Abstract
Aims
We evaluated the involvement of cytochrome P450 (CYP) isoforms 2C9 and 2C19 in chlorpropamide 2-hydroxylation in vitro and in chlorpropamide disposition in vivo.
Methods
To identify CYP isoforms(s) that catalyse 2-hydroxylation of chlorpropamide, the incubation studies were conducted using human liver microsomes and recombinant CYP isoforms. To evaluate whether genetic polymorphisms of CYP2C9 and/or CYP2C19 influence the disposition of chlorpropamide, a single oral dose of 250 mg chlorpropamide was administered to 21 healthy subjects pregenotyped for CYP2C9 and CYP2C19.
Results
In human liver microsomal incubation studies, the formation of 2-hydroxychlorpropamide (2-OH-chlorpropamide), a major chlorpropamide metabolite in human, has been best described by a one-enzyme model with estimated Km and Vmax of 121.7 ± 19.9 µm and 16.1 ± 5.0 pmol min−1 mg−1 protein, respectively. In incubation studies using human recombinant CYP isoforms, however, 2-OH-chlorpropamide was formed by both CYP2C9 and CYP2C19 with similar intrinsic clearances (CYP2C9 vs. CYP2C19: 0.26 vs. 0.22 µl min−1 nmol−1 protein). Formation of 2-OH-chlorpropamide in human liver microsomes was significantly inhibited by sulfaphenazole, but not by S-mephenytoin, ketoconazole, quinidine, or furafylline. In in vivo clinical trials, eight subjects with the CYP2C9*1/*3 genotype exhibited significantly lower nonrenal clearance [*1/*3 vs.*1/*1: 1.8 ± 0.2 vs. 2.4 ± 0.1 ml h−1 kg−1, P < 0.05; 95% confidence interval (CI) on the difference 0.2, 1.0] and higher metabolic ratios (of chlorpropamide/2-OH-chlorpropamide in urine: *1/*3 vs. *1/*1: 1.01 ± 0.19 vs. 0.56 ± 0.08, P < 0.05; 95% CI on the difference −0.9, −0.1) than did 13 subjects with CYP2C9*1/*1 genotype. In contrast, no differences in chlorpropamide pharmacokinetics were observed for subjects with the CYP2C19 extensive metabolizer vs. poor metabolizer genotypes.
Conclusions
These results suggest that chlorpropamide disposition is principally determined by CYP2C9 activity in vivo, although both CYP2C9 and CYP2C19 have a catalysing activity of chlorpropamide 2-hydroxylation pathway.
Keywords: chlorpropamide, CYP2C19, CYP2C9, in vitro, in vivo
Introduction
Chlorpropamide is a sulphonylurea oral hypoglycaemic agent used in the treatment of Type 2 diabetes mellitus. It has a long half-life and duration of hypoglycaemic action, and thus is usually given as a once-daily dose [1, 2]. Chlorpropamide is extensively metabolized in human liver: as much as 80% of a single chlorpropamide dose and several of its metabolites, including 2-hydroxychlorpropamide (2-OH-chlorpropamide), 3-hydroxychlorpropamide, p-chlorobenzene-sulphonylurea, and p-chlorobenzene-sulphonamide, were identified in human urine after oral dosing [3, 4]. In humans, 2-hydroxylation is the principal metabolic fate of chlorpropamide; 2-OH-chlorpropamide recovered in urine accounts for approximately 43–69% of the daily dose [5].
Chlorpropamide has a higher incidence of hypoglycaemic, adverse drug reactions after normal dosing than do other sulphonylureas, which is partially due to its longer duration and higher potency [6, 7]. In addition, the apparently unpredictable occurrence of adverse reactions after dosing may be due to broad individual variability in chlorpropamide disposition. Up to a 30-fold difference in steady-state serum chlorpropamide concentrations and plasma clearances subsequent to therapeutic dosing in patients has been reported [8, 9].
Metabolic drug interactions appear to be another important factor in individual variability in chlorpropamide disposition. Rifampin, a known inducer of CYP enzymes, has been reported to decrease significantly the plasma concentration of coadministered chlorpropamide, and thus increase the effective chlorpropamide dosage, after concomitant administration [10]. Furthermore, plasma chlorpropamide concentrations increase in the presence of coadministered drugs known to inhibit CYP enzymes [5]. These observations suggest that the metabolic fate of chlorpropamide contributes markedly to its interindividual pharmacokinetic variation in human, and genetic polymorphisms of the CYP isoform(s) participating in the chlorpropamide metabolism may also be responsible for the wide individual variability in chlorpropamide disposition. However, we are unaware of any studies that directly address this issue.
In our previous clinical trial to evaluate chlorpropamide metabolism, a major metabolite peak was found in a high-performance liquid chromatography (HPLC) chromatogram of urine sample from a human taking chlorpropamide. We confirmed that this peak was consistent with 2-OH-chlorpropamide by liquid chromatography/mass spectrometry (LC/MS) and sought to identify the CYP isoforms(s) that catalyse 2-hydroxylation of chlorpropamide during its catabolism. We found that both CYP2C9 and CYP2C19 were involved in chlorpropamide 2-hydroxylation in human liver microsomes. Since both CYP isoforms exhibit genetic polymorphisms [11, 12], we further determined whether these genetic polymorphisms influenced the disposition of chlorpropamide. In addition, we analysed the inhibition of human liver CYP isoforms by chlorpropamide in order to facilitate prediction of inhibitory interactions of chlorpropamide with concomitantly administered drugs that are CYP substrates.
Methods
Chemicals
Chlorpropamide, tolbutamide, dextromethorphan, dextrorphan, phenacetin, acetaminophen, sulfaphenazole, ketoconazole, quinidine, β-NADP, EDTA, MgCl2, glucose-6-phosphate, and glucose-6-phosphate dehydrogenase were purchased from Sigma Chemical Co. (St Louis, MO, USA). Midazolam, 4-hydroxytolbutamide, 1-hydroxymidazolam, S-mephenytoin, furafylline, and 4′-hydroxymephenytoin were obtained from Ultrafine Chemical Co. (Manchester, UK). Acetonitrile, chloroform, and methanol were purchased from Fisher Scientific (Pittsburgh, PA, USA).
Human liver microsomes and recombinant CYP isoforms
Human liver tissue samples (HL-20, HL-21, and HL-23) were obtained from partial hepatectomies performed to remove metastatic tumours. Patients who had taken any known CYP inhibitors or inducers in the week before the surgical operation were excluded. All liver tissues were identified to have the wild genotype of CYP2C9*1/*1 and CYP2C19*1/*1 that were determined from the extracted genomic DNAs from frozen liver tissue using using the DNeasy® tissue kit (Qiagen, Hilden, Germany). Human liver microsomes were prepared from histopathologically normal, nontumour-bearing parenchymal tissue, using differential ultracentrifugation, as described previously [13]. The microsomal pellets were resuspended in 100 m m phosphate buffer (pH 7.4) containing 1.0 m m EDTA and 5.0 m m MgCl2. Approval by the Institutional Review Board of Busan Paik Hospital, Busan, Korea and donor patient's consent were obtained for the use of human liver tissue. Human recombinant CYP isoforms (Supersomes®, cDNA-expressed microsomes obtained from a baculovirus expression system), CYP 1A2, 2A6, 2C8, 2C9, 2C19, 2D6, 2E1, and 3A4, were purchased from Gentest Corporation (Woburn, MA, USA). Protein concentrations and CYP contents were supplied by the manufacturer.
