouring at least one variant allele (P = 0.047). Patients harbouring the wild type ABCG2 CTCA genotype were associated with significantly higher values for relative extent of conversion (REC) of irinotecan to SN-38 compared with patients carrying at least one deletion CTCA allele (P = 0.019).
Conclusions
The present exploratory study shows that genetic polymorphisms in drug transporter genes, particularly in ABCB1 and ABCG2 genes, may be important in influencing the pharmacokinetics of irinotecan and its metabolites. The predictive value of the identified allelic variants in the ABCG2 and ABCB1 genes on irinotecan disposition should be further investigated in a larger patient population as well as in other ethnic populations.
Keywords: ABC transporters, drug metabolizing enzymes, single nucleotide polymorphisms
Introduction
A clear understanding of interindividual variability in drug response is important to allow optimization of therapy for the individual patient. This is particularly important in cancer therapy as the therapeutic window between efficacy and toxicity is very narrow and the interpatient variability to drug responses is very large. Much of the variabilities in drug response are speculated to be due at least in part to differences in activities of drug metabolizing enzymes or the functions of drug transporters that affect the pharmacokinetics of various drugs as well as alterations in drug targets that may influence the pharmacodynamic characteristics of a drug molecule. Polymorphisms in genes encoding these proteins are partly responsible for the observed heterogeneity in drug responses between different individuals as well as between different ethnic groups. Also, the influence of environmental factors (e.g. nutrition, concurrent medications) cannot be excluded.
Irinotecan (CPT-11) is a topoisomerase I inhibitor and is widely used in the treatment of metastatic colorectal cancer either in combination therapy with 5-fluorouracil and leucovorin or as monotherapy in the second line setting [1]. It has an extremely complex metabolic pathway involving the participation of both phase I and II drug metabolizing enzymes as well as numerous transporters belonging to the adenosine triphosphate binding cassette (ABC) family of transporters. Following intravenous infusion, it is hydrolyzed by carboxylesterases to 7-ethyl-10-hydroxycamptothecin (SN-38), which is 100- to 1000-fold more active than irinotecan [2]. SN-38 undergoes hepatic glucuronidation by uridine diphosphate glucuronosyltransferase 1A1 (UGT1A1) to form an inactive glucuronide, SN-38G [3, 4]. Irinotecan also undergoes oxidative metabolism by the CYP3A4/5 enzymes to form the major plasma metabolite 7-ethyl-10-[4-N-(5-aminopentanoic acid) -1 -piperidino] -carbonyloxycamptothecin (APC) and 7-ethyl-10-[4-amino-1-piperidino]-carbonyloxycamptothecin (NPC) [5, 6]. Both APC and NPC lack cytotoxic activity but NPC may be functionally important as it can be hydrolyzed by carboxylesterase to SN-38 [6, 7].
In the clinical setting, the pharmacokinetics-pharmacodynamics of irinotecan show wide interindividual variation in response as well as occurrence of toxic side-effects. Two major toxicities of irinotecan are diarrhoea and myelosuppression, which can be life threatening, and have been the topic of much research in recent years [1, 8]. The incidence of severe diarrhoea ranges from 11 to 36.4% in colorectal cancer patients and has been associated with a genetic defect in the promoter region of the UGT1A1 gene [9, 10]. The latter is polymorphic and cancer patients with seven TA repeats in the TATA box region of the UGT1A1 gene have decreased glucuronidating capacity [11]. UGT1A1 activity varies significantly in human livers with a 17-fold difference between minimum and maximum rates of SN-38 glucuronidator phenotype [9]. In addition to UGT1A1, UGT1A7 and UGT1A9 are involved in the glucuronidation of SN-38 [12, 13] and the participation of these two isoforms may obscure the predictive role of the UGT1A1*28 genotype [18]. The metabolism of irinotecan is further complicated by the involvement of polymorphic CYP3A4/5 mediated oxidative reactions. Besides the involvement of drug metabolizing enzymes, several transporters belonging to the ABC superfamily are involved in mediating the elimination of irinotecan and its metabolites [14–17]. Genetic polymorphisms in the ABC transporters have been shown to be associated with altered transport of their respective substrates, thereby affecting the pharmacokinetics-pharmacodynamics of their drug substrates.
To date, most studies have investigated the influence of UGT1A1 promoter polymorphisms on the pharmacodynamics of irinotecan [8–11]. Few studies have been done to investigate the co-operative influence of multiple single nucleotide polymorphisms (SNPs) in the various drug metabolizing enzymes and ABC transporters on the pharmacokinetics-pharmacodynamics of irinotecan and its metabolites. In a recent study by Mathijssen et al. [18], the authors explored the relationships between the disposition of irinotecan and its metabolites and several allelic variants of genes involved in irinotecan's metabolic pathway. Of the 18 genetic variants identified in nine genes, only the homozygous T allele in exon 12 (1236C > T) was found to be significantly associated with increased exposure to irinotecan and SN-38 and no pharmacokinetic relationships were found with other variant genotypes. The authors postulated that genotyping for ABCB11236C > T SNP in cancer patients may assist in dose optimization of irinotecan. Since there are ethnic differences in the prevalence of these allelic variants involved in the irinotecan disposition pathway, it would be useful to identify relevant marker SNPs in cancer patients belonging to other ethnic populations as well. Thus the objective of this exploratory study was to investigate the associations between genetic polymorphisms in candidate genes involved in irinotecan disposition and individual irinotecan pharmacokinetic parameters in Asian nasopharyngeal carcinoma patients.
