Abstract
Aim
The aim of this study was to quantify the mRNA expression of three cytochromes P450 (CYP) and P-glycoprotein (P-gp) in the human gastrointestinal (GI) tract.
Method
Biopsies were obtained from gastric, duodenal, colonic and rectal mucosa during routine gastro-colonoscopy in 27 patients. The biopsies were snap-frozen in liquid nitrogen. Real-time reverse transcriptase-polymerase chain reaction (RT-PCR) was used for the quantitative analyses of mRNA expressed by the CYP2E1, CYP3A4 and CYP3A5 genes, and the MDR1 gene coding for P-gp protein. The mRNA expression of b-actin was used as an internal standard for comparisons between samples.
Results
All CYP genes were expressed at all locations throughout the GI tract, although all showed substantial interindividual variation. CYP2E1 had the highest expression at all locations (P < 0.05 to P < 0.0001), except in the right colon. CYP3A4 and CYP3A5 had their highest mRNA expression in the duodenum (P < 0.001 and P < 0.000 001, respectively) and CYP2E1 in the stomach (P < 0.01). MDR1 mRNA concentrations increased along the GI tract with the highest expression being in the left colon (P < 0.000001).
Conclusion
Multiple sampling within the same individual enabled us to study the intraindividual variation in expression of CYP and MDR1 genes along the GI tract. We find that CYP2E1 mRNA expression is higher than that of the other CYPs. CYP3A expression is highest in the duodenum and that of MDR1 increases from stomach and duodenum to colon.
Keywords: Cytochrome P450, Drug metabolism, GI-tract
Introduction
Over the past decade the important role of the gastrointestinal (GI) tract and especially the small intestine in the presystemic metabolism of orally administered drugs has become apparent [1–3]. It has been shown that the metabolism of some orally administered drugs is unaffected in patients with impaired liver function [4]. Kolars et al. [5] showed that metabolites of cyclosporine were detected in portal venous blood during the anhepatic phase of liver transplantation when the drug was administered directly to the small bowel. Furthermore, Paine et al. [6] have shown that midazolam is metabolized in the gut wall.
The cytochromes P450 (CYP) enzymes belong to a superfamily of membrane-bound haeme-containing proteins responsible for the metabolism of several endogenous compounds such as steroids and fatty acids, as well as drugs and other xenobiotics [7, 8]. Three CYP gene families are predominantly involved in drug metabolism, namely CYP1, CYP2 and CYP3, many members of which are induced or inhibited by a range of compounds [9].
Another determinant of the oral bioavailability of drugs is P-glycoprotein (P-gp) which is encoded for by the multidrug resistance gene (MDR1). P-gp is a membrane-bound transporter protein, which was first identified as being responsible for the development of multidrug resistance [10]. P-gp acts as a cellular efflux pump in an ATP-dependent process [11]. P-gp is present in mucosal cells of the GI tract, in the biliary canaliculi of the liver, and in the brush border of the proximal tubules of the kidney, where it contributes to the excretion of substances into the gut, bile and urine, respectively [12]. Schwab et al. [13] suggested that one of the polymorphisms of the MDR1 gene, C3435T, which leads to decreased P-gp expression, is associated with increased susceptibility to ulcerative colitis. Furthermore, it has been shown that mdr1a knockout mice develop a form of this disease, which can be prevented by antibiotics [14]. These findings suggest that P-gp plays a protective role against intestinal bacteria and toxins.
The use of molecular techniques and especially the real-time reverse transcription-polymerase chain reaction (RT-PCR) makes it possible to detect and quantify RNA in very small samples [15].
The aim of this study was to study the expression of CYP2E1, CYP3A4, CYP3A5 and MDR1 mRNA along the GI tract within the same individual.
Patients and methods
Patients
Patients referred to the endoscopy unit, University Hospital of Uppsala, were asked to participate in the study. Demographic and clinical features of the patients are shown in Table 1. The ethics committee of the University of Uppsala approved the project. After informed consent, biopsies from five different locations in the GI tract (stomach, duodenum, right colon, left colon and rectum) were collected in 27 patients together with biopsies for routine clinical investigation. Samples were obtained from macroscopically normal mucosa where possible, and snap-frozen in liquid nitrogen and stored at −70°C until preparation of mRNA.