In vitro experiments
Microsomal incubations
Incubation of chlorpropamide with human liver microsomes yielded one major metabolite, hydroxychlorpropamide, which was produced linearly with an incubation time of up to 120 min and up to 2.0 mg of microsomal protein per incubation. The optimal conditions for microsomal incubation were determined in the linear range. In all experiments, chlorpropamide was dissolved and diluted serially in methanol, an aliquot of the chlorpropamide was transferred to a 1.5-ml tube, and the methanol was removed by evaporating to dryness with a vacuum centrifuge (Speedvac AES 2010; Savant Instruments, Holbrook, NY, USA). Reaction mixtures (250 µl) containing microsomes (1 mg ml−1) and various concentrations of chlorpropamide (10–1000 µm) reconstituted in 50 m m potassium phosphate buffer (pH 7.4) were preincubated for 5 min at 37 °C. Reactions were then initiated by addition of an NADPH generating system (1.3 m m NADP, 3.3 m m glucose-6-phosphate, 0.5 U ml−1 glucose-6-phosphate dehydrogenase, and 3.3 m m MgCl2). After 60 min incubation at 37 °C, reactions were terminated by placing the incubation tube on ice and by the immediate addition of 100 µl of ice-cold 10% (w/v) perchloric acid. All experiments using human liver microsomes were performed in duplicate.
Activity of human recombinant CYP isoforms
To identify the specific CYP isoforms responsible for chlorpropamide 2-hydroxylation, chlorpropamide (100 µm) was incubated with 20 pmol of human recombinant CYP isoforms: CYP1A2, 2A6, 2C8, 2C9, 2C19, 2D6, 2E1 or 3A4, or a control vector. Since CYP2C9 and CYP2C19 appeared to be the major enzymes catalysing chlorpropamide 2-hydroxylation, further studies of the kinetics of formation of 2-hydroxy metabolite by these CYP isoforms were performed at chlorpropamide concentrations up to 2000 µm. The experiments using CYP isoforms were performed in duplicate.
Selective chemical inhibition of CYP isoforms
To characterize the role of individual CYP isoforms in the chlorpropamide 2-hydroxylation observed in microsomal incubation studies, the effects of selective or semiselective chemical inhibitors were evaluated. The rate of formation of 2-OH-chlorpropamide from chlorpropamide was estimated in the absence (control) and concentrations beyond the window of inhibitory selectivity of the following known CYP isoform-specific inhibitors: furafylline (CYP1A2, 1 and 10 µm), sulfaphenazole (CYP2C9, 1 and 10 µm), quinidine (CYP2D6, 1 and 10 µm), S-mephenytoin (CYP2C19, 10 and 100 µm), and ketoconazole (CYP3A4, 1 and 5 µm) [13]. In this study, a chlorpropamide concentration of 100 µm was chosen because the Km values for chlorpropamide 2-hydroxylation in microsomes ranged from 100 to 140 µm. Inhibitors were used at =50 µm. In inhibition experiments with furafylline, a mechanism-based selective inhibitor of CYP1A2 [14], the furafylline was preincubated in the liver microsome mixture containing the NADPH-generating system for 15 min, and the reaction was then initiated by adding chlorpropamide. All other inhibitors tested were coincubated with chlorpropamide and microsomes, and the reactions were then initiated by adding the NADPH-generating system.
Inhibition of CYP isoforms by chlorpropamide
The inhibitory effect of chlorpropamide (at up to 250 µm) on the five human CYP isoforms was tested in human liver microsomes by CYP-specific metabolic pathway probes that are routinely used in our laboratory [13, 15]. The reaction probes used were phenacetin O-deethylation for CYP1A2 [16], tolbutamide 4-hydroxylation for CYP2C9 [17], S-mephenytoin 4-hydroxylation for CYP2C19 [18], dextromethorphan O-demethylation for CYP2D6 [19], and midazolam 1-hydroxylation for CYP3A4 [20].
Enzyme kinetic analysis
Enzyme kinetics of 2-OH-chlorpropamide formation were analysed first by graphical methods, such as Eadie–Hofstee plots, and ultimately by nonlinear least squares regression analysis, in which the data were fitted to various models. The best model describing 2-OH-chlorpropamide formation in human liver microsomes was selected according to the Akaike Information Criteria (AIC), a measure of goodness-of-fit, based on maximum likelihood. The apparent kinetic parameters for the formation of 2-OH-chlorpropamide (Km and Vmax) were determined by fitting the Michaelis–Menten equation to unweighted data (mean value of estimates obtained from duplicated experiments metabolite formation rate and chlorpropamide concentration) by nonlinear least squares regression, using WinNonlin® Vol. 2.0 (Pharsight Corporation, Mountain View, CA, USA).
In incubation studies with human recombinant CYP isoforms, a simple Michaelis–Menten model was fitted to the actual data (mean value of estimates obtained from duplicated experiments) for major metabolite formation to estimate the enzyme kinetic parameters. Intrinsic clearance (CLint) was calculated from the estimated kinetic parameters as CLint = Vmax/Km. The extent of inhibition of chlorpropamide 2-hydroxylation by CYP isoform-selective inhibitors and of CYP isoform-specific probe reactions by chlorpropamide was described as percentage of control activity. The inhibitor concentration producing a 50% reduction in reaction velocity (IC50) was estimated by nonlinear fitting of percentage control activity data to the simple inhibition model.
In vivo clinical studies
To evaluate the contributions of CYP2C9 and CYP2C19 to the disposition of chlorpropamide in vivo, the pharmacokinetics of chlorpropamide disposition and 2-hydroxy metabolite formation were compared among human subjects with wild-type or mutant CYP2C9 and CYP2C19 genotypes. The mutant genotypes are associated with decreased enzyme activity. In this study, we enrolled 21 healthy Korean subjects of known CYP2C9 and CYP2C19 genotypes. Thirteen of these subjects were homozygous for CYP2C9*1; six of these had a CYP2C19 extensive metabolizer (EM) genotype (i.e. CYP2C19*1/*1), and seven had a CYP2C19 poor metabolizer (PM) genotype (i.e. CYP2C19*2/*2, *3/*3, or *2/*3). Eight of the subjects were heterozygous for CYP2C9*3 and homozygous for the CYP2C19 EM genotype. The subjects consisted of 19 males and two females; they were from 20 to 29 years old, and in good health, as shown by physical examinations and routine laboratory tests for liver and renal function. No differences in weight, height, or other demographic data were found to correlate with different CYP2C9 and/or CYP2C19 genotypes among the subjects. This study was approved by the Institutional Review Board of Busan Paik Hospital, Busan, Korea, and all subjects gave their written consent before participating this study.