Methods
Patient selection
Patients were enrolled onto this study if they fulfilled the following selection criteria: a histologic or cytologic diagnosis of nasopharyngeal carcinoma; age between 18 and 70 years; a performance status of 0, 1, or 2 on the Eastern Cooperative Oncology Group (ECOG) scale; no prior use of irinotecan; a life expectancy of at least 3 months; adequate bone marrow function (total WBC count ≥3500 µl−1, absolute neutrophil count ≥1500 µl−1, platelet count ≥100,000 µl−1, and haemoglobin level ≥9 g dl−1), adequate hepatic function (total bilirubin levels ≤2.0 mg dl−1, AST and ALT levels ≤2.5 times the upper limit of normal), adequate renal function (creatinine level ≤1.5 g dl−1 or 133 µmol l−1 and 24 h creatinine clearance ≥40 ml min−1), and written informed consent to the study. Patients were ineligible if they experienced the following: had serious infectious diseases or other severe complications such as pre-existing cardiac disease, uncontrolled diabetes, bleeding or colitis; had active concurrent malignancies; had symptomatic brain metastases; were lactating or pregnant women, or were willing to be pregnant; or had other medical problems severe enough to prevent compliance with the protocol. None of the patients was receiving drugs known to interact with irinotecan. The study was approved by the ethics committee and review board of the National Cancer Centre, Singapore. All patients gave informed consent prior to enrolment into the study.
Drug administration
Irinotecan was supplied by Aventis Pharma as 5 ml vials containing 100 mg of the drug and was diluted in 250 ml of normal saline for administration. It was then administered to the patient as a 90 min intravenous infusion on days 1, 8, and 15. The regimen was repeated every 28 days. The weekly dosage regimen was fixed at 100 mg m−2. All concomitant medications taken by the patients during the study were recorded. Subsequent doses of irinotecan were withheld if the patients had an absolute neutrophil count ≤1000 µl−1, platelet count ≤100,000 µl−1, or diarrhoea of grade 3 or 4.
Pharmacokinetic study
Pharmacokinetic blood sampling (3 ml) was done on day 1 of cycle 1 and blood was obtained from the opposite arm of each patient at the following times: immediately before infusion; 15, 30, 45 and 90 min after start of infusion, and at 5, 20 and 45 min, and 1.5, 2, 2.5, 3, 3.5, 4, 6, 8, 12, 24 and 48 h after end of infusion. The blood samples were immediately centrifuged and the plasma was stored at −20°C until analysis.
Plasma concentrations of total irinotecan and SN-38 were determined by reversed-phase high-performance liquid chromatography with fluorescence detection developed by Barilero et al. that allowed simultaneous determination of both compounds in the same run [19]. Briefly, plasma was extracted using a solid-phase extraction step. The extracts were chromatographed on a C18 reversed-phase analytical column using a mobile phase composed of 34% acetonitrile and 66% potassium dihydrogen phosphate (0.1 m) containing 3 m m sodium 1-heptanesulphonate (pH 4). Irinotecan, SN-38 and the internal standard (camptothecin) were determined by a fluorescence detector with excitation wavelength at 323 nm and emission at 428 nm for irinotecan and 380 nm and 540 nm for SN-38. The concentrations of irinotecan and SN-38 were determined from peak area ratios of either compound to the internal standard. Total SN-38G concentrations were determined as the increase in SN-38 concentration following incubation with β-glucuronidase. The limits of quantification for irinotecan, SN-38 and SN-38G were 3, 0.6 and 6 ng ml−1, respectively. The calibration curves were linear over the concentration ranges tested, i.e. 3–10000 ng ml−1 for irinotecan (r2 = 0.9996), and 0.6–2000 ng ml−1 for SN-38 (r2 = 0.9996), and 6–20000 ng ml−1 for SN-38G (r2 = 0.9997), in plasma. The coefficients of variation for the intraday and interday reproducibility were 7.0% and 6.4% for irinotecan, 4.4% and 3.0% for SN-38, and 2.5% and 2.0% for SN-38G, respectively. Concentrations of the CYP3A4-mediated irinotecan metabolites APC and NPC in plasma were not measured in the currently studied patients because of limited sample supply that precluded an additional analysis on the same material.
Pharmacokinetic analysis
Pharmacokinetic parameters for total irinotecan, its metabolite (SN-38) and SN-38G were determined by noncompartmental methods using a nonlinear regression program, WinNonLin version 2.1 (Pharsight Inc, Mountain View, CA, USA). Peak plasma concentrations (Cmax) and time to peak concentration (tmax) were identified from individual subject concentration-time curves. Area under the plasma concentration-time curve from time zero to the time (t) of the last detectable concentration AUC(0,t) was calculated using the trapezoidal rule. The area was extrapolated to infinity AUC(0,∞) by adding Ct/λz to AUC(0,t), where Ct was the last detectable plasma concentration. Apparent elimina-tion rate constants (λz) were estimated by least-squares regression of values in the terminal log-linear region of the plasma irinotecan, SN-38 and SN-38G concentration-time curves, whereas half-life (t1/2,z) was calculated as ln(2)/λz. Total clearance (CL) of irinotecan was calculated as the ratio of dose to AUC(0,∞).