Table 1. Clinical features of the patients.
| Patient | Age (years) | Sex | Smoking, cigarettes/day | Alcohol last week | Medication (most important) | Clinical diagnoses |
|---|---|---|---|---|---|---|
| 1 | 74 | M | No | 32 g | None | HP1-gastritis, duodenitis |
| 2 | 69 | M | No | 110 g | None | Anaemia HP-gastritis |
| 3 | 46 | F | No | No | Steroids, metronidazole | Gastric erosions, proctitis |
| 4 | 34 | M | No | 58 g | None | Anaemia—normal PAD2 |
| 5 | 36 | F | No | No | Steroids, mesalazine | IBD3—normal PAD |
| 6 | 30 | F | No | 82 g | None | Microscopic colitis |
| 7 | 37 | F | 20 | 45 g | Paracetamol, salicylic acid | Focal intestinal gastric metaplasia |
| 8 | 52 | M | 20–30 | 10 g | Omeprazole | Erosive gastritis, duodenitis |
| 9 | 24 | F | No | No | Sulphasalazine, citalopram cetirizin | Inactive proctitis |
| 10 | 58 | F | No | No | None | IBS4 |
| 11 | 82 | M | Pipe | No | None | Anaemia, gastric ulcer, HP-neg. |
| 12 | 59 | F | 1–2 | 20 g | Paracetamol, calcium | IBS |
| 13 | 36 | F | 5–10 | 148 g | None | IBS |
| 14 | 31 | F | No | 6 g | Oral contraceptives | Caeliac disease—villous atrophy |
| 15 | 41 | M | 10–20 | 40 g | Paracetamol, fluconazole, theophylline | Chronic gastritis, lymphocytic colitis |
| 16 | 47 | F | No | No | Metoprolol | Atrophic gastritis |
| 17 | 43 | F | No | 50 g | Promethazine | HP-gastritis, IBS |
| 18 | 25 | F | No | No | Salicylic acid, NSAID oral contraceptives | IBS |
| 19 | 79 | F | No | No | Digoxin, furosemide, clopidogrel, lanzoprazole | Collagenous colitis |
| 20 | 60 | M | No | 35 g | Metoprolol, felodipine | HP-gastritis, IBS |
| 21 | 20 | F | 20 | No | Oral contraceptives | Suspected Crohn's disease |
| 22 | 47 | F | 10–15 | No | Ventoline | Chronic gastritis, IBS |
| 23 | 30 | M | No | 80 g | None | IBS |
| 24 | 22 | M | No | No | None | IBS |
| 25 | 59 | M | No | 72 g | Data missing | IBS |
| 26 | 65 | M | No | Data missing | Allopurinol, digoxin, spironolactone | Caecal adenoma |
| 27 | 70 | M | No | No | Hydroxyurea, omeprazole | Data missing |
HP, Helicobacter pylori.
PAD, Peripheral arterial disease.
IBD, inflammatory bowel disease.
IBS, irritable bowel syndrome.
Purification of mRNA
mRNA was obtained from the samples (wet weight about 5 mg) using the Quickprep micro mRNA purification kit according to the instructions of the manufacturer (Amersham Biosciences, SE 75184, Uppsala, Sweden).
Subsequentely, mRNA was precipitated in 95% ethanol at −20°C for 1 h. The pellet obtained was then resuspended in 50 µl Rnase-free water. The amount of mRNA was determined by UV spectroscopy at 260 nm, with correction for background at 320 nm.
CDNA synthesis
Purified mRNA (50 ng) was used for cDNA synthesis using a First strand cDNA synthesis kit (Amersham Biosciences). Less than 50 ng mRNA was obtained from 15 out of the 135 samples, which was corrected for in the analysis of the data.
Real-time PCR
The cDNA was diluted 1 : 10 with Millipore filtered water before real-time RT-PCR. Primer pairs and fluorescent Taqman™ probes specific for CYP2E1, CYP3A4, CYP3A5 and MDR1 were used, the design of which was described earlier [16, 17]. The house-keeping b-actin gene was used as an internal control to enable comparison between samples. The b-actin probe was labelled with FAM (6-carboxyfluorescein) and purchased from Applied Biosystems.
Ten microlitres of the diluted cDNA mix, containing 3.3 ng mRNA, 300 n m each of the forward and reverse primers, 50 n m of the probe and a ready-made mastermix with heat-activated Taq polymerase, uracil-N-glycosylase (UNG) and nucleotides (Applied Biosystems) were used in the PCR reaction.
Real-time PCR was performed using an ABI PRISM™ SDS 7700 instrument (Perkin-Elmer 549 Albany Street, Boston, MA, USA). UNG inactivation for 2 min at 50°C and a Taq polymerase activation step at 94°C for 10 min were followed by 50 cycles (each at 95°C 15 s; 60°C 30 s). All samples were run in duplicates. At least two nontemplate controls were included in all PCR runs.
The cycles to threshold (Ct) value was determined during the period when the different PCR reactions were in the early logarithmic phase. The limit of detection of mRNA was defined in an earlier study [16] at a Ct value of 38.
The variation in Ct values for duplicates of all samples (n = 135) varied between 0 and 47% with a median of 1.7% and a mean of 3.6%. The mean coefficient of variation (CV) for one sample run on four occasions varied between 0.1 and 4.6% (mean = 2.8%) in five samples from one patient.
