Abstract
Aims
To characterize the kinetics of S-naproxen (‘naproxen’) acyl glucuronidation and desmethylnaproxen acyl and phenolic glucuronidation by human liver microsomes and identify the human UGT isoform(s) catalysing these reactions.
Methods
Naproxen and desmethylnaproxen glucuronidation were investigated using microsomes from six and five livers, respectively. Human recombinant UGTs were screened for activity towards naproxen and desmethylnaproxen. Where significant activity was observed, kinetic parameters were determined. Naproxen and desmethylnaproxen glucuronides were measured by separate high-performance liquid chromatography methods.
Results
Naproxen acyl glucuronidation by human liver microsomes followed biphasic kinetics. Mean apparent Km values (±SD, with 95% confidence interval in parentheses) for the high- and low-affinity components were 29 ± 13 µm (16, 43) and 473 ± 108 µm (359, 587), respectively. UGT 1A1, 1A3, 1A6, 1A7, 1A8, 1A9, 1A10 and 2B7 glucuronidated naproxen. UGT2B7 exhibited an apparent Km (72 µm) of the same order as the high-affinity human liver microsomal activity, which was inhibited by the UGT2B7 selective ‘probe’ fluconazole. Although data for desmethylnaproxen phenolic glucuronidation by human liver microsomes were generally adequately fitted to either the single- or two-enzyme Michaelis–Menten equation, model fitting was inconclusive for desmethylnaproxen acyl glucuronidation. UGT 1A1, 1A7, 1A9 and 1A10 catalysed both the phenolic and acyl glucuronidation of desmethylnaproxen, while UGT 1A3, 1A6 and 2B7 formed only the acyl glucuronide. Atypical glucuronidation kinetics were variably observed for naproxen and desmethylnaproxen glucuronidation by the recombinant UGTs.
Conclusion
UGT2B7 is responsible for human hepatic naproxen acyl glucuronidation, which is the primary elimination pathway for this drug.
Keywords: desmethylnaproxen, human drug glucuronidation, S-naproxen, UDP-glucuronosyltransferase, UGT2B7
Introduction
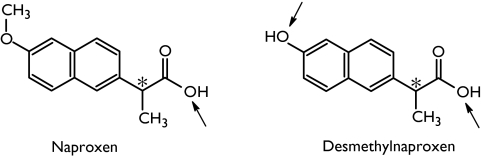
S-Naproxen (‘naproxen’) is an aryl propionic acid derivative (Figure 1) which continues to find widespread use as a nonsteroidal anti-inflammatory agent. For example, in excess of one million prescriptions were dispensed for this drug in England in 2003 [1]. The principal route of elimination of naproxen in humans is formation of an acyl glucuronide. This pathway is responsible for approximately 60% of total clearance [2, 3]. Demethylation, to form desmethylnaproxen, apparently accounts for the remainder of naproxen metabolism [4]. Cytochromes P450 1A2 and 2C9 mediate human hepatic naproxen demethylation [5]. Once formed, however, desmethylnaproxen is glucuronidated. Unlike naproxen, which forms only an acyl glucuronide, desmethylnaproxen may form both acyl and phenolic glucuronides (Figure 1). Renal excretion of both unchanged naproxen and desmethylnaproxen is minor [3].
Figure 1.
Structures of naproxen and desmethylnaproxen, showing sites of glucuronidation
Glucuronidation reactions are facilitated by the enzyme UDP-glucuronosyltransferase (UGT). UGT catalyses the covalent linkage of glucuronic acid, derived from the cofactor UDP-glucuronic acid (UDPGA), to a substrate bearing a suitable acceptor group (most commonly hydroxyl, carboxylic acid or amine). Consistent with its broad substrate profile, UGT exists as an enzyme ‘superfamily’[6]. Fifteen functional UGT isoforms have been identified to date; UGT 1A1, 1A3, 1A4, 1A6, 1A7, 1A8, 1A9, 1A10, 2A1, 2B4, 2B7, 2B10, 2B15, 2B17 and 2B28 [7]. The majority of these isoforms are expressed in liver, although UGT2A1 is mainly localized in nasal epithelium and UGT 1A7, 1A8 and 1A10 apparently occur only in the gastrointestinal tract [8]. Another isoform, UGT2B11, is considered an ‘orphan’ enzyme since, despite considerable effort, substrates have not been identified to date. Available evidence suggests that the individual UGTs exhibit distinct, but overlapping, substrate and inhibitor selectivities and differ in terms of regulation [7].
Despite the widespread use of naproxen, the kinetics of naproxen and desmethylnaproxen glucuronidation by human liver are poorly characterized. Similarly, there has been no systematic investigation of the UGT isoforms contributing to naproxen and desmethylnaproxen glucuronidation. This study aimed to characterize the kinetics of naproxen acyl glucuronidation and desmethylnaproxen acyl and phenolic glucuronidation by human liver microsomes and, using a ‘panel’ of recombinant UGTs expressed in cell culture, identify the human isoform(s) responsible for the glucuronidation of naproxen and desmethylnaproxen. Additionally, the work provided an opportunity to investigate the kinetic behaviour of human UGTs.
Materials and methods
Materials
The glucuronides of S-naproxen and S-desmethylnaproxen (acyl and phenolic) were provided by AstraZeneca (Sodertalje, Sweden). S-Naproxen and desmethylnaproxen were obtained from Syntex Discovery Research (Palo Alto, CA, USA) and fluconazole from Pfizer Ltd (Sydney, Australia). Alamethicin, UDPGA and β-glucuronidase were purchased from Sigma-Aldrich (St Louis, MO, USA). All other chemicals and reagents were of analytical reagent grade.