Pharmacogenetic analysis
Genomic DNA was extracted from lymphocytes according to standard methods using proteinase K digestion followed by phenol/chloroform extractions. The following SNPs were analyzed: CYP3A4 (CYP3A4*1B,*4, *5 and *6); CYP3A5 (CYP3A5*3 and *6); UGT1A1 (T-3263G and UGT1A1*28); ABCB1 (C1236T, G2677T/A and C3435T); ABCC2 (C-24T and C334–49T) and ABCG2 (5′-UTR: −19572–19569 CTCA deletion, G-19202C, T-18845C, −18604 A deletion; G34A, C376T, C421A, A1244G and G1245C). PCR for CYP3A4, CYP3A5, UGT1A1, ABCB1 and ABCC2 were based on previously published methods [20–23].
The primer sequences for ABCG2 SNPs in the 5′ flanking region (–19572–19569delCTCA, G-19202C, T-18845C, −18604delA), G34A, C376T, C421A, A1444G and G1445C were created based on published sequences (GenBank Accession Nos: AC084732 for the 5′ flanking region, and NM004827 for the exonic region). The PCR amplifications were carried out in an MJ Research PTC-100 thermal cycler (MJ Research Inc., Waltham, MA, USA) with the following reagents in a 20 µl reaction mixture: 100 ng genomic DNA, 0.25 µm of each primer, 10 x PCR buffer containing 10 m m Tris and 50 m m KCl, 1.5 m m MgCl2, 200 µm each of dNTP and 1 U Taq DNA polymerase. The PCR conditions consisted of an initial denaturation at 94 °C for 1 min, followed by amplification using 33 cycles of 94 °C for 30 s, 55 °C for 30 s (G34A, C376T, A1444G and G1445C) or 58 °C for 30 s (–19572–19569delCTCA, G-19202C, T-18845C, −18604delA and C421A), and elongation at 72 °C for 30 s, followed by a final extension at 72 °C for 5 min. The amplified PCR products for ABCG2–19572–19569delCTCA, G-19202C, T-18845C, −18604delA were purified using the QIAquick PCR Purification kit (QIAGEN Inc. Valencia, CA, USA), and sequenced on a Beckman Coulter CEQ 2000XL DNA Analysis System (Beckman Coulter, Inc., Fullerton, CA, USA).
PCR products for ABCG2 G34A were subjected to MegaBACE™ SNuPe™ genotyping analysis according to the manufacturer's protocol: to 5 µl of the amplified DNA fragment, 10 units of exonuclease I and 2 units of shrimp alkaline phosphatase were added. The treated PCR products were then incubated at 37 °C. The enzymes were deactivated at 80 °C for 20 min 4 µl of the SNuPe™ premix (Amersham Biosciences Corp, Piscataway, NJ, USA) and 2 pmol of the SNP-specific primer were then added to 5 µl of the treated PCR product, giving a final reaction volume of 10 µl. The SNuPe products were generated at 96 °C for 10 s, 50 °C for 5 s and 60 °C for 10 s for a total of 25 cycles. The SNuPe reaction products were purified by gel filtration using AutoSeq96™ Dye Terminator Clean-up Kit (Amersham Biosciences Corp, Piscataway, NJ, USA). 5 µl of MegaBACE SNuPe Multiple Injection Marker was added to the purified product prior to analysis.
The amplified reaction products of exons 4, 5 and 12 were purified using QIAquick PCR purification kit followed by digestion with 10% RsaI (C376T), MseI (C421A), NcoI (A1444G) or FokI (G1445C) for a minimum of 2 h at 37 °C. The digested PCR products of exons 4 (349 bp) and 12 (342 bp) were analyzed by electrophoretic separation on 2% agarose gel, and those of exon 5 (131 bp) on 15% polyacrylamide gel, followed by direct visualization under UV light. Genotype verifications were carried out by direct sequencing of the PCR product using the CEQ Dye Terminator Cycle Sequencing Quick Start Kit and a CEQ 2000 automated sequencer (Beckman Coulter, Inc., Fullerton, CA, USA).
Statistical analysis
Fischer's exact test was used to assess Hardy–Weinberg equilibrium between the genotype frequencies. The nonparametric Kruskal–Wallis test was used to test for genotype–phenotype associations in the nasopharyngeal carcinoma patients. As this was an exploratory study, no Bonferroni correction was applied for multiple comparisions. Statistical significance was set at P < 0.05. All statistical analyses were done using Stata (STATA Statistical Software release 7.0, Stata Corporation, College Station, TX, USA).
Results
Irinotecan pharmacokinetics in nasopharyngeal carcinoma patients
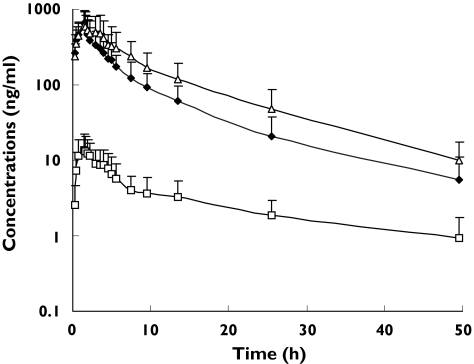
Pharmacokinetic evaluations were done in 29 nasopharyngeal carcinoma patients (25 males and four females). The median age of the patients was 50 years (range 27–71) and the median BSA was 1.6 m2 (range 1.3–1.9). All patients had ECOG performance status of ≤2. Twenty-seven Chinese, one Malay and one Indian patients were enrolled in the study. The plasma concentration-time profiles of irinotecan, SN-38 and SN-38G are shown in Figure 1 and the summarized pharmacokinetic parameters (mean ± SD) are listed in Table 1. Irinotecan, SN-38 and SN-38G concentrations peaked rapidly after the end of infusion and the decline of irinotecan paralleled that of its metabolites (Figure 1). The CL of irinotecan was 32.5 ± 12.0 l h−1 m−2 and was higher than the irinotecan CL values reported in other ethnic populations (range: 12–20 l h−1 m−2) during short infusions of 30–90 min at varying schedules of administrations [24–27]. The volume of distribution at steady state (Vss) was 262.0 ± 109.8 l m−2. There were also wide interpatient variabilities for the various AUC ratios. The relative extent of conversion (REC) of irinotecan to SN-38 and the relative extent of glucuronidation (REG) of SN-38 to SN-38G showed 8- and 28-fold variations, respectively (Table 1).