The standard curves for CYP2E1, CYP3A4, CYP3A5, and MDR1 cDNA were generated from three or four samples containing different known concentrations of cDNA. The standards were run at the same time as the samples.
It has previously been shown by Bowen et al. [18] that different enzymes give rise to the same standard curve in real-time RT-PCR, given that all reactions are optimized. Therefore, the concentration of b-actin mRNA was calculated from a calibration curve that was a composite of 10 different standard curves from four different CYP enzymes [16].
The amplicons of each reaction have previously been sequenced for confirmation of identity [17].
Statistical analysis
For comparison between paired samples the Friedman anova with Kendall coefficient of variation and Wilcoxon matched pair test were used, and for comparison between unpaired samples the Mann-Whitney U-test was used (Statistica®] (StatSoft Inc., 2300 East 14th Street, Tulsa, OK, USA).
Results
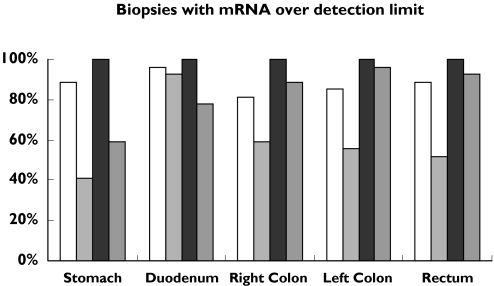
The percentage of biopsies with measurable amounts of each mRNA is shown in Figure 1. CYP2E1 mRNA expression was detected in 81–96% of the samples at the different sites along the GI tract. CYP3A5 mRNA was detected in all samples at all locations, whereas CYP3A4 mRNA detection varied between 41% and 96% and P-gp mRNA between 59% and 96%. However, the data from all samples, and also those with a Ct value above 38, are included in Figure 2, since they all expressed measurable concentrations of b-actin-specific mRNA, that is, a Ct value below 38. Because of the small size of the forceps used, biopsies contained only mucosal tissue. The majority of the patients were diagnosed with irritable bowel disease (Table 1) and as expected there was no inflammation in these biopsies. When all samples were considered, no difference in the median b-actin mRNA expression was observed in those that showed inflammation compared with those that did not.
Figure 1.
The number of biopsies containing mRNA above the detection limit (Ct over 38) expressed as a percent of all biopsies. CYP2E1 (□), CYP3A4 ( ), CYP3A5 (
), CYP3A5 ( ), P-gp (
), P-gp ( )
)
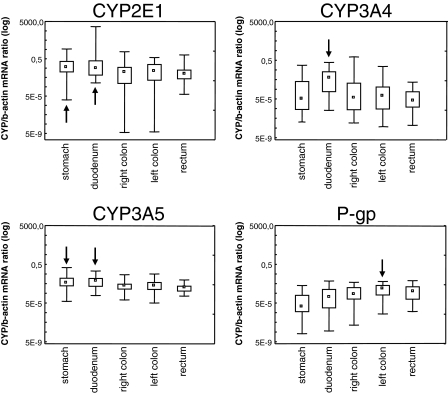
Figure 2.
Box plots of the CYP-specific/b-actin mRNA ratio for each enzyme at different locations of the human gastrointestinal tract (data from all samples included). Arrows indicate the location with the highest mRNA expression for each enzyme. Median (□), 25%–75% ( ), Min-Max (
), Min-Max ( )
)
The expression of CYP2E1 mRNA was highest at all sites (P < 0.05 to P < 0.0001) except in right colon, where it was equal to the expression of CYP3A5 mRNA (Figure 2 and Table 2). The highest expression of CYP2E1 mRNA was observed in the stomach and duodenum (P < 0.01), but there was a substantial interindividual variation (Kendall coefficient of concordance = 0.16). In the duodenum the expression of CYP2E1 mRNA exceeded that of CYP3A4, CYP3A5 and P-gp by approximately seven-, six- and 300-fold, respectively.
Table 2. Median (25–75% quartile) mRNA/â-actin ratios in different parts of the gastrointestinal tract.
| CYP2E1 | CYP3A4 | CYP3A5 | P-gp | |
|---|---|---|---|---|
| Stomach | 0.061 (0.017–0.29) | 0.000053 (0.0000040–0.0036) | 0.0058 (0.0026–0.020) | 0.00002 (0.0000060–0.00034) |
| Duodenum | 0.053 (0.0078–0.33) | 0.0072 (0.00026–0.036) | 0.0095 (0.0024–0.019) | 0.00019 (0.000014–0.0014) |
| Right colon | 0.017 (0.00010–0.06) | 0.00006 (0.0000030–0.0023) | 0.0028 (0.0012–0.0048) | 0.00042 (0.00011–0.0022) |
| Left colon | 0.023 (0.0022–0.13) | 0.0001 (0.0000040–0.00095) | 0.0029 (0.0011–0.0074) | 0.0015 (0.00030–0.0036) |
| Rectum | 0.012 (0.0032–0.034) | 0.000032 (0.0000060–0.00031) | 0.0018 (0.00076–0.0030) | 0.00073 (0.00011–0.0026) |
The expression of CYP3A4 mRNA was highest in the duodenum (P < 0.001), with no or low expression in the stomach and colon (Figure 2 and Table 2), and showed great interindividual variability (Kendall coefficient of concordance = 0.25).