Human liver microsomes
Human livers (H6, H10, H12, H13, H29 and H40) were obtained from the human liver ‘bank’ of the Department of Clinical Pharmacology, Flinders Medical Centre. Approval was obtained from the Flinders Medical Centre Research Ethics Committee and from the donor next-of-kin for the procurement and use of human liver tissue in xenobiotic metabolism studies. Microsomes were prepared by differential centrifugation. Liver portions in 0.1 m phosphate buffer (pH 7.4) containing 1.15% w/v potassium chloride were homogenized sequentially with a Janke and Kunkle Ultra Turax (24 000 r.p.m.) and a Potter-Elvehjem homogenizer (mechanical drive at 1480 r.p.m.). The homogenate was centrifuged at 700 g for 10 min and then at 10 000 g for a further 10 min. The supernatant fraction was aspirated and centrifuged at 105 000 g for 60 min at 4 °C. The resulting pellet was re-suspended in 0.1 m phosphate buffer (pH 7.4) containing 1.15% w/v potassium chloride and centrifuged at 105 000 g for 60 min at 4 °C. The microsomal pellet was suspended in 0.1 m phosphate buffer (pH 7.4) containing 20% glycerol and stored at −80 °C until use. Microsomal protein concentrations were determined by the method of Lowry et al. [9] using bovine serum albumin as standard. Human liver microsomes used in incubations were activated with the pore forming peptide alamethicin (50 µg mg−1 microsomal protein) by preincubation on ice for 30 min [10].
Expression of recombinant human UGT isoforms in cell culture
The UGT 1A1, 1A3, 1A4, 1A6, 1A7, 1A8, 1A9, 1A10, 2B4, 2B7, 2B15 and 2B17 cDNAs were expressed in a human embryonic kidney cell line (HEK293) as described by Uchaipichat et al. [11]. Briefly, cells were separately transfected with the individual cDNAs cloned in the pEF-IRES-puro6 expression vector. Following transfection, cells were incubated at 37 °C in Dulbecco's modified Eagle's medium, which contained puromycin (1.5 mg l−1), 10% fetal calf serum, and gentamicin (160 mg l−1) in a humidified incubator with an atmosphere of 5% CO2. Puromycin-resistant colonies were pooled and re-plated in the above media. After growth to at least 80% confluency, cells were harvested and washed in phosphate-buffered saline. Cells were subsequently lysed by sonication using a Heat Systems Ultrasonics sonicator set at a microtip limit of 4. Lysates were centrifuged at 12 000 g for 5 min, and the supernatant fraction was separated and stored in phosphate buffer (0.1 m, pH 7.4). Activity of all recombinant UGTs (except UGT1A4) was confirmed using the nonselective substrate 4-methylumbelliferone [11]. The activity of UGT1A4 was confirmed using trifluoperazine as substrate [12].
Chromatography
Chromatography was performed using an Agilent 1100 series high-performance liquid chromatography (Agilent Technologies, North Ryde, NSW, Australia). The system comprised a quaternary solvent delivery module with in-line degassing, auto injector, temperature-controlled column oven, and variable wavelength uv-vis detector. The column temperature was 25 °C for both the naproxen and desmethylnaproxen glucuronidation assays.
Naproxen glucuronidation assay
Incubation mixtures, in a total volume of 0.5 ml, contained activated human liver microsomes (0.25 mg), naproxen (10–12 concentrations in the range 5–4000 µm), MgCl2 (4 m m) and phosphate buffer (0.1 m, pH 7.4). Separate ‘blank’ incubations were performed in the absence of UDPGA and human liver microsomes. After a 5-min preincubation, reactions were initiated by the addition of UDPGA such that the final concentration of cofactor was 5 m m. Incubations were performed in air at 37 °C (shaking water bath), and terminated after 20 min by the addition of an equal volume of ice-cold 4% v/v acetic acid in methanol. The mixtures were vortex mixed, placed on ice, and then centrifuged at 5000 g for 10 min to precipitate microsomal protein. An aliquot (30 µl) of the supernatant fraction was injected onto a Waters Novapak C18 column (150 × 3.9 mm, 4 µm; Milford, MA, USA), which was eluted with 30 : 70 acetonitrile–water (containing 0.12% v/v acetic acid) at a flow rate of 1.5 ml min−1. Analytes were detected by UV absorption at 225 nm. Under these conditions, retention times for naproxen acyl glucuronide and naproxen were 2.4 and 11.2 min, respectively. Incubations performed using recombinant UGTs followed a similar procedure, but with the following modifications: incubation volume, 0.2 ml; HEK293 cell lysate protein, 0.3 mg; and incubation time, 40 min.
The identity of the naproxen glucuronide peak formed by incubations of human liver microsomes and recombinant UGTs was confirmed by reference to an authentic standard and by hydrolysis with β-glucuronidase. However, the naproxen acyl glucuronide standard was hygroscopic. Although useful for metabolite identification, it was considered unsuitable for standard curve construction. Hence, unknown concentrations of naproxen acyl glucuronide were determined by reference to a standard curve constructed for naproxen in the concentration range 0.5–10 µm. Naproxen standard curves were linear over this concentration range, with r2 values >0.99. The lower limit of quantification was 0.02 µm. Overall assay within-day precision was assessed by measuring naproxen glucuronide formation for eight separate incubations with the same batch of human liver microsomes at low (20 µm) and high (2000 µm) substrate concentrations. The within-day coefficients of variation were 5.4% and 5.2% at the low and high substrate concentrations, respectively. Linearity of product formation with respect to incubation time and microsomal protein concentration was determined for substrate concentrations of 100 and 1000 µm. The formation of naproxen acyl glucuronide was linear with incubation times to at least 60 min and microsomal protein concentrations to at least 2 mg ml−1.