Figure 1.
Plasma concentration-time profiles of irinotecan and its metabolites, SN-38 and SN-38G. CPT-11 (♦), SN-38 (□), SN-38G (▵)
Table 1.
Pharmacokinetic parameters of irinotecan and its two metabolites, SN-38 and SN-38G (n = 29)
| Pharmacokinetic parameters | Mean ± SD |
|---|---|
| Irinotecan | |
| ″″″″Cmax (µg l−1) | 712.9 ± 336.4 |
| ″″″″AUC(0,∞) (µg l−1 h) | 3600 ± 1577 |
| ″″″″t1/2,z (h) | 8.28 ± 1.99 |
| ″″″″CL (l h−1 m−2) | 32.5 ± 12.0 |
| ″″″″Vss (l m−2) | 262.0 ± 109.8 |
| SN-38 | |
| ″″″″Cmax (µg l−1) | 16.1 ± 8.7 |
| ″″″″AUC(0,∞) (µg l−1 h) | 167.3 ± 88.8 |
| ″″″″t1/2,z (h) | 17.3 ± 8.8 |
| SN-38G | |
| ″″″″Cmax (µg l−1) | 676.9 ± 403.5 |
| ″″″″AUC(0,∞) (µg l−1 h) | 5558 ± 2871 |
| ″″″″t1/2,z (h) | 8.91 ± 1.81 |
| AUC ratios | |
| ″″″″REC | 0.050 ± 0.024 |
| ″″″″REG | 47.6 ± 50.1 |
| ″″″″BI (µg l−1 h) | 144.2 ± 138.5 |
Genotype–phenotype associations
Six candidate genes involved in the disposition of irinotecan and its metabolites were genotyped. Twenty-one SNPs and one dinucleotide repeat were analyzed. All genotype frequencies were in Hardy–Weinberg equilibrium. The estimated variant genotype frequencies were as follows: CYP3A5*3/*3 (11%), UGT1A1[T-3263G (36%) and *28 (7%)], ABCB1[C1236T, G2677T/A, C3435T (25–39%)], ABCC2 C-24T (4%) and C334–49T (7%), ABCG2[CTCAdel (4%), G34A (4%), C421A (11%)]. All patients were wild-type for CYP3A4 SNPs.
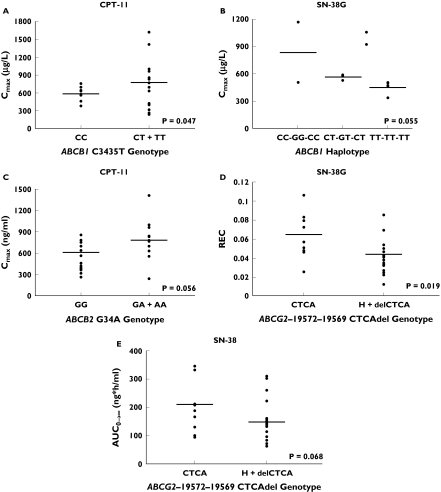
Selected genotype–phenotype associations are shown in Table 2 and Figures 2A–E. Our study revealed that SNPs in ABCB1 and ABCG2 genes seem to be particularly important in influencing the pharmacokinetics of irinotecan and its metabolites. Nasopharyngeal carcinoma patients with wild-type CC genotype at exon 26 of the ABCB1 gene had significantly lower Cmax values of irinotecan compared with the combined group of patients who were heterozygous or homozygous for the variant genotype (Figure 2A, P = 0.047). Since the SNPs at the 3 loci of the ABCB1 gene are in strong linkage disequilibrium (18), association analysis was carried out between the three-point substitution genotypes of the ABCB1 gene and the pharmacokinetic parameters of irinotecan, SN-38 and SN-38G. As shown in Figure 2B, a significant genotypic-phenotypic trend was only noted for SN-38G Cmax. Patients with the TT-TT-TT three-point substitution genotype had lower SN-38G Cmax values compared with patients who were wild type or heterozygous at the 3 loci. A trend for a genotypic-phenotypic correlation was present between the ABCC2 C-24T SNP in the 5′-untranslated region (CC vs CT vs TT, P = 0.055) and irinotecan AUC(0,∞). Nasopharyngeal carcinoma patients with wild type genotype had the lowest irinotecan AUC(0,∞) compared with patients with CT or TT genotypes. Of the nine SNPs analyzed for the ABCG2 gene, variant genotypes were only found for the −19572–19569delCTCA, G34A and C421A SNPs. Two of these SNPs were found to correlate significantly with irinotecan AUC(0,∞) and the REC. Patients who were wild type for the G34A SNP at exon 2 showed a trend towards lower systemic exposures to irinotecan (i.e. Cmax) compared with patients with one or two variant alleles (Figure 2C, P = 0.056). Also the REC values were significantly different between patients with wild type CTCA genotype and the combined group of patients with at least one deleted CTCA allele (0.064 ± 1.17 vs 0.043 ± 0.02, P = 0.019, Figure 2D). The cancer patients with wild-type CTCA genotype had greater REC of irinotecan to SN-38 compared with patients who were heterozygous or homozygous for the variant deletion allele (0.064 ± 0.023 vs 0.043 ± 0.020). The higher REC of irinotecan to SN-38 in wild-type patients was associated with an approximately 30% increase in AUC(0,∞) of SN-38 (Figure 2E). No statistically significant relationships were observed among the other variants in ABC transporters and pharmacokinetic parameters of irinotecan and its metabolites (Table 2).