Genomic DNA for CYP2C9 and CYP2C19 genotyping was extracted from leucocytes of peripheral venous blood using a QIAamp® DNA Blood Mini Kit (Qiagen, Hilden, Germany). CYP2C9 and CYP2C19 genotypes were determined by a polymerase chain reaction-restriction fragment length polymorphism method, described previously [11, 12].
The subjects were prohibited from ingesting any drugs, alcohol, caffeine-containing foods, or grapefruit juice during the study period. On the day of sample collection, each subject received a single, 250-mg oral dose of chlorpropamide after overnight fasting. Blood samples were drawn before dosing and at 0.5, 1.0, 1.5, 2, 3, 4, 6, 8, 12, 24, 48, 72, and 96 h after oral administration. Blood samples were collected in heparinized glass tubes (Vacutainer®; Becton Dickinson, Fanklin Lakes, NJ, USA) and centrifuged at 1000 × g for 10 min to separate plasma. The blood glucose level was measured repeatedly up to 4 h before diet after chlorpropamide administration to monitor the possible development of a hypoglycaemic state. In addition, urine samples were collected 24 h after dosing. Both blood and urine samples were stored at −80 °C until assayed.
Chlorpropamide and 2-hydroxychlorpropamide assays
The concentrations of chlorpropamide and 2-OH-chlorpropamide in plasma, urine, and microsomal incubates were determined by the reverse-phase HPLC method of Csilag et al. with modification [21]. Plasma or urine (0.25 ml) was added to a 10-ml glass tube containing an internal standard (2 µg tolbutamide), 250 ml of 1 m hydrochloric acid, and 3 ml of chloroform. After vigorous stirring for 3 min on a vortex mixer, the aqueous phase was separated by centrifugation (1000 × g for 10 min) and discarded. The remaining organic phase was subsequently evaporated to dryness in a vacuum centrifuge and reconstituted with 100 µl of acetonitrile. A 10-µl aliquot of the reconstituted organic phase was injected onto a LiChrosorb RP-8 HPLC column (250 × 4 mm internal diameter, 10 µm particle size; Merck®, Darmstadt, Germany) attached to a Gilson HPLC system consisting of a model 307 pump, a model 234 autoinjector, and a model 118 UV detector (Villiers Le Bel, France). The mobile phase consisted of 1% acetic acid/acetonitrile (70/30, v/v) adjusted to pH 4.0 with 4 m NaOH, and the flow rate was 1.4 ml min−1. The eluate was monitored by UV detection at a wavelength of 235 nm. The retention times for chlorpropamide and hydroxychlorpropamide were approximately 13.3 and 3.6 min, respectively. The lower limit of quantification for chlorpropamide was 0.1 µg ml−1, which is sufficient for routine pharmacokinetics monitoring. Using these methods, the daily coefficients of variation were estimated to be 5.1 and 6.2% at chlorpropamide concentrations of 1 and 20 µg ml−1, respectively.
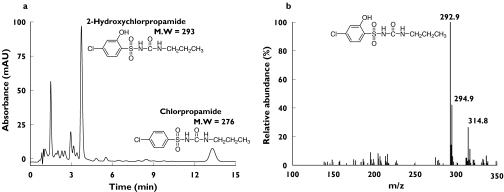
Since a 2-OH-chlropropamide standard was not available, derived kinetic parameters such as Vmax and CLint and urinary metabolic ratio are apparent values estimated from the fractional conversion of chlorpropamide calibration curve. First, the 2-OH-chlorpropamide HPLC peak was identified using LC/MS. Samples of the microsomal incubates at 100 µm chlorpropamide and of urine extracts from subjects who took chlorpropamide were subjected to LC/MS, and the MH+ ion at m/z 293 and the [M + Na+] adduct ion at m/z 315 were consistent with 2-OH-chlorpropamide structure (Figure 1). LC/MS was carried out by coupling an Agilent 1100 series HPLC system (Agilent, Palo Alto, CA, USA) to an API 3000 triple quadrupole tandem mass spectrometer equipped with a Turbo Ionspray ionization source (Applied Biosystems, Foster City, CA, USA). The mobile phase was acetonitrile/water (2/8, v/v) with 0.1% formic acid at a flow rate of 0.2 ml min−1. The source temperature and ionspray voltage were kept at 375°C and 5.5 kV, respectively. By this analysis, the 2-OH-chlropropamide peak was found to appear at 3.9 min in the HPLC system used in this study.
Figure 1.
(a) Representative high-performance liquid chromatography elution profile of chloroform extracts of human urine collected after a single 250-mg oral dose of chlorpropamide. (b) Electrospray mass spectrum (positive-ion mode) and structure of 2-OH-chlorpropamide. Mass peaks with m/z 292.9 and 314.8 correspond to MH+ and [M + Na+] adduct ions, respectively. Experimental conditions were as described under Methods
In microsomal incubation studies, an internal standard (20 µl of 50 µm tolbutamide) was added to the supernatant fraction obtained by centrifugation of incubation mixtures, which was then directly injected into the HPLC system. All other methods for the assay of 2-OH-chlorpropamide in microsomal incubates were the same as those described above. Calibration standards of chorpropamide were prepared at six concentrations, ranging from 0.01 to 10 µm, in a blank microsomal incubation mixture. Similarly, two quality control (QC) samples were made with concentration levels at 0.2 and 5 µm. The interassay precision values for QC samples were less than 7.8%.
Pharmacokinetic and data analysis
The pharmacokinetic parameters for chlorpropamide were estimated by noncompartmental analysis, using WinNonlin® Vol. 2.0 (Pharsight Corporation). The elimination rate constant (k) was estimated from the least-square regression slope of terminal plasma concentrations. The total area under the plasma concentration–time curve (AUC) was calculated by the linear trapezoidal rule. The AUC from zero to infinity (AUC∞) was calculated as AUCinf = AUC + Ct/k, where Ct is the last plasma concentration measured. The half-life (t1/2) of chlorpropamide was calculated as 0.693/k. The oral clearance (CLF) was calculated as CLF = Dose/AUCinf. The renal clearance (CLR) was calculated as CLR =Ae0−24/AUC0−24, in which Ae0−24 represents the amount of chlorpropamide excreted in urine between 0 and 24 h. The nonrenal clearance (CLNR) was calculated as CLNR = CLF − CLR. The apparent volume of distribution (Vd/F) was calculated as Vd/F = Dose·AUMC/(AUCinf)2. The maximum plasma concentration (Cmax) and time to reach Cmax (Tmax) were estimated directly from the observed plasma concentration–time data. The metabolic ratio, defined as the chlorpropamide/2-OH-chlorpropamide molar ratio measured from the 24-h urine, was estimated in order to compare the extent of chlorpropamide 2-hydroxylation between subjects with different CYP2C9 and CYP2C19 genotypes.
All in vivo results are expressed as the mean ± SE. The unpaired t-test was used to analyse pharmacokinetic differences between groups with different CYP2C9 or CYP2C19 genotypes. SAS® software (V6.12; SAS Institute, Cary, NC, USA) was used for all statistical analyses and P < 0.05 was considered to be statistically significant.