Desmethylnaproxen glucuronidation assay
Incubation mixtures, in a total volume of 0.2 ml, contained activated human liver microsomes (0.2 mg) or HEK293 cell lysate (0.4 mg), desmethylnaproxen (10–12 concentrations in the range 80–7000 µm), MgCl2 (4 m m) and phosphate buffer (0.1 m, pH 7.4). Separate ‘blank’ incubations were performed in the absence of UDPGA and human liver microsomes. After a 5-min preincubation, reactions were initiated by the addition of UDPGA (final concentration 5 m m). Incubations were performed in air at 37 °C, and terminated after 45 min by the addition of an equal volume of ice-cold 4% v/v acetic acid in methanol. Mixtures were vortex mixed, placed on ice, and then centrifuged at 5000 g for 10 min to precipitate the microsomal protein. An aliquot (30 µl) of the supernatant fraction was injected onto a Waters Novapak C18 column (150 × 3.9 mm, 4 µm), which was eluted with 18 : 82 acetonitrile–water (containing 0.12% v/v acetic acid) at a flow rate of 1.5 ml min−1. Analytes were detected by UV absorption at 225 nm. Retention times for desmethylnaproxen phenolic glucuronide, desmethylnaproxen acyl glucuronide, and desmethylnaproxen were 2.4, 4.4 and 11.2 min, respectively. The identity of the desmethylnaproxen acyl and phenolic glucuronide peaks formed by incubations of human liver microsomes and recombinant UGTs was confirmed by reference to authentic standards (which were stable on storage at −80 °C under N2) and by hydrolysis with β-glucuronidase.
Standard curves prepared using the authentic desmethylnaproxen acyl and phenolic glucuronides were linear over the concentration ranges 1.0–25 µm and 0.5–5 µm, respectively, with r2 values >0.99. The lower limit of quantification for both desmethylnaproxen glucuronides was 0.05 µm. Overall assay within-day precision was assessed by measuring desmethylnaproxen acyl and phenolic glucuronide formation for 10 separate incubations with the same batch of human liver microsomes at substrate concentrations of 100 and 2000 µm. Within-day coefficients of variation were in the range 2.2–2.7%. Linearity of product formation with respect to incubation time and microsomal protein concentration was determined for substrate concentrations of 100 and 1000 µm. The formation of both desmethylnaproxen glucuronides was linear with incubation times to at least 60 min and microsomal protein concentrations to at least 2 mg ml−1.
Data analysis
All incubations were performed in duplicate. Data points represent the mean (<10% variance) of the duplicate estimations. Kinetic constants for naproxen and desmethylnaproxen glucuronidation were obtained by fitting untransformed experimental data to equations 1 to 4 using EnzFitter (Biosoft, Cambridge, UK). Goodness of fit was assessed from comparison of the parameter SE of fit and 95% confidence limits, coefficient of determination (r2), and F-statistic.
The Michaelis–Menten equation,
| (1) |
where v is the rate of metabolite formation, Vmax is the maximum velocity (as pmol product per min per mg microsomal or cell lysate protein), Km is the Michaelis constant (substrate concentration at 0.5 Vmax), and [S] is the substrate concentration.
The two-enzyme Michaelis–Menten equation,
| (2) |
where the subscripts 1 and 2 represent the high and low affinity components, respectively. Substrate inhibition model [13],
| (3) |
where Ksi is the constant describing the substrate inhibition interaction.
The Hill equation, which describes sigmoidal kinetics [14]
| (4) |
where S50 is the substrate concentration resulting in 50% of Vmax (analogous to Km in previous equations) and n is the Hill coefficient.
Results
Naproxen glucuronidation by human liver microsomes and recombinant UGTs
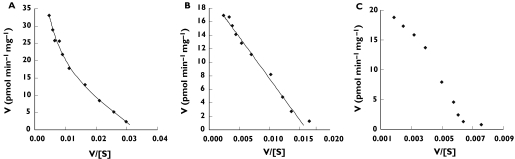
Naproxen acyl glucuronidation exhibited biphasic kinetics in all six livers investigated (Figure 2A). Data were well fitted to the two-enzyme Michaelis–Menten model, and derived kinetic constants for the high- and low-affinity components of the reaction are summarized in Table 1. As noted previously, the naproxen glucuronide was hygroscopic and unsuitable for standard curve construction. Thus, standard curves were prepared using naproxen. Rates of naproxen acyl glucuronide formation and Vmax values for this metabolite should therefore be considered ‘apparent’. (It should be noted that the slope of a standard curve prepared using desmethylnaproxen was 50% higher than the slope of the standard curve for desmethylnaproxen acyl glucuronide and 30% higher than the slope of the standard curve for desmethylnaproxen phenolic glucuronide.) The mean (± SD) microsomal ‘apparent’ intrinsic clearances (calculated as Vmax/Km) for the high- and low-affinity components of naproxen acyl glucuronidation were 9.1 ± 3.9 µl min−1 mg−1 and 1.3 ± 0.5 µl min−1 mg−1, respectively. It should be noted that naproxen exhibits minor nonspecific binding to human liver microsomes [15], and hence this factor will not significantly influence the kinetic analysis of naproxen glucuronidation in vitro.
Figure 2.
Representative Eadie–Hofstee plots for the conversion of naproxen to naproxen acyl glucuronide using the following enzyme sources: (A) human liver microsomes (H12); (B) UGT2B7; (C) UGT1A9; and (D) UGT1A10. Units of V/[S] are pmol glucuronide µm−1 min−1 mg−1. Points show experimentally determined values. Curves are the computer-generated curves of best fit
Table 1.