Table 2.
Summary of genotype–phenotype associations for genes in which variant alleles were observed
| Polymorphism | Pharmacokinetic parameters |
|---|---|
| ABCB1 C3435T (CC vs CT + TT) | CPT-11 Cmax (P = 0.047) |
| ABCB1 haplotypes | |
| ″″″″(CC-GG-CC vs. CT-GT-CT vs TT-TT-TT) | SN-38G: Cmax (P = 0.055) |
| ″″″″ABCC2 C-24T (CC vs CT vs TT) | CPT-11 AUC(0,∞) (P = 0.055) |
| ″″″″ABCG2–19572–19569 CTCA del (CTCA vs Het + CTCA del) | REC (AUCSN-38/AUCCPT-11) (P = 0.019) |
| ″″″″ABCG2 G34A (GG vs GA + AA) | CPT-11 Cmax (P = 0.056) |
Figure 2.
Genotype–phenotype associations
Discussion
Genetic polymorphisms of genes that encode proteins involved in biochemical pathways are becoming increasingly important in explaining the interpatient variabilities commonly observed with drugs with narrow therapeutic indices. Most chemotherapeutic agents are often characterized as having large interpatient variations in their pharmacokinetic-pharmacodynamic profiles and the use of pharmacogenetic information to explain these variabilities may be a useful therapeutic guide to dosage optimization. Furthermore, polygenic rather than monogenic traits are often responsible for the observed differences in pharmacokinetics-pharmacodynamics of a therapeutic agent. This is particularly true if the disposition of a drug involves a complex metabolic pathway such as that of irinotecan.
Because of the multiplicity of transporters involved in the disposition of irinotecan and its metabolites, the variabilities observed with the pharmacokinetics of irinotecan and its metabolites may vary widely. The contributions of individual transporters to the disposition of irinotecan and its metabolites may differ greatly between patients as well as between populations belonging to different ethnic groups. Furthermore, due to the polygenic nature of irinotecan metabolism and differences in allelic frequencies in different populations, different genetic polymorphisms may be responsible for differences in the pharmacokinetics of irinotecan and its metabolites in different ethnic populations. Identification of relevant marker SNPs that may be predictive of altered pharmacokinetics-pharmacodynamics of irinotecan and its metabolites may be useful in its dosage optimization.
The present exploratory study investigated the influence of SNPs in various drug metabolizing and ABC transporter genes on the disposition of irinotecan and its metabolites in Asian nasopharyngeal carcinoma patients. Amongst the various SNPs that were investigated, only those in the ABC transporter genes were significantly correlated with the pharmacokinetics of irinotecan and its metabolites. No significant associations were noted with SNPs in genes encoding the drug metabolizing enzymes CYP3A4/5 and UGT1A1; in the case of variants in the CYP3A4 gene, phenotypic consequences cannot be ruled out completely because in the studied cohort of patients no variant alleles were observed. With regards to the transporter SNPs, the association between the ABCG2–19572–19569delCTCA SNP and REC was highly significant (P = 0.019). The higher REC values were associated with higher exposures to SN-38 although this relationship was statistically nonsignificant. This appears to be the first in vivo finding demonstrating the clinical significance of an ABCG2 SNP in cancer patients receiving irinotecan. Previous studies have shown decreased transcriptional activity of the ABCG2 gene following deletion of the region 1285 and 628 bp upstream of the transciptional start site [28]. Since the –19572–19569delCTCA variant is located −655 to −652 bp from the transcription start site, a possible effect on ABCG2 transcriptional level cannot be ruled out. No phenotypic associations were found between the ABCG2 G34A (exon 2) and C421A (exon 5) SNPs and exposure levels to SN-38. This was in contrast to other in vitro findings suggesting increased sensitivity of cell lines harbouring these SNPs to SN-38, in particular the C421A SNP [29, 30].
The association between the ABCB1 C3435T SNP (located at exon 26) and exposure level to irinotecan was also significant in the cancer patients (P = 0.047). In a similar exploratory study done by Mathijssen et al. [18], a significant association was only found between the ABCB1 C1236T SNP in exon 12 and increased exposures to irinotecan and SN-38. The C3435T SNP is a wobble SNP and was initially shown by Hoffmeyer et al. [31] to be associated with decreased intestinal expression of ABCB1 and an increase in exposure levels to digoxin. This finding, however, has been in conflict with other studies and the exact role of the C3435T SNP in influencing the pharmacokinetics of ABCB1 substrates is unclear. It is possible that the C3435T SNP may influence exposure levels of drugs through its haplotypic associations with two other high frequency SNPs, namely, ABCB1 C1236T and G2677T/A SNPs at exons 12 and 21, respectively [20]. Recently, Sai et al. [32] showed that haplotypic associations between these three SNPs were responsible for reduced renal clearance of irinotecan and its metabolites in Japanese cancer patients. In the present study, a trend towards significant association was only observed between the three-point substitution genotypes of the ABCB1 gene and SN-38G Cmax (Figure 2B). Owing to the small sample size of this study, the exact relevance of this finding is not clear and a more detailed analysis of the influence of ABCB1 haplotypes on pharmacokinetics of irinotecan and its metabolites is warranted in our population.