Results
CYP isoforms involved in chlorpropamide 2-hydroxylation in vitro
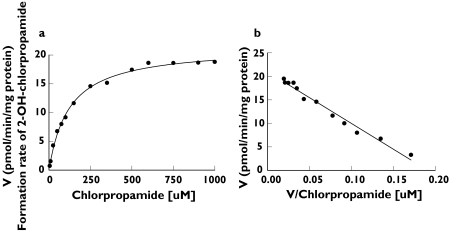
The formation of 2-OH-chlorpropamide from chlorpropamide (1–1000 µm) in human liver microsomes displayed typical Michaelis–Menten kinetics. When the data were fitted to several models, the best fit was obtained from the one-enzyme model, rather than from the two-enzyme model associated with the smaller value of AIC. The fit to the one-enzyme model is also shown in an Eadie–Hofstee plot (Figure 2). The mean kinetic parameters Km and Vmax were estimated to be 121.7 ± 19.9 µm and 16.1 ± 5.0 pmol min−1 mg−1 protein (Table 1).
Figure 2.
(a) Formation of 2-OH-chlorpropamide and (b) Eadie–Hofstee plot for formation of 2-OH-chlorpropamide from a 60-min incubation of a representative human liver microsome (HL-20) with chlorpropamide. Each point represents the mean of duplicate incubations
Table 1.
Apparent kinetic parameters for chlorpropamide 2-hydroxylation in human liver microsomes and human recombinant CYP isoforms
| Km (± SE)* | Vmax (± SE)* | CLint | |
|---|---|---|---|
| (µm) | (pmol min−1 mg−1) | (µl min−1 mg−1) | |
| Microsome preparation | |||
| HL-20 | 100.5 (± 21.8) | 11.8 (± 0.9) | 0.12 |
| HL-21 | 124.7 (± 8.8) | 21.6 (± 0.4) | 0.17 |
| HL-23 | 140.0 (± 8.2) | 15.0 (± 0.3) | 0.11 |
| Mean ± SD | 121.7 ± 19.9 | 16.1 ± 5.0 | 0.13 ± 0.04 |
| (µm) | (pmol min−1 pmol−1) | (µl min−1 nmol−1) | |
| Human recombinant CYP isoform | |||
| CYP2C9 | 249.6 (± 15.8) | 64.5 (± 1.1) | 0.26 |
| CYP2C19 | 584.9 (± 101.8) | 131.2 (± 9.1) | 0.22 |
SE of kinetic value calculated by fitting program.
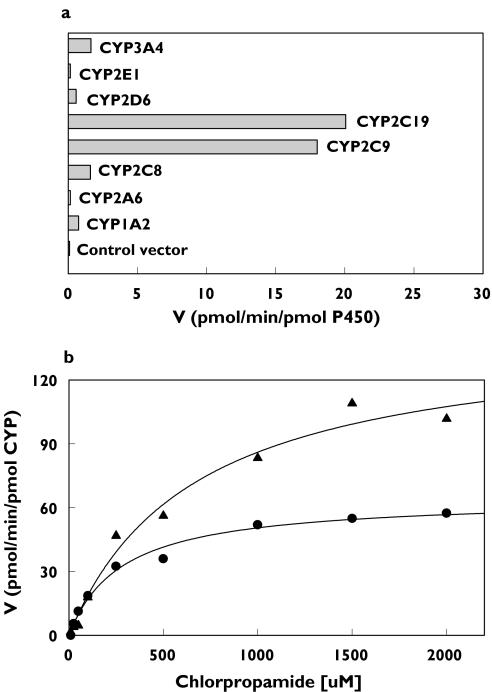
However, in the incubation study with eight human recombinant CYP isoforms, significant formation of 2-OH-chlorpropamide was observed for two CYP isoforms (CYP2C9 and CYP2C19) (Figure 3A). The estimated Km and Vmax values for CYP2C9 were one-half of those for CYP2C19, resulting in similar intrinsic clearances for the two CYP isoforms (0.26 µl min−1 nmol−1 for CYP2C9 and 0.22 µl min−1 nmol−1 for CYP2C19) (Figure 3B and Table 1). The human recombinant CYP1A2, CYP2A6, CYP2C8, CYP2D6, CYP2E1, and CYP3A4 isoforms produced only minimal or undetectable 2-OH-chlorpropamide at 100 µm chlorpropamide.
Figure 3.
(a) Formation of 2-OH-chlorpropamide from chlorpropamide (100 µm) by various human recombinant CYP isoforms. (b) Michaelis–Menten plots for the kinetics of chlorpropamide 2-hydroxylation obtained from human recombinant CYP2C9 (•) and CYP2C19 (▴) supersomes. Each point represents the mean of duplicate incubations
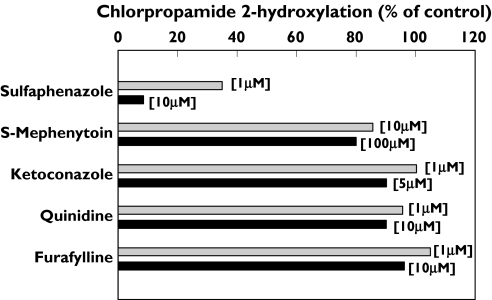
Sulfaphenazole, a selective inhibitor of CYP2C9, almost abolished the formation of 2-OH-chlorpropamide from chlorpropamide in the human microsomal incubation (Figure 4). S-Mephenytoin had only a minimal inhibitory effect at the highest concentration tested, and chlorpropamide 2-hydroxylation was not inhibited by treatment with any other known selective inhibitors of CYP isoforms, such as furafylline, quinidine, and ketoconazole.
Figure 4.
Inhibition of chlorpropamide 2-hydroxylation in human liver microsomes by CYP isoform-specific inhibitors including furafylline (CYP1A2), sulfaphenazole (CYP2C9), S-mephenytoin (CYP2C19), quinidine (CYP2D6), and ketoconazole (CYP3A4). Each bar represents the mean value obtained from three different liver microsomes (HL-20, -21, and -23)
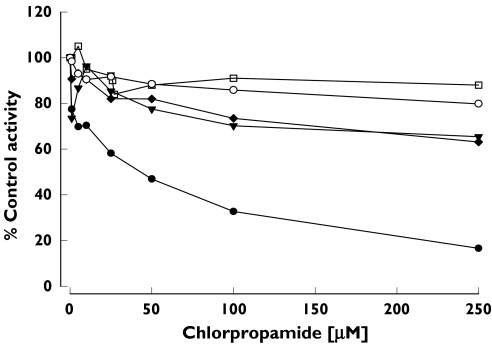
In this study, we also evaluated the inhibitory effect of chlorpropamide on five CYP isoform-specific metabolic pathway probes. Chlorpropamide inhibited CYP2C9-catalysed tolbutamide 4-hydroxylation with an estimated IC50 of 37.4 ± 19.2 µm; this activity was decreased to 30% of control activity in the presence of 100 µm chlorpropamide (Figure 5). In contrast, probe reactions representing other CYP isoform-specific activities were not significantly inhibited by chlorpropamide at concentrations up to 250 µm.