Derived two-enzyme Michaelis–Menten kinetic parameters for the formation of naproxen acyl-glucuronide by human liver microsomes
| Liver number | Km1 (µm) | Vmax1 (pmol min−1 mg−1) | Km2 (µm) | Vmax2 (pmol min−1 mg−1) |
|---|---|---|---|---|
| H6 | 14 | 114 | 293 | 473 |
| H10 | 32 | 237 | 605 | 332 |
| H12 | 19 | 247 | 411 | 847 |
| H13 | 36 | 476 | 537 | 665 |
| H29 | 49 | 142 | 496 | 520 |
| H40 | 26 | 254 | 495 | 566 |
| Mean (± SD) | 29 ± 13 | 245 ± 127 | 473 ± 108 | 567 ± 176 |
| 95% CI | 16, 43 | 111, 379 | 359, 587 | 383, 752 |
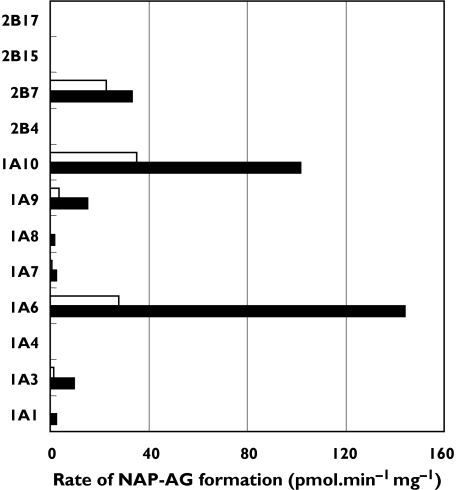
UGT 1A1, 1A3, 1A4, 1A6, 1A7, 1A8, 1A9, 1A10, 2B4, 2B7, 2B15 and 2B17 were screened for naproxen acyl glucuronidation activity at substrate concentrations of 100 and 1000 µm. All isoforms except UGT 1A4, 2B4, 2B15 and 2B17 metabolized naproxen (Figure 3). Naproxen glucuronidation kinetics were characterized for those isoforms with activities >10 pmol min−1 mg−1 at a substrate concentration of 1000 µm (viz. UGT 1A3, 1A6, 1A9, 1A10 and 2B7). Kinetic data (parameter ± parameter SE) for UGT1A3 (Km 4375 ± 6 µm, Vmax 49 ± 0.1 pmol min−1 mg−1), UGT1A6 (Km 855 ± 22 µm, Vmax 264 ± 2 pmol min−1 mg−1) and UGT2B7 (Km 72 ± 0.1 µm, Vmax 33 ± 0.1 pmol min−1 mg−1) were best described by the Michaelis–Menten equation. Naproxen glucuronidation by UGT1A9 was best fitted to the Hill equation with negative cooperativity (S50 1036 ± 56 µm, Vmax 30 ± 1 pmol min−1 mg−1, n = 0.82 ± 0.02), while data for UGT1A10 were consistent with weak substrate inhibition (Km 350 ± 33 µm, Ksi 13440 ± 2950 µm, Vmax 176 ± 8 pmol min−1 mg−1). Representative Eadie–Hofstee plots for each kinetic model (UGT2B7, UGT1A9 and UGT1A10) are shown in Figure 2B–D.
Figure 3.
Formation of naproxen acyl glucuronide by recombinant human UDP-glucuronosyltransferases at substrate (naproxen; NAP) concentrations of 100 (□) and 1000 µm (▪). Results represent the means of duplicate estimations
Effects of the UGT2B7 selective inhibitor fluconazole were investigated to confirm that UGT2B7 was the high-affinity enzyme responsible for naproxen glucuronidation by human liver microsomes. Experiments were conducted with pooled alamethicin-activated microsomes (from livers H6, H10, H12, H13, H29 and H40) at naproxen concentrations of 10 and 2000 µm. Substitution of the mean Km and Vmax values for the high- and low-affinity components of naproxen glucuronidation by human liver microsomes (Table 1) in the two-enzyme Michaelis–Menten equation indicates that the high-affinity enzyme(s) is responsible for 84% of activity at a substrate concentration of 10 µm and 34% of activity at a substrate concentration of 2000 µm. Fluconazole, 2.5 m m, inhibited human liver microsomal naproxen glucuronidation by 66% and 26% at the low and high substrate concentrations, respectively. By comparison, fluconazole (2.5 m m) inhibited UGT2B7 catalysed naproxen glucuronidation by 72% at a substrate concentration of 75 µm (the approximate Km for this enzyme).
Desmethylnaproxen acyl and phenolic glucuronidation by human liver microsomes and recombinant UGTs
Desmethylnaproxen phenolic glucuronidation exhibited apparent biphasic kinetics for four of the five livers investigated (viz. H10, H12, H13, and H40) (Figure 4A). Data for these livers were adequately fitted to the two-enzyme Michaelis–Menten model (Table 2). It should be noted, however, that fitting to the Hill equation generally provided similar goodness of fit estimates. Kinetic constants [mean ± SD, with 95% confidence interval (CI) in parentheses] for these four livers derived using the Hill equation were: S50 2557 ± 1624 µm (444, 4187); n 0.83 ± 0.08 (0.74, 0.97); and Vmax 47 ± 10 pmol min−1 mg−1 (22, 60). In contrast, desmethylnaproxen phenolic glucuronidation by liver H29 was best described by single-enzyme Michaelis–Menten kinetics (Figure 4B and Table 2).
Figure 4.
Representative Eadie–Hofstee plots for the conversion of desmethylnaproxen to desmethylnaproxen phenolic glucuronide using the following enzyme sources: (A) human liver microsomes (H12); (B) human liver microsomes (H29); and (C) UGT1A9. Units of V/[S] are pmol glucuronide µm−1 min−1 mg−1. Points show experimentally determined values. Curves are the computer-generated curves of best fit
Table 2.
Derived kinetic parameters for the formation of desmethylnaproxen phenolic glucuronide by human liver microsomes
| Liver number | Km1 (µm) | Vmax1 (pmol min−1 mg−1) | Km2 (µm) | Vmax2 (pmol min−1 mg−1) |
|---|---|---|---|---|
| H10 | 57 | 1.4 | 1237 | 27 |
| H12 | 579 | 17 | 9935 | 43 |
| H13 | 287 | 8 | 4924 | 40 |
| H40 | 203 | 12 | 2776 | 48 |
| Mean (± SD) | 282 ± 220 | 10 ± 7 | 4718 ± 3792 | 40 ± 12 |
| 95% CI | −68, 631 | 0, 20 | −1317, 10753 | 25, 53 |
| H29 | 1224 | 20 | – | – |
Data for livers H10, H12, H13 and H14 were derived from fitting to the two-enzyme Michaelis–Menten equation. Data for liver H29 were derived from fitting to the single-enzyme Michaelis–Menten equation.