The glucuronidation pathway catalyzed by UGT1A1 is the main detoxification pathway for SN-38 and genetic polymorphisms in the UGT1A1 gene have been previously shown to be associated with altered glucuronidation of SN-38 [33]. The presence of seven TA repeats (*28) in the TATA box of the UGT1A1 gene is associated with decreased transcriptional activity and decreased glucuronidating capacity. Patients harbouring the UGT1A1*28 genotype had decreased glucuronidation of SN-38 and higher incidences of SN-38 induced diarrhoea compared with patients harbouring six TA repeats (wild-type) [33]. The frequency of the UGT1A1*28 allele varies in different ethnic populations and amongst Asian populations, it has been reported to be highest in the Indians and low in Chinese and Malays [21]. There are several possible reasons for the lack of association between UGT1A1*28 and pharmacokinetic parameters in the present study. Firstly, it could have been due to the low frequency of this allele in our local patients who were mainly of Chinese origin. Secondly, the presence of relevant coding region SNPs or haplotypes in the UGT1A1 gene that may be of functional importance but which were not analyzed in the present study cannot be ruled out. Thirdly, there may be a dose dependent effect of UGT1A1*28 genotype on glucuronidation capacity. The present study utilized a weekly schedule of irinotecan administration whereas most other studies have used a three weekly regimen. Glucuronide conjugation metabolism may be saturable at higher doses of irinotecan although this phenomenon has never been shown in the clinical setting due to the inherent toxicity associated with administering irinotecan beyond the maximum tolerated dose levels. A similar dose-dependent effect of CYP3A5*3 genotype on the pharmacokinetics of the ABT-773 was recently reported by Katz et al. [34] and it may be hypothesized that irinotecan glucuronidation rate may also be dose dependent and the genotypic effect of UGT1A1*28 may be masked at the usual clinical doses that are used. However, despite the lack of genotypic–phenotypic associations, the median values for the REG of SN-38 were higher in our cancer patients compared with other studies [18, 35] (Table 3). It is of interest to note that the Asian patients with the UGT1A1*28 genotype had higher values for REG than the Caucasian patients (Table 2). This suggests that the relative glucuronidating capacity in patients with homozygous UGT1A1*28 genotype may be much higher in Asians compared with Caucasians which may explain the lack of severe irinotecan induced diarrhoea in our patients. However, the sample size in all three studies is too small to give any precise estimate for the UGT1A1*28 genotypic group. A currently ongoing comparative study in Asian and Caucasian patients with cancer will further assess the inter-relationship between the relative extent of glucuronidation of SN-38 and the UGT1A1*28 genotype.
Table 3.
Effect of UGT1A1*28 on REG
| Genotype | Reference | n | REG (mean ± SD) (median; range) |
|---|---|---|---|
| Wild-type | This study | 17 | 33.4 ± 2.5 (25.6; 9.3–259.2) |
| Mathijssen et al. [18] | 32 | 7.6 ± 4.1 (6.6) | |
| Iyer et al. [35] | 9 | 9.3 ± 11 | |
| Heterozygous | This study | 10 | 36.8 ± 1.7 (34.3; 17.7–93.7) |
| Mathijssen et al. [18] | 19 | 7.1 ± 3.6 (6.6) | |
| Iyer et al. [35] | 7 | 4.0 ± 1.7 | |
| Variant | This study | 2 | 33.8 ± 1.3 (33.8; 27.7–41.2) |
| Mathijssen et al. [18] | 2 | 2.2 ± 5.2 (3.7) | |
| Iyer et al. [35] | 4 | 2.4 ± 1.1 |
No apparent associations were found between CYP3A4 or CYP3A5 polymorphisms and the pharmacokinetics of irinotecan and its metabolites. Although the role of CYP3A5 in irinotecan metabolism is unclear, CYP3A4 mediated oxidative metabolism of irinotecan is important as one of the minor metabolites, NPC, can be hydrolyzed to form active SN-38. Since most Asians are wild-type for CYP3A4 [20], this may partly contribute to the high clearance of irinotecan observed in our patients (Table 1).
In summary, the present exploratory study showed that SNPs in ABCB1 and ABCG2 genes may be functionally important with regards to altered pharmacokinetics of irinotecan and its metabolites. The predictive value of the identified SNPs in these two transporter genes on irinotecan pharmacokinetics is currently being investigated in a larger patient population of Asian origin as well as in other ethnic populations including a cohort of Caucasian cancer patients.