Figure 5.
Effect of chlorpropamide on phenacetin O-deethylation for CYP1A2 (○, acetaminophen), tolbutamide 4-hydroxylation for CYP2C9 (•, 4-hydroxytolbutamide), S-mephenytoin 4′-hydroxylation for CYP2C19 (▾, 4′-hydroxymephenytoin), dextromethorphan O-demethylation for CYP2D6 (□, dextrorphan), and midazolam 1-hydroxylation for CYP3A4 (♦, 1-hydroxymidazolam). Each data point represents the mean value obtained from three different liver microsomes (HL-20, -21, and -23)
Disposition of chlorpropamide in vivo in relation to CYP2C9 and CYP2C19 genetic polymorphism
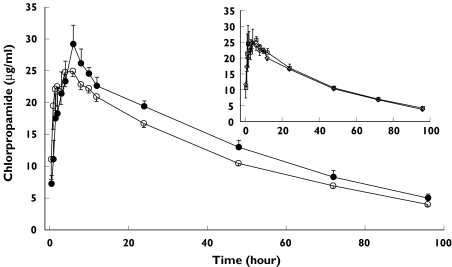
After a single oral dose of 250 mg chlorpropamide, mean plasma chlorpropamide concentrations in subjects heterozygous for the mutant CYP2C9*3 allele tended to be higher than those in subjects with the wild-type CYP2C9*1 allele (Figure 6). However, no difference in plasma chlorpropamide concentrations was found for the CYP2C19 EM vs. CYP2C19 PM genotypes (Figure 6, inset).
Figure 6.
Mean plasma chlorpropamide concentration–time profile after a single oral administration of chlorpropamide (250 mg) in 13 healthy subjects with CYP2C9*1/*3:CYP2C19 extensive metabolizer (EM) or poor metabolizer (PM) genotypes (○) vs. eight subjects with CYP2C9*1/*1:CYP2C19 EM genotypes (•). (Inset) Mean plasma chlorpropamide concentration–time profile for six subjects with CYP2C9*1/*1:CYP2C19 EM genotypes (□) vs. seven subjects with CYP2C9*1/*1:CYP2C19 PM genotypes (◊). Each bar indicates the mean ± SE for a specific CYP2C9 and CYP2C19 genotype
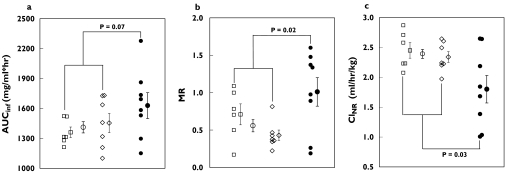
Because significant individual variation in pharmacokinetic parameters was observed, especially in subjects with the CYP2C9*1/*3 allele, parameter values for subjects with different CYP2C9 genotypes overlapped (Figure 7). The estimated nonrenal clearance was significantly decreased in the eight CYP2C9*1/*3 subjects, compared with the 13 homozygous CYP2C9*1/*1 subjects [1.8 ± 0.2 vs. 2.4 ± 0.1 ml h−1 kg−1, respectively, P < 0.05; 95% confidence interval (CI) on the difference 0.2, 1.0] (Table 2). The metabolic ratio of chlorpropamide/2-OH-chlorpropamide in urine for 24 h was also significantly higher in the CYP2C9*1/*3 subjects than in the CYP2C9*1/*1 subjects (1.01 ± 0.19 vs. 0.56 ± 0.08, respectively, P < 0.05; 95% CI on the difference −0.9, −0.1) (Table 2). Although the AUCinf of chlorpropamide was higher in CYP2C9*1/*3 subjects than in CYP2C9*1/*1 subjects, the difference was not statistically significant in this study (*1/*1 vs.*1/*3: 1411.0 ± 56.6 vs. 1628.0 ± 129.6 µg ml−1 h−1, P = 0.07; 95% CI on the difference −476.1, 42.0). Other pharmacokinetic parameters showed no remarkable differences between the two genotype groups of CYP2C9. There was no significant difference in any parameter between groups with CYP2C19 EM and CYP2C19 PM genotypes (Table 2).
Figure 7.
Scatter plots of (a) AUCinf, (b) metabolic ratio (MR = molar amount of chlorpropamide/2-hydroxychlopropamide in urine for 24 h after dosing), and (c) nonrenal clearance (CLNR) estimated from a single, 250-mg oral dose of chlorpropamide in subjects with different CYP2C9 and CYP2C19 genotypes. □, CYP2C9*1/*1:CYP2C19 extensive metabolizer (EM) genotype; s, CYP2C9*1/*1:CYP2C19 poor metabolizer (PM) genotype; ••, mean value for subjects with CYP2C9*1/*1 genotypes; and •, CYP2C9*1/*3:CYP2C19 EM genotype. Each symbol with bar indicates the mean ± SE of a group. The P-value was determined by unpaired t-test between subjects with different CYP2C9 genotypes and was considered to be significant if P < 0.05
Table 2.
Comparison of pharmacokinetic parameters of chlorpropamide after single oral administration of 250 mg chlorpropamide to subjects with different CYP2C9 and CYP2C19 genotypes
| CYP2C19 | CYP2C9 | |||
|---|---|---|---|---|
| EMa (n = 6) | PMb (n = 7) | *1/*1 (n = 13) | *1/*3 (n = 8) | |
| Cmax (µg ml−1) | 27.9 ± 1.6 | 31.0 ± 2.6 | 29.6 ± 1.6 | 31.7 ± 1.8 |
| t1/2 (h) | 35.8 ± 2.4 | 38.9 ± 2.2 | 37.5 ± 1.6 | 39.4 ± 4.2 |
| CL/F (ml h−1 kg−1) | 2.9 ± 0.1 | 2.7 ± 0.1 | 2.8 ± 0.1 | 2.7 ± 0.3 |
| CLNR (ml h−1 kg−1)c | 2.5 ± 0.1 | 2.3 ± 0.1 | 2.4 ± 0.1 | 1.8 ± 0.2* |
| Vd/F (ml kg−1) | 146.4 ± 3.7 | 151.6 ± 7.2 | 149.2 ± 4.2 | 141.8 ± 11.0 |
| AUCinf (µg ml−1 h−1) | 1360.7 ± 53.0 | 1454.2 ± 96.6 | 1411.0 ± 56.6 | 1628.0 ± 129.6 |
| Metabolic ratiod | 0.71 ± 0.14 | 0.43 ± 0.07 | 0.56 ± 0.08 | 1.01 ± 0.19* |
Each value indicates mean ± SE.
EM of CYP2C19; CYP2C19*1/*1.
PM of CYP2C19; CYP2C19*2/*2, *3/*3, or *2/*3.
CLNR, Nonrenal clearance.
P < 0.05; unpaired t-test between subjects with different CYP2C9 genotypes.