Model fitting of kinetic data for desmethylnaproxen acyl glucuronidation was also inconclusive. Eadie–Hofstee plots for all livers were suggestive of nonMichaelis–Menten kinetics (Figure 5A; nonfitted experimental data shown), although goodness of fit parameters for fitting to the single-enzyme Michaelis–Menten equation were similar to the other models. Fitting data to the two-enzyme Michaelis–Menten equation gave mean (± SD, with 95% CI in parenthesis) Km values of 489 ± 347 µm (58, 920) and 4436 ± 3117 µm (565, 8307), and mean Vmax values of 127 ± 118 pmol min−1 mg−1 (−19, 273) and 1196 ± 488 pmol min−1 mg−1 (590, 1802) for the respective high- and low-affinity components. Fitting data to the single-enzyme Michaelis–Menten equation provided mean Km and Vmax values 2526 ± 861 µm (1457, 3596) and 1155 pmol min−1 mg−1 (634, 1676), respectively. Kinetic parameters derived using the Hill equation were: S50 3649 ± 1993 µm (1174, 6124); n 0.91 ± 0.05 (0.85, 0.96); and Vmax 1344 ± 538 pmol min−1 mg−1 (667, 2003). Although Km (or S50) values for the two pathways of desmethylnaproxen glucuronidation were similar (irrespective of the model used to generate kinetic constants), Vmax values for acyl glucuronidation were one to two orders of magnitude higher than for phenolic glucuronidation.
Figure 5.
Representative Eadie–Hofstee plots for the conversion of desmethylnaproxen to desmethylnaproxen acyl glucuronide using the following enzyme sources: (A) human liver microsomes (H13); (B) UGT2B7; (C) UGT1A6; and (D) UGT1A9. Units of V/[S] are pmol glucuronide µm−1 min−1 mg−1. Points show experimentally determined values. Curves are the computer-generated curves of best fit
UGT 1A1, 1A3, 1A6, 1A7, 1A9, 1A10 and 2B7, but not UGT 1A4, 2B15 or 2B17, converted desmethylnaproxen to its acyl glucuronide (Figure 6). In contrast, UGT 1A3, 1A6 and 2B7 did not form the phenolic glucuronide (Figure 7). Desmethylnaproxen acyl glucuronidation by UGT1A3 (Km 3384 ± 16 µm, Vmax 11 ± 0.1 pmol min−1 mg−1), UGT1A10 (Km 595 ± 49 µm, Vmax 179 ± 4 pmol min−1 mg−1) and UGT2B7 (Km 3133 ± 179 µm, Vmax 80 ± 2 pmol min−1 mg−1) was adequately modelled using the Michaelis–Menten equation (Figure 5B), whereas data for UGT1A6 were marginally better fitted to the Hill equation (S50 609 ± 36 µm; n 0.95 ± 0.02; Vmax 285 ± 7 pmol min−1 mg−1) (Figure 5C) Although inspection of the Eadie–Hofstee plot for desmethylnaproxen acyl glucuronidation by UGT1A9 suggests deviation from Michaelis–Menten kinetics, fitting to the Hill (S50 6003 ± 36 µm; n 1.07 ± 0.01; Vmax 29 ± 0.1 pmol min−1 mg−1) and Michaelis–Menten (Km 7724 ± 418 µm, Vmax 34 ± 1 pmol min−1 mg−1) equations provided similar goodness of fit parameters (Figure 5D; nonfitted experimental data shown). Similarly, desmethylnaproxen phenolic glucuronidation by UGT1A10 (Km 868 ± 46 µm, Vmax 146 ± 2 pmol min−1 mg−1) was adequately modelled using the Michaelis–Menten equation, although the Hill equation (with negative cooperativity) provided a better model for UGT1A9 (S50 4795 ± 21 µm; n 0.90 ± 0.01; Vmax 30 ± 0.1 pmol min−1 mg−1) (Figure 4C; nonfitted experimental data shown).
Figure 6.
Formation of desmethylnaproxen acyl glucuronide by recombinant human UDP-glucuronosyltransferases at substrate (desmethylnaproxen; DMN) concentrations of 500 (□) and 5000 µm (▪). Results represent the means of duplicate estimations
Figure 7.
Formation of desmethylnaproxen phenolic glucuronide by recombinant human UDP-glucuronosyltransferases at substrate (desmethylnaproxen; DMN) concentrations of 500 (□) and 5000 µm (▪). Results represent the means of duplicate estimations
Discussion
Naproxen glucuronidation exhibited biphasic kinetics in human liver microsomes, characteristic of the involvement of high- and low-affinity UGTs in this pathway. The mean Km value for the high-affinity component determined from fitting to the two-enzyme Michaelis–Menten equation was 16-fold lower than the Km for the low-affinity reaction, and ‘apparent’ intrinsic clearances differed sevenfold. Assuming an unbound fraction in plasma of 0.018 [3], the maximum plasma unbound concentration of naproxen following a single oral dose of 500 mg or on chronic dosing with 250 mg b.d. is approximately 5 µm[3, 16]. Following a single oral dose of 1000 mg, the maximum plasma unbound concentration of naproxen may be estimated as 9 µm[4]. Hence, the low-affinity UGT(s) would be expected to make only a minor contribution (<15%) to naproxen clearance in patients receiving this drug.