Acknowledgments
Competing interests: None declared
References
- 1.Saltz LB, Cox JV, Blanke C, et al. Irinotecan plus fluorouracil and leucovorin for metastatic colorectal cancer. Irinotecan Study Group. N Engl J Med. 2000;343:905–14. doi: 10.1056/NEJM200009283431302. [DOI] [PubMed] [Google Scholar]
- 2.Kawato Y, Aonuma M, Hirota Y, Kuga H, Sato K. Intracellular roles of SN-38, a metabolite of the camptothecin derivative CPT-11, in the antitumour effect of CPT-11. Cancer Res. 1991;51:4187–91. [PubMed] [Google Scholar]
- 3.Rivory LP, Robert J. Identification and kinetics of a beta-glucuronide metabolite of SN-38 in human plasma after administration of the camptothecin derivative irinotecan. Cancer Chemother Pharmacol. 1995;36:176–9. doi: 10.1007/BF00689205. [DOI] [PubMed] [Google Scholar]
- 4.Haaz MC, Rivory L, Jantet S, Ratanasavanh D, Robert J. Glucuronidation of SN-38, the active metabolite of irinotecan, by human hepatic microsomes. Pharmacol Toxicol. 1997;80:91–6. doi: 10.1111/j.1600-0773.1997.tb00289.x. [DOI] [PubMed] [Google Scholar]
- 5.Rivory LP, Riou JF, Haaz MC, et al. Identification and properties of a major plasma metabolite of irinotecan (CPT-11) isolated from the plasma of patients. Cancer Res. 1996;56:3689–94. [PubMed] [Google Scholar]
- 6.Dodds HM, Haaz MC, Riou JF, Robert J, Rivory LP. Identification of a new metabolite of CPT-11 (irinotecan): Pharmacological properties and activation of SN-38. J Pharmacol Exp Ther. 1998;286:578–83. [PubMed] [Google Scholar]
- 7.Rivory LP. Metabolism of CPT-11. Impact on activity. Ann N Y Acad Sci. 2000;922:205–15. doi: 10.1111/j.1749-6632.2000.tb07039.x. [DOI] [PubMed] [Google Scholar]
- 8.Mathijssen RH, van Alphen RJ, Verweij J, et al. Clinical pharmacokinetics and metabolism of irinotecan (CPT-11) Clin Cancer Res. 2001;7:2182–94. [PubMed] [Google Scholar]
- 9.Iyer L, Hall D, Das S, et al. Phenotype-genotype correlation of in vitro SN-38 (active metabolite of irinotecan) and bilirubin glucuronidation in human liver tissue with UGT1A1 promoter polymorphisms. Clin Pharmacol Ther. 1999;65:576–82. doi: 10.1016/S0009-9236(99)70078-0. [DOI] [PubMed] [Google Scholar]
- 10.Ando Y, Saka H, Asai G, Sugiura S, Shimokata K, Kamataki T. UGT1A1 genotypes and glucuronidation of SN-38, the active metabolite of irinotecan. Ann Oncol. 1998;9:845–7. doi: 10.1023/a:1008438109725. [DOI] [PubMed] [Google Scholar]
- 11.Iyer L, King CD, Whitington PF, et al. Genetic predisposition to the metabolism of irinotecan (CPT-11). Role of uridine diphosphate glucuronosyltransferase isoform 1A1 in the glucuronidation of its active metabolite (SN-38) in human liver microsomes. J Clin Invest. 1998;101:847–54. doi: 10.1172/JCI915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ciotti M, Basu N, Brangi M, Owens IS. Glucuronidation of 7-ethyl-10-hydroxycamptothecin (SN-38) by the human UDP-glucuronosyltransferases encoded at the UGT1 locus. Biochem Biophys Res Commun. 1999;260:19–202. doi: 10.1006/bbrc.1999.0453. [DOI] [PubMed] [Google Scholar]
- 13.Villeneuve L, Girard H, Fortier LC, Gagne JF, Guillemette C. Novel functional polymorphisms in the UGT1A7 and UGT1A9 glucuronidating enzymes in Caucasian and African-American subjects and their impact on the metabolism of 7-ehtyl-10-hydroxycamptothecin and flavopiridol anticancer drugs. J Pharmacol Exp Ther. 2003;307:117–28. doi: 10.1124/jpet.103.054072. [DOI] [PubMed] [Google Scholar]
- 14.Iyer L, Ramirez J, Shepard DR, et al. Biliary transport of irinotecan and metabolites in normal and P-glycoprotein-deficient mice. Cancer Chemother Pharmacol. 2002;49:336–41. doi: 10.1007/s00280-001-0420-4. [DOI] [PubMed] [Google Scholar]
- 15.Chen ZS, Furukawa T, Sumizawa T, et al. ATP-dependent efflux of CPT-11 and SN-38 by the multidrug resistance protein (MRP) and its inhibition by PAK-104P. Mol Pharmacol. 1999;55:921–8. [PubMed] [Google Scholar]
- 16.Chu XY, Kato Y, Niinuma K, Sudo KI, Hakusui H, Sugiyama Y. Multispecific organic anion transporter is responsible for the biliary excretion of the camptothecin derivative irinotecan and its metabolites in rats. J Pharmacol Exp Ther. 1997;281:304–14. [PubMed] [Google Scholar]
- 17.Nakatomi K, Yoshikawa M, Oka M, Ikegami Y, Hayasaka S, Sano K, et al. Transport of 7-ethyl-10-hydroxycamptothecin (SN-38) by breast cancer resistance protein ABCG2 in human lung cancer cells. Biochem Biophys Res Commun. 2001;288:827–32. doi: 10.1006/bbrc.2001.5850. [DOI] [PubMed] [Google Scholar]
- 18.Mathijssen RHJ, Marsh S, Karlsson MO, et al. Irinotecan pathway genotype analysis to predict pharmacokinetics. Clin Cancer Res. 2003;9:3246–53. [PubMed] [Google Scholar]