Metabolic ratio = chlorpropamide/2-OH-chlorpropamide molar amount in urine for 24 h after dosing.
Discussion
In this study, we demonstrated that chlorpropamide 2-hydroxylation is catalysed by the two CYP isoforms CYP2C9 and CYP2C19, and that the CYP2C9 isoform is responsible for the majority of this 2-hydroxylation activity in human liver microsomes. Experiments with human recombinant CYP isoforms clearly showed that CYP2C9 and CYP2C19 enzymes catalysed 2-hydroxylation of chlorpropamide with similar intrinsic clearance. In addition, specific inhibitors of CYP2C9 and CYP2C19 both decreased the formation of 2-hydroxychlorpropamide. However, the extent of inhibition by S-mephenytoin, a CYP2C19-specific inhibitor [15], was much smaller than that by sulfaphenazole, a potent CYP2C9-specific inhibitor [22]. Nor did microsomal incubation studies result in clear evidence that two kinds of CYP isoforms are involved in chlorpropamide 2-hydroxylation. The incubation data was best fitted to a one-enzyme model rather than to a two-enzyme model, and visual inspection of an Eadie–Hofstee plot of the data also supported the involvement of only one enzyme in this enzymatic reaction. These results imply that a kind of enzyme, CYP2C9, plays a major role in chlorpropamide hydroxylation in human liver.
Similar results were observed for the 4-methylhydroxylation of tolbutamide, a well-characterized CYP2C9-specific substrate. This compound has a chemical structure very similar to that of chlorpropamide, and it has been used extensively as a selective probe for CYP2C9 catalytic activity [23]. Formation of 4-methylhydroxy-tolbutamide has long been known to be principally determined by CYP2C9 activity both in vitro [24–26] and in vivo [27, 28]. However, several in vitro experiments demonstrated that, in addition to CYP2C9, CYP2C19 is also involved in tolbutamide 4-methylhydroxylation: Venkatakrishnan et al. [29] and Desta et al. [30] reported that recombinant CYP2C19 catalyses this reaction with intrinsic clearance, similar to that of recombinant CYP2C9. According to Wester et al. [31], CYP2C19 is responsible for 14–22% of tolbutamide metabolism, as determined by an in vitro inhibition study using a mono-specific anti-CYP2C9 antibody in human liver microsomes with and without CYP2C19. In this study, we could not quantify the contribution of CYP2C19 to chlorpropamide 2-hydroxylation, but CYP2C19 appeared to have a minor role in the formation of 2-OH-chlorpropamide, and CYP2C9 appeared to be the major CYP isoform catalysing this pathway, as it does in the 4-methylhydroxylation of tolbutamide [31, 32]. In human liver tissue, CYP2C9 is present at a much higher level than CYP2C19 [33], with molar ratios of CYP2C9 to CYP2C19 reported as 17 : 1 [34] or 13 : 1 [32]. This suggests that the lower level of human hepatic CYP2C19 expression than that of CYP2C9 may preclude its essential role for catalysing hepatic tolbutamide and chlorpropamide metabolism.
Further evidence that CYP2C9 is the major CYP isoform catalysing chlorpropamide 2-hydroxylation in human was also obtained through in vivo studies of the effects of CYP2C9 and CYP2C19 genotypes on chlorpropamide pharmacokinetics. In this study, the disposition of chlorpropamide was influenced by CYP2C9 genotype, but not by CYP2C19 genotype. The nonrenal clearance of chlorpropamide was significantly lower in subjects with the CYPC2C9*1/*3 variant than in subjects with the wild-type CYP2C9*1/*1. In addition, the metabolic ratio of urinary chlorpropamide/2-OH-chlropropamide was about twice as high in CYP2C9*1/*3 subjects, compared with CYP2C9*1/*1 subjects. In contrast, no remarkable differences in any pharmacokinetic parameters were found to correlate with CYP2C19 EM vs. PM genotype. These findings suggest that individual variations in chlorpropamide disposition are at least partially due to differences in CYP2C9 genotype, whereas CYP2C19 genotype is not a significant factor. Overall, these results are quite similar to those reported by Shon et al. [35] and Kirchheiner et al. [36], who studied tolbutamide disposition in vivo in relation to CYP2C9 and CYP2C19 genetic polymorphisms. In their studies, 4-methylhydroxylation of tolbutamide was found to be significantly influenced by CYP2C9 genotype but not appreciably influenced by CYP2C19 genotype.
Despite screening more than 1000 Korean subjects, we were unable to find any Korean subjects with a homozygous CYP2C9*3/*3 genotype. This circumstance is not surprising in light of the extremely low frequency of the CYP2C9*3 allele, which is estimated by the Hardy–Weinberg Law to be present in only one in 7700 Korean subjects (Yoon et al. [12]). In a study by Kirchheiner et al. [36], the mean oral clearance of tolbutamide observed for homozygous CYP2C9*3/*3 subjects was only 27% of that observed for CYP2C9*1/*3 subjects (150 vs. 560 ml h−1). Therefore, the CYP2C9*3/*3 mutant genotype would be expected to hinder chlorpropamide disposition dramatically in these subjects.
It has been well-known that genetic polymorphism of CYP2C9 markedly affects the disposition of sulfonylureas, such as tolbutamide, glyburide, glimepiride, and glipizide and especially the CYP2C9*3 allele is responsible for poor metabolizer of those. However, in our clinical study, the difference in chlorpropamide disposition for the CYP2C9*1/*3 vs. CYP2C9*1/*1 genotypes was minor compared with the difference in the pharmacokinetic disposition of those sulfonylurea drugs for the two genotypes [35–38]. Additionally, pharmacokinetic characteristics varied even among subjects with the same CYP2C9 and CYP2C19 genotype. The pharmacokinetic characteristics of chlorpropamide may be partially responsible for this observation, since renal excretion is highly influenced by urinary pH and the percent of a chlorpropamide dose excreted into urine is reported to vary from 6 to 60%, depending on the urine pH [5, 39]. With urine acidification of subjects, indicating that renal excretion would not contribute to interindividual diversity of chlorpropamide kinetics, the differences in plasma concentration and pharmacokinetic parameters between groups of CYP2C9 genotype seem to be more evident.
In a previous study of the CYP isoforms involved in chlorpropamide disposition, Kallio et al. [39] proposed the possible involvement of CYP2D6 based on the relationship between chlorpropamide disposition and CYP2D6 genetic polymorphisms. They found that debrisoquine oxidation phenotype correlated with the metabolic ratio of chlorpropamide/hydroxy metabolite of chlorpropamide in urine. In contrast, our in vitro studies using recombinant CYP2D6 and with quinidine, a selective CYP2D6 inhibitor [19], clearly showed that CYP2D6 does not mediate 2-hydroxylation of chlorpropamide. However, CYP2D6 may be involved in the formation of other metabolites, such as 3-hydroxychlorpropamide, p-chlorobenzene-sulphonylurea, and p-chlorobenzene-sulphonamidem [3, 4].