Recombinant UGT 1A1, 1A3, 1A6, 1A7, 1A8, 1A9, 1A10 and 2B7 glucuronidated naproxen. Of the hepatically expressed enzymes (viz. UGT 1A1, 1A3, 1A6, 1A9 and 2B7), highest activity was observed with UGT1A6 and UGT2B7. Notably, however, the Km value for naproxen glucuronidation by UGT2B7 (72 µm) was close to the Km values for the high-affinity component of human liver microsomal naproxen glucuronidation (14–49 µm). Recent work in this laboratory (V. Uchaipichat, P. I. Mackenzie and J. O. Miners, manuscript submitted for publication) using a panel of recombinant human UGTs demonstrated that fluconazole is a selective competitive inhibitor of UGT2B7. At an added concentration of 2.5 m m, fluconazole inhibited UGT2B7 by approximately 70% but other isoforms by <25%. In the present study, fluconazole inhibited naproxen glucuronidation by UGT2B7 and human liver microsomes to a similar extent (approximately 70%). Collectively, the results indicate that UGT2B7 is the high-affinity enzyme involved in hepatic naproxen glucuronidation. It is likely that the low-affinity component of hepatic naproxen glucuronidation comprises several isoform activities, since numerous UGTs have the capacity to glucuronidate naproxen. In particular, UGT 1A6 and 1A9 have Km or S50 values of similar order to the low-affinity component of human liver microsomal naproxen glucuronidation.
Desmethylnaproxen phenolic glucuronidation exhibited apparent biphasic kinetics in four of the five livers investigated and Michaelis–Menten kinetics in the other. However, model fitting to the Hill equation (with negative cooperativity) provided similar goodness of fit estimates to those obtained with the two-enzyme model. Eadie–Hofstee plots for desmethylnaproxen acyl glucuronidation were also suggestive of atypical kinetics, although goodness of fit parameters for the Michaelis–Menten equation were again similar to the other models. The capacity of several UGTs to convert desmethylnaproxen to its acyl glucuronide suggests multiple UGTs could contribute to desmethylnaproxen glucuronidation in human liver. However, none of the derived Km values for the recombinant enzymes matched the Km values of the high-affinity reactions from fitting to the two-enzyme Michaelis–-Menten equation. Of the hepatically expressed UGTs, only UGT 1A1 and 1A9 formed desmethylnaproxen phenolic glucuronide. The involvement of as yet unidentified UGTs in the metabolism of naproxen and desmethylnaproxen cannot be discounted. In addition, UGT 2A1, 2B10, 2B11 and 2B28 were not investigated here. However, UGT2A1 is expressed primarily in the nasal epithelium while UGT 2B10, 2B11 and 2B28 are devoid of activity or exhibit very low capacity for xenobiotic glucuronidation [7, 8].
Recent reports from this and other laboratories [11, 17–19] have demonstrated that ‘atypical’, or nonMichaelis–Menten, kinetics may occur for reactions catalysed by recombinant human UGTs. In particular, it was demonstrated using 4-methylumbelliferone and 1-naphthol as the substrates that kinetic models varied with substrate (for the same isoform) and from isoform to isoform (with the same substrate) [11]. Consistent with these observations, naproxen glucuronidation by UGT 1A3, 1A6 and 2B7 followed Michaelis–Menten kinetics whereas UGT1A9 and UGT1A10 exhibited negative cooperativity and substrate inhibition, respectively. Similarly, desmethylnaproxen acyl glucuronidation by UGT1A6 and UGT1A9 and desmethylnaproxen phenolic glucuronidation by UGT1A9 were suggestive of cooperativity, although these reactions catalysed by other isoforms were modelled adequately by the Michaelis–Menten equation. The Hill and substrate inhibition models imply binding of more than one substrate molecule in the active site, and atypical glucuronidation kinetic data may alternatively be analysed using multisite models [11]. Artefactual sources of atypical kinetics in vitro[13] can be discounted in the present study, but it is possible that atypical glucuronidation kinetics is solely an in vitro phenomenon. It is noteworthy, however, that atypical kinetic behaviour in vitro is not associated with all substrates of a particular UGT, which is consistent with results reported for other enzymes (e.g. CYP3A4 [13]) that exhibit nonhyperbolic kinetics.
As indicated previously, it may be predicted from the results presented here that UGT2B7 is responsible for naproxen acyl glucuronidation, the major elimination pathway for this drug, in vivo. UGT2B7 has the capacity to glucuronidate numerous clinically used drugs, including other nonsteroidal anti-inflammatory drugs [20], opioids [17, 21], zidovudine [22], and dimethylxanthenone acetic acid [23]. The capacity of UGT2B7 to glucuronidate the carboxylic acid function of naproxen and desmethylnaproxen, but not the phenolic group of desmethylnaproxen, is consistent with the reported ability of this enzyme to metabolize carboxylic acid-containing drugs [20]. Drugs reported to inhibit UGT2B7 activity include amitriptyline, diclofenac, fluconazole and probenecid [11, 16, 24, 25], and it is possible that these compounds may interact with naproxen in vivo. Although inhibition of naproxen elimination is unlikely to have important clinical consequences, naproxen may alternately act as a competitive inhibitor of other UGT2B7 substrates in vivo. While this appears not to have been investigated, naproxen has been shown to inhibit UGT2B7 activity in vitro[23]. UGT2B7 exhibits genetic polymorphism. Substitution of His for Tyr at residue 268 results from a C to T transversion at nucleotide 802 of the UGT2B7 coding region [26]. However, available evidence suggests that this polymorphism has little effect on drug glucuronidation [21, 26], but an influence on naproxen elimination cannot be discounted.
This study focused primarily on the hepatic glucuronidation of naproxen and desmethylnaproxen. However, UGT1A10, which is expressed only in the gastrointestinal tract, metabolized both compounds. Indeed, of the enzymes investigated, UGT1A10 exhibited the highest affinity for desmethylnaproxen. Two other isoforms expressed only in the gastrointestinal tract, UGT 1A7 and 1A8, also had the capacity to glucuronidate naproxen. Although almost the entire dose of naproxen can be accounted for in urine [27], this does not preclude hydrolysis of any naproxen glucuronide synthesized in the gastrointestinal tract and subsequent absorption of the re-formed parent drug.