- 19.Barilero I, Gandia D, Armand JP, et al. Simultaneous determination of the camptothecin analogue CPT-11 and its active metabolite SN-38 by high-performance liquid chromatography – application to plasma pharmacokinetic studiesin cancer patients. J Chromatogr. 1992;575:275–80. doi: 10.1016/0378-4347(92)80156-k. [DOI] [PubMed] [Google Scholar]
- 20.Chowbay B, Cumaraswamy S, Cheung YB, Zhou Q, Lee EJ. Genetic polymorphisms in MDR1 and CYP3A4 genes in Asians and the influence of MDR1 haplotypes on cyclosporin disposition in heart transplant patients. Pharmacogenetics. 2003;13:89–95. doi: 10.1097/00008571-200302000-00005. [DOI] [PubMed] [Google Scholar]
- 21.Balram C, Sabapathy K, Fei G, Khoo KS, Lee EJ. Genetic polymorphisms of UDP-glucuronosyltransferase in Asians: UGT1A1*28 is a common allele in Indians. Pharmacogenetics. 2002;12:81–3. doi: 10.1097/00008571-200201000-00012. [DOI] [PubMed] [Google Scholar]
- 22.Balram C, Zhou Q, Cheung YB, Lee EJ. CYP3A5*3 and *6 single nucleotide polymorphisms in three distinct Asian populations. Eur J Clin Pharmacol. 2003;59:123–6. doi: 10.1007/s00228-003-0594-2. [DOI] [PubMed] [Google Scholar]
- 23.Ito S, Ieiri I, Tanabe M, Suzuki A, Higuchi S, Otsubo K. Polymorphism of the ABC transporter genes, MDR1, MRP1 and MRP2/cMOAT, in healthy Japanese subjects. Pharmacogenetics. 2001;11:175–84. doi: 10.1097/00008571-200103000-00008. [DOI] [PubMed] [Google Scholar]
- 24.de Jonge MJ, Verweij J, de Bruijn P, et al. Pharmacokinetic, metabolic and pharmacodynamic profiles in a dose-escalating study of irinotecan and cisplatin. J Clin Oncol. 2000;18:195–203. doi: 10.1200/JCO.2000.18.1.195. [DOI] [PubMed] [Google Scholar]
- 25.de Forni M, Bugat R, Chabot GG, et al. Phase I and pharmacokinetic study of the camptothecin derivative irinotecan administered on a weekly schedule in cancer patients. Cancer Res. 1994;54:4347–54. [PubMed] [Google Scholar]
- 26.Chabot GG. Clinical pharmacokinetics of irinotecan. Clin Pharmacokinet. 1997;33:245–59. doi: 10.2165/00003088-199733040-00001. [DOI] [PubMed] [Google Scholar]
- 27.Negoro S, Fukuoka M, Masuda N, et al. Phase I study of weekly intravenous infusions of CPT-11, a new derivative of camptothecin in the treatment of advanced non-small cell lung cancer. J Natl Cancer Inst. 1991;83:1164–8. doi: 10.1093/jnci/83.16.1164. [DOI] [PubMed] [Google Scholar]
- 28.Bailey-Dell KJ, Hassel B, Doyle LA, Ross DD. Promoter characterisation and genomic organization of the human breast cancer resistance protein (ATP-binding cassette transporter G2) gene. Biochim Biophys Acta. 2001;1520:234–41. doi: 10.1016/s0167-4781(01)00270-6. [DOI] [PubMed] [Google Scholar]
- 29.Imai Y, Nakane M, Kage K, et al. C421A polymorphism in the human breast cancer resistance protein gene is associated with low expression of Q141K protein and low level drug resistance. Mol Cancer Ther. 2002;1:611–6. [PubMed] [Google Scholar]
- 30.Mizuarai S, Aozasa N, Kotani H. Single nucleotide polymorphisms results in impaired membrane localisation and reduced ATPase activity in multidrug transporter ABCG2. Int J Cancer. 2004;109:238–46. doi: 10.1002/ijc.11669. [DOI] [PubMed] [Google Scholar]
- 31.Hoffmeyer S, Burk O, van Richter O, et al. Functional polymorphisms of the human multidrug-resistance gene: multiple sequence variations and correlation of one allele with P-glycoprotein expression and activity in vivo. Proc Natl Acad Sci USA. 2000;97:3473–8. doi: 10.1073/pnas.050585397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sai K, Kaniwa N, Itoda M, et al. Haplotype analysis of ABCB1/MDR1 blocks in a Japanese population reveals genotype-dependent renal clearance of irinotecan. Pharmacogenetics. 2003;13:741–57. doi: 10.1097/00008571-200312000-00005. [DOI] [PubMed] [Google Scholar]
- 33.Hanioka K, Ozawa S, Jinno H, Ando M, Saito Y, Sawada J. Human liver UDP-glucuronosyltransferase isoforms involved in the glucuronidation of 7-ethyl-10-hydroxycamptothecin. Xenobiotica. 2001;31:687–99. doi: 10.1080/00498250110057341. [DOI] [PubMed] [Google Scholar]
- 34.Katz DA, Grimm DR, Cassar SC, et al. CYP3A5 genotype has a dose-dependent effect on ABT-773 plasma levels. Clin Pharmacol Ther. 2004;75:516–28. doi: 10.1016/j.clpt.2004.01.013. [DOI] [PubMed] [Google Scholar]
- 35.Iyer L, Das S, Janisch L, et al. UGT1A1*28 polymorphism as a determinant of irinotecan disposition and toxicity. Pharmacogenomics J. 2002;2:43–7. doi: 10.1038/sj.tpj.6500072. [DOI] [PubMed] [Google Scholar]