In this study, we also assessed chlorpropamide for its potential as an inhibitor of CYP isoforms. As expected, this compound was a potent inhibitor of tolbutamide 4-hydroxylation, with an estimated mean IC50 of 37.4 µm in human liver microsomes. No significant inhibition of reactions catalysed by other CYP isoforms, such as CYP1A2-catalysed phenacetin O-deethylation, CYP2C19-catalysed S-mephenytoin 4-hydroxylation, CYP2D6-catalysed dextromethorphan N-demethylation, and CYP3A-catalysed midazolam 1-hydroxylation, was observed in vitro. At the plasma concentrations (25–250 µm) obtained from usual therapeutic doses, chlorpropamide may have significant potential for interaction with other coadministered CYP2C9 substrate therapeutics such as phenytoin, S-warfarin, losartan, torasemide, celecoxib, and fluvastatin, and many nonsteroidal anti-inflammatory drugs including diclofenac, flurbiprofen, and ibuprofen [40]. Further evaluation of the potential interactions of chlorpropamide in vivo are clearly needed.
In conclusion, these in vitro microsomal incubation experiments and in vivo clinical trial demonstrated that the sulphonylurea oral hypoglycaemic agent chlorpropamide is a substrate of CYP2C9 and has a potential for being a CYP2C9 inhibitor. CYP2C9 appears to be the major CYP isoform catalysing chlorpropamide 2-hydroxylation, and genetic polymorphism of CYP2C9 has a significant influence on the disposition of chlorpropamide in vivo. Although CYP2C19 is involved in chlorpropamide 2-hydroxylation in vitro, the extent of its involvement appears to be low enough for CYP2C19 genetic polymorphism not to contribute to individual variations in chlorpropamide disposition.
Acknowledgments
This study was supported by a grant from the Ministry of Science and Technology of Korea (National Research Laboratory Program)
References
- 1.Sarior G, Melander A, Schersten B, Wahlin-Boll E. Comparative single-dose kinetics and effects of four sulfonylureas in healthy volunteers. Acta Med Scand. 1980;208:301–307. doi: 10.1111/j.0954-6820.1980.tb01199.x. [DOI] [PubMed] [Google Scholar]
- 2.Huupponen R, Lammintausta R. Chlorpropamide bioavailability and pharmacokinetics. Int J Clin Pharmacol. 1981;19:331–333. [PubMed] [Google Scholar]
- 3.Brotherton PM, Grieveson P, McMartin C. A study of the metabolic fate of chlorpropamide in man. Clin Pharmacol Ther. 1969;10:505–514. doi: 10.1002/cpt1969104505. [DOI] [PubMed] [Google Scholar]
- 4.Taylor JA. Pharmacokinetics and biotransformation of chlorpropamide in man. Clin Pharmacol Ther. 1972;13:710–718. doi: 10.1002/cpt1972135part1710. [DOI] [PubMed] [Google Scholar]
- 5.Campbell RK, Hansten PD. Metabolism of chlorpropamide. Diabetes Care. 1981;4:332. doi: 10.2337/diacare.4.2.332. [DOI] [PubMed] [Google Scholar]
- 6.Shorr RI, Ray WA, Daugherty JR, Griffin MR. Individual sulfonylureas and serious hypoglycemia in older people. J Am Geriatr Soc. 1996;44:751–755. doi: 10.1111/j.1532-5415.1996.tb03729.x. [DOI] [PubMed] [Google Scholar]
- 7.Harrower AD. Comparative tolerability of sulphonylureas in diabetes mellitus. Drug Saf. 2000;22:313–320. doi: 10.2165/00002018-200022040-00004. [DOI] [PubMed] [Google Scholar]
- 8.Bergman U, Chritenson I, Jansson B, Wiholm B-E, Ostman J. Wide variation in serum chlorpropamide concentration in outpatients. Eur J Clin Pharmacol. 1980;18:165–169. doi: 10.1007/BF00561585. [DOI] [PubMed] [Google Scholar]
- 9.Huupponen R, Viikari J, Saarimaa H. Chlorpropamide and glibenclamide serum concentrations in hospitalized patients. Ann Clin Res. 1982;14:119–122. [PubMed] [Google Scholar]
- 10.Self TH, Morris T. Interaction of rifampin and chlorpropamide. Chest. 1980;77:800–801. doi: 10.1378/chest.77.6.800. [DOI] [PubMed] [Google Scholar]
- 11.Nasu K, Kubota T, Ishizaki T. Genetic analysis of CYP2C9 polymorphism in a Japanese population. Pharmacogenetics. 1997;7:405–409. doi: 10.1097/00008571-199710000-00011. [DOI] [PubMed] [Google Scholar]
- 12.Yoon YR, Shon JH, Kim MK, et al. Frequency of cytochrome P450 CYP2C9 mutant alleles in a Korean population. Br J Clin Pharmacol. 2000;51:277–280. doi: 10.1046/j.1365-2125.2001.00340.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shin JG, Park JY, Kim MJ, et al. Inhibitory effects of tricyclic antidepressants (TCAs) on human cytochrome P450 enzymes in vitro: mechanism of drug interaction between TCAs and phenytoin. Drug Metab Dispos. 2002;30:1102–1107. doi: 10.1124/dmd.30.10.1102. [DOI] [PubMed] [Google Scholar]
- 14.Sesardic D, Boobis AR, Murray BP, et al. Furafylline is a potent and selective inhibitor of cytochrome P450IA2 in man. Br J Clin Pharmacol. 1990;29:651–663. doi: 10.1111/j.1365-2125.1990.tb03686.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim KA, Kim MJ, Park JY, et al. Stereoselective metabolism of lansoprazole by human liver cytochrome P450 enzymes. Drug Metab Dispos. 2003;31:1227–1234. doi: 10.1124/dmd.31.10.1227. [DOI] [PubMed] [Google Scholar]
- 16.Tassaneeyakul W, Birkett DJ, Veronese ME, et al. Specificity of substrate and inhibitor probes for human cytochromes P450 1A1 and 1A2. J Pharmacol Exp Ther. 1993;265:401–407. [PubMed] [Google Scholar]
- 17.Knodell RG, Hall SD, Wilkinson GR, Guengerich FP. Hepatic metabolism of tolbutamide: characterization of the form of cytochrome P-450 involved in methyl hydroxylation and relationship to in vivo disposition. J Pharmacol Exp Ther. 1987;241:1112–1119. [PubMed] [Google Scholar]
- 18.Wrighton SA, Stevens JC. The human hepatic cytochromes P450 involved in drug metabolism. Crit Rev Toxicol. 1992;22:1–21. doi: 10.3109/10408449209145319. [DOI] [PubMed] [Google Scholar]