UGT2B7, the principal enzyme shown here to be involved in human liver microsomal naproxen acyl glucuronidation, is known to be expressed in kidney [8]. UGT 1A3, 1A6 and 1A9, which additionally had the capacity to glucuronidate naproxen, are also expressed in kidney. Consistent with this observation, results from this laboratory (P. Tsoutsikos, J. O. Miners and K. M. Knights, unpublished data) indicate that specific activities for naproxen glucuronidation by human kidney microsomes are similar to those reported here for human liver microsomes. Comparable activities for drug glucuronidation by human kidney and liver tissue have been noted in other studies [7]. It is therefore likely that renal drug glucuronidation may have an important ‘local’ detoxification role [28]. However, when microsomal scaling factors are applied to the two tissues, whole organ intrinsic clearance for the liver is substantially higher than that for the kidney and hence the latter tissue contributes to metabolic clearance to a minor extent only [7, 29].
In summary, results presented here indicate that UGT2B7 is likely to be the principal enzyme responsible for S-naproxen elimination in vivo, whereas multiple UGTs appear to be involved in desmethylnaproxen glucuronidation. As reported for other glucuronidated compounds, nonMichaelis–Menten kinetics is a feature of naproxen and desmethylnaproxen glucuronidation by several UGT isoforms. Thus, the characterization of drug glucuronidation in vitro necessarily requires the investigation of a wide range of substrate concentrations and analysis of data using appropriate kinetic models.
Acknowledgments
This work was supported by grants-in-aid from the National Health & Medical Research Council of Australia and AstraZeneca Pharmaceuticals. The authors are grateful to Professor Ron Dickinson and Drs Russell Addison and Karine Mardon for their assistance in establishing the HPLC assays.
References
- 1.Prescription cost analysis data. Drugs used in rheumatic diseases and gout. http://www.publications.doh.gov.uk/prescriptionstatistics/
- 2.Upton RA, Buskin JN, Williams RL, Holford NHG, Riegelman S. Negligible excretion of unchanged ketoprofen, naproxen and probenecid in urine. J Pharm Sci. 1980;69:1254–7. doi: 10.1002/jps.2600691105. [DOI] [PubMed] [Google Scholar]
- 3.Vree TB, van den Biggelaar-Martea M, Verwey-van Wissen CPWGM, Vree ML, Guelen PJM. The pharmacokinetics of naproxen, its metabolite O-desmethylnaproxen, and their acyl glucuronides in humans. Effects Cimetidine Br J Clin Pharmacol. 1993;35:467–72. doi: 10.1111/j.1365-2125.1993.tb04171.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Runkel R, Chaplin M, Sevelius H, Ortega E, Segre E. Pharmacokinetics of naproxen overdoses. Clin Pharmacol Ther. 1976;24:269–77. doi: 10.1002/cpt1976203269. [DOI] [PubMed] [Google Scholar]
- 5.Miners JO, Coulter S, Tukey RH, Veronese ME, Birkett DJ. Cytochromes P450 1A2 and 2C9 are responsible for the human hepatic O-demethylation of R- and S-naproxen. Biochem Pharmacol. 1996;51:1003–8. doi: 10.1016/0006-2952(96)85085-4. [DOI] [PubMed] [Google Scholar]
- 6.Mackenzie PI, Owens IS, Burchell B, Bock KW, Bairoch I, Belanger A, Fournel-Gigleux S, Green M, Hum DW, Iyanagi T, Lancet D, Louisot P, Magdalou J, Roy Chowdhury J, Ritter JK, Schachter H, Tephly TR, Tipton KF, Nebert DW. The UDP-glycosyltransferase gene superfamily: recommended nomenclature update based on evolutionary divergence. Pharmacogenetics. 1997;7:255–69. doi: 10.1097/00008571-199708000-00001. [DOI] [PubMed] [Google Scholar]
- 7.Miners JO, Smith PA, Sorich MJ, McKinnon RA, Mackenzie PI. Predicting human drug glucuronidation parameters: Application of in vitro and in silico modelling approaches. Ann Rev Pharmacol Toxicol. 2004;44:1–25. doi: 10.1146/annurev.pharmtox.44.101802.121546. [DOI] [PubMed] [Google Scholar]
- 8.Tukey RH, Strassburg CP. Human UDP-glucuronosyltransferases: metabolism, expression and disease. Ann Rev Pharmacol Toxicol. 2000;40:581–616. doi: 10.1146/annurev.pharmtox.40.1.581. [DOI] [PubMed] [Google Scholar]
- 9.Lowry OH, Rosebrough NJ, Farr AL, Randall RJ. Protein measurement by the Folin phenol reagent. 1951. pp. 265–75. [PubMed]
- 10.Boase S, Miners JO. In vitro – in vivo correlations for drugs eliminated by glucuronidation: investigations with the model substrate zidovudine. Br J Clin Pharmacol. 2002;54:493–503. doi: 10.1046/j.1365-2125.2002.01669.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Uchaipichat V, Mackenzie PI, Guo X-H, Gardner-Stephen D, Galetin A, Houston JB, Miners JO. Human UDP-glucuronosyltransferases. Isoform selectivity and kinetics of 4-methylumbelliferone and 1-naphthol glucuronidation, effects of organic solvents, and inhibition by diclofenac and probenecid. Drug Metab Disp. 2004;32:413–23. doi: 10.1124/dmd.32.4.413. [DOI] [PubMed] [Google Scholar]
- 12.Dehal SS, Gagne PV, Crespi CL, Patten CJ. Characterization of a probe substrate and an inhibitor of UDP glucuronosyltransferase (UGT) 1A4 activity in human liver microsomes (HLM) and cDNA-expressed UGT-enzymes. http://www.bdbiosciences.com/discovery_labware/gentest/products/pdf/1A4_AAPS_S01T056R.
- 13.Houston JB, Kenworthy KE. In vitro – in vivo scaling of CYP kinetic data not consistent with the classical Michaelis–Menten model. Drug Metab Disp. 2000;28:246–54. [PubMed] [Google Scholar]
- 14.Cornish-Bowden A. Fundamentals of Enzyme Kinetics. London: Portland Press; 1995. Control of enzyme activity; pp. 203–16. [Google Scholar]
- 15.McLure JA, Miners JO, Birkett DJ. Nonspecific binding of drugs to human liver microsomes. Br J Clin Pharmacol. 2000;49:453–61. doi: 10.1046/j.1365-2125.2000.00193.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Runkel R, Mroszczak E, Chaplin M, Sevelius H, Segre E. Naproxen–probenecid interaction. Clin Pharmacol Ther. 1978;24:706–13. doi: 10.1002/cpt1978246706. [DOI] [PubMed] [Google Scholar]
- 17.Stone AN, Mackenzie PI, Galetin A, Houston JB, Miners JO. Isoform selectivity and kinetics of morphine 3- and 6-glucuronidation by human UDP-glucuronosyltransferases: evidence of atypical glucuronidation kinetics by UGT2B7. Drug Metab Disp. 2003;31:1086–9. doi: 10.1124/dmd.31.9.1086. [DOI] [PubMed] [Google Scholar]
- 18.Williams JA, Ring BJ, Cantrell VE, Campanale K, Jones DR, Hall SD, Wrighton SA. Differential modulation of UDP-glucuronosyltransferase 1A1 (UGT1A1) catalyzed estradiol 3-glucuronidation by the addition of UGT1A1 substrates and other compounds to human liver microsomes. Drug Metab Disp. 2002;30:1266–73. doi: 10.1124/dmd.30.11.1266. [DOI] [PubMed] [Google Scholar]
- 19.Soars MG, Ring BJ, Wrighton SA. The effect of incubation conditions on the enzyme kinetics of UDP-glucuronosyltransferase. Drug Metab Disp. 2003;31:762–7. doi: 10.1124/dmd.31.6.762. [DOI] [PubMed] [Google Scholar]
- 20.Jin C-J, Miners JO, Lillywhite KJ, Mackenzie PI. Complementary deoxyribonucleic acid cloning and expression of a human liver uridine diphosphate glucuronosyltransferase glucuronidating carboxylic acid containing drugs. J Pharmacol Exp Ther. 1993;264:475–9. [PubMed] [Google Scholar]
- 21.Coffman BL, King CD, Rios GR, Tephly TR. The glucuronidation of opioids, xenobiotics and androgens by human UGT2B7(Y268) and UGT2B7(H268) Drug Metab Disp. 1998;26:73–7. [PubMed] [Google Scholar]
- 22.Barbier O, Turgeon D, Girard C, Green MD, Tephly TR, Belanger A. 3′-Azido-3′-deoxythymidine is glucuronidated by human UDP-glucuronosyltransferase 2B7 (UGT2B7) Drug Metab Disp. 2000;28:497–502. [PubMed] [Google Scholar]
- 23.Miners JO, Valente L, Lillywhite KJ, Mackenzie PI, Burchell B, Baguley BC, Kestell P. Preclinical prediction of factors influencing the elimination of 5,6-dimethylxanthenone-4-acetic acid, a new anticancer drug. Cancer Res. 1997;57:284–9. [PubMed] [Google Scholar]
- 24.Yue Q, von Bahr C, Odar-Cederlof I, Sawe J. Glucuronidation of codeine and morphine in human liver and kidney microsomes: effect of inhibitors. Pharmacol Toxicol. 1990;66:221–6. doi: 10.1111/j.1600-0773.1990.tb00737.x. [DOI] [PubMed] [Google Scholar]
- 25.Sahai J, Gallicano K, Pakuts A, Cameron WD. Effect of fluconazole on zidovudine pharmacokinetics in patients infected with human immunodeficiency virus. J Infect Dis. 1994;169:1103–7. doi: 10.1093/infdis/169.5.1103. [DOI] [PubMed] [Google Scholar]
- 26.Bhasker CR, McKinnon W, Stone A, Lo ACT, Kubota T, Ishizaki T, Miners JO. Genetic polymorphism of UDP-glucuronosyltransferase 2B7 (UGT2B7) at amino acid 268: ethnic diversity of alleles and potential clinical significance. Pharmacogenetics. 2000;10:679–85. doi: 10.1097/00008571-200011000-00002. [DOI] [PubMed] [Google Scholar]
- 27.Runkel R, Forchielli E, Sevelius H, Chaplin M, Segre E. Nonlinear plasma level response to high doses of naproxen. Clin Pharmacol Ther. 1974;20:261–6. doi: 10.1002/cpt1974153261. [DOI] [PubMed] [Google Scholar]
- 28.Tsoutsikos P, Miners JO, Stapleton A, Thomas T, Sallustio BC, Knights KM. Evidence that unsaturated fatty acids are potent inhibitors of renal UDP-glucuronosyltransferases (UGT): kinetic studies using human cortical microsomes and recombinant UGT1A9 and UGT2B7. Biochem Pharmacol. 2004;67:191–9. doi: 10.1016/j.bcp.2003.08.025. [DOI] [PubMed] [Google Scholar]
- 29.Bowalgaha K, Miners JO. The glucuronidation of mycophenolic acid by human liver, kidney and jejunum microsomes. Br J Clin Pharmacol. 2001;52:605–9. doi: 10.1046/j.0306-5251.2001.01487.x. [DOI] [PMC free article] [PubMed] [Google Scholar]