- 19.Broly F, Libersa C, Lhermitte M, Bechtel P, Dupuis B. Effect of quinidine on the dextromethorphan O-demethylase activity of microsomal fractions from human liver. Br J Clin Pharmacol. 1989;28:29–36. doi: 10.1111/j.1365-2125.1989.tb03502.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thummel KE, Shen DD, Podoll TD, et al. Use of midazolam as a human cytochrome P450 3A probe. I. In vitro–in vivo correlations in liver transplant patients. J Pharmacol Exp Ther. 1994;271:549–556. [PubMed] [Google Scholar]
- 21.Csillag K, Vereczkey L, Gachalyi B. Simple high-performance liquid chromatographic method for the determination of tolbutamide and its metabolites in human plasma and urine using photodiode-array detection. J Chromatogr. 1989;490:355–363. doi: 10.1016/s0378-4347(00)82793-0. [DOI] [PubMed] [Google Scholar]
- 22.Baldwin SJ, Bloomer JC, Smith GJ, Ayrton AD, Clarke SE, Chenery RJ. Ketoconazole and sulphaphenazole as the respective selective inhibitors of P4503A and 2C9. Xenobiotica. 1995;25:261–270. doi: 10.3109/00498259509061850. [DOI] [PubMed] [Google Scholar]
- 23.Miners JO, Birkett DJ. The use of tolbutamide as a substrate probe for human hepatic cytochrome P450 2C9. Meth Enzymol. 1996;272:139–145. doi: 10.1016/s0076-6879(96)72017-7. [DOI] [PubMed] [Google Scholar]
- 24.Miners JO, Smith KJ, Robson RA, McManus ME, Veronese ME, Birkett DJ. Tolbutamide hydroxylation by human liver microsomes. Kinetic characterization and relationship to other cytochrome P-450 dependent xenobiotic oxidations. Biochem Pharmacol. 1988;37:1137–1144. doi: 10.1016/0006-2952(88)90522-9. [DOI] [PubMed] [Google Scholar]
- 25.Relling MV, Aoyama T, Gonzalez FJ, Meyer UA. Tolbutamide and mephenytoin hydroxylation by human cytochrome P450s in the CYP2C subfamily. J Pharmacol Exp Ther. 1990;252:442–447. [PubMed] [Google Scholar]
- 26.Srivastava PK, Yun CH, Beaune PH, Ged C, Guengerich FP. Separation of human liver microsomal tolbutamide hydroxylase and (S)-mephenytoin 4′-hydroxylase cytochrome P-450 enzymes. Mol Pharmacol. 1991;40:69–79. [PubMed] [Google Scholar]
- 27.Back DJ, Tjia J, Monig H, Ohnhaus EE, Park BK. Selective inhibition of drug oxidation after simultaneous administration of two probe drugs, antipyrine and tolbutamide. Eur J Clin Pharmacol. 1988;34:157–163. doi: 10.1007/BF00614553. [DOI] [PubMed] [Google Scholar]
- 28.Veronese ME, Miners JO, Randles D, Gregov D, Birkett DJ. Validation of the tolbutamide metabolic ratio for population screening with use of sulfaphenazole to produce model phenotypic poor metabolizers. Clin Pharmacol Ther. 1990;47:403–411. doi: 10.1038/clpt.1990.46. [DOI] [PubMed] [Google Scholar]
- 29.Venkatakrishnan K, von Moltke LL, Greenblatt DJ. Relative quantities of catalytically active CYP 2C9 and 2C19 in human liver microsomes: application of the relative activity factor approach. J Pharm Sci. 1998;87:845–853. doi: 10.1021/js970435t. [DOI] [PubMed] [Google Scholar]
- 30.Desta Z, Soukhova NV, Shin JG, Flockhart DA. Involvement of cytochrome P450 2C19 in methylhydroxylation of tolbutamide in vitro. Clin Pharmacol Ther. 2000;67:167. [Google Scholar]
- 31.Wester MR, Lasker JM, Johnson EF, Raucy JL. CYP2C19 participates in tolbutamide hydroxylation by human liver microsomes. Drug Metab Dispos. 2000;28:354–359. [PubMed] [Google Scholar]
- 32.Lasker JM, Wester MR, Aramsombatdee E, Raucy JL. Characterization of CYP2C19 and CYP2C9 from human liver: respective roles in microsomal tolbutamide, S-mephenytoin, and omeprazole hydroxylations. Arch Biochem Biophys. 1998;353:16–28. doi: 10.1006/abbi.1998.0615. [DOI] [PubMed] [Google Scholar]
- 33.Shimada Y, Yamazaki H, Mimura M, Inui Y, Guengerich FP. Interindividual variations in human liver cytochrome P450 enzymes involved in the oxidation of drugs, carcinogens and toxic chemicals: studies with liver microsomes of 30 Japanese and 30 Caucasians. J Pharmacol Exp Ther. 1994;270:414–423. [PubMed] [Google Scholar]
- 34.Inoue K, Yamazaki H, Imiya K, Akasaka S, Guengerich FP, Shimada T. Relationship between CYP2C9 and 2C19 genotypes and tolbutamide methyl hydroxylation and S-mephenytoin 4′-hydroxylation activities in livers of Japanese and Caucasian populations. Pharmacogenetics. 1997;7:103–113. doi: 10.1097/00008571-199704000-00003. [DOI] [PubMed] [Google Scholar]
- 35.Shon JH, Yoon YR, Kim KA, et al. Effects of CYP2C19 and CYP2C9 genetic polymorphisms on the disposition of and blood glucose lowering response to tolbutamide in humans. Pharmacogenetics. 2002;12:111–119. doi: 10.1097/00008571-200203000-00005. [DOI] [PubMed] [Google Scholar]
- 36.Kirchheiner J, Bauer S, Meineke I, et al. Impact of CYP2C9 and CYP2C19 genetic polymorphisms on tolbutamide kinetics and the insulin and glucose response in healthy volunteers. Pharmacogenetics. 2002;12:101–109. doi: 10.1097/00008571-200203000-00004. [DOI] [PubMed] [Google Scholar]
- 37.Kidd RS, Straughn AB, Meyer MC, Blaisdell J, Goldstein JA, Dalton JT. Pharmacokinetics of chlorpheniramine, phenytoin, glipizide and nifedipine in an individual homozygous for the CYP2C9*3 allele. Pharmacogenetics. 1999;9:71–80. doi: 10.1097/00008571-199902000-00010. [DOI] [PubMed] [Google Scholar]
- 38.Niemi M, Cascorbi I, Timm R, Kroemer HK, Neuvonen PJ, Kivisto KT. Glyburide and glimepiride pharmacokinetics in subjects with different CYP2C9 genotypes. Clin Pharmacol Ther. 2002;72:326–332. doi: 10.1067/mcp.2002.127495. [DOI] [PubMed] [Google Scholar]
- 39.Kallio J, Huupponen R, Pyykko K. The relationship between debrisoquine oxidation phenotype and the pharmacokinetics of chlorpropamide. Eur J Clin Pharmacol. 1990;39:93–95. doi: 10.1007/BF02657068. [DOI] [PubMed] [Google Scholar]
- 40.Miners JO, Birkett DJ. Cytochrome P4502C9: an enzyme of major importance in human drug metabolism. Br J Clin Pharmacol. 1998;45:525–538. doi: 10.1046/j.1365-2125.1998.00721.x. [DOI] [PMC free article] [PubMed] [Google Scholar]