The expression of CYP3A5 mRNA was highest in the duodenum and stomach (P < 0.000 001) and was less in the colon (Kendall coefficient of concordance = 0.34) (Figure 2 and Table 2).
To validate our finding that the expression of CYP3A5 mRNA was generally higher than that of CYP3A4, we analysed expression of these CYP genes in four different liver samples and one additional intestinal sample, obtained at surgery. In addition, another laboratory analysed the same livers and intestine using a similar method and found comparable results.
For P-gp, mRNA expression increased from stomach and duodenum to left colon (P < 0.000 001] a Kendall coefficient of concordance = 0.34) (Figure 2 and Table 2).
The interindividual variability in expression at each site in the GI tract was again very high. Each individual showed a similar expression pattern for the investigated CYP enzymes and P-gp throughout the GI tract.
We did not find any difference in mRNA expression between smokers (n = 8) and nonsmokers (n = 19), or between those who had an alcohol intake of less than 50 g compared with those who consumed more than that (n = 7) during the week before investigation, for any of the CYPs studied. Furthermore, there were no sex differences in expression (F = 15, M = 12), or differences between patients on regular medication (n = 16) compared with patients not taking medication (n = 11).
Discussion
To our knowledge, this is the first study to characterize the expression of CYP enzymes and P-gp along the length of the GI tract of the same individual in a large patient population. We determined the mRNA expression of three important CYP enzymes and the MDR1 gene at five different sites in 27 patients. The methods used were sensitive enough for several analyses of the very small amount of biopsy material (about 5 mg wet weight) that can be obtained during the endoscopic investigation. However, there was insufficient to study these CYPs or P-gp at a functional or protein level.
We found that CYP2E1 had the highest mRNA expression of the CYPs studied in all parts of the GI tract. Our findings are consistent with those of other investigators who have demonstrated the presence of CYP2E1 in the terminal ileum and colon of patients with irritable bowel disease, Crohn's disease and ulcerative colitis, respectively [19]. However, because of post-transcriptional regulation [20], different authors have failed to find a relationship between CYP2E1 mRNA and protein concentration [21, 22]. This suggests that there is no clear association between CYP2E1 mRNA expression and metabolic activity.
It has been claimed that there is a relationship between mRNA and protein expression for CYP3A4 and CYP1A2 [21, 22], although the data are conflicting, and our group has not been able to confirm this assertion (unpublished data).
Watkins and coworkers [23] have published data on the relative abundance of CYP3A4 in the intestinal mucosa, and protein expression and catalytic activity have been reported to decrease longitudinally along the small intestine [24]. We found higher CYP3A4 mRNA expression in the duodenum than in the stomach and the colon, which is in agreement with earlier reports based on human transplant donors [25, 26].
It has been shown that CYP3A4 in the gut is an important determinant of the bioavailability of cyclosporine, because approximately one-half to two-thirds of its metabolism occurs presystemically when the drug is orally administered [1, 27]. Furthermore, other drugs such as midazolam are metabolized to a varying degree by CYP enzymes in the gut [28]. It has also been shown that plasma concentrations of digoxin (with which two of the patients were treated) decrease with concomitant administration of rifampicin. This may be explained by an increase in intestinal P-gp during treatment with this inducing agent [3].
CYP3A5 mRNA expression was found in all the samples studied. Earlier studies [29, 30] demonstrated the presence of CYP3A5 mRNA in the small intestine, but CYP3A5 protein was not detected. In contrast, Lown et al. [31] found CYP3A5 protein in the small bowel of 70% of the patients studied, and they suggested that this enzyme may be expressed more in the intestine than in the liver. Kolars et al. [26] detected CYP3A5 and CYP3A4 mRNA in all regions of the GI tract using microsomes from a human organ donor.
The median concentration of CYP3A5 mRNA was highest in duodenal mucosa but the difference was not as pronounced as for CYP3A4 mRNA. Although CYP3A4 is considered to be the major CYP enzyme in the small intestine [26]. Lown et al. [32] described similar concentrations of CYP3A4 and CYP3A5 protein in enterocytes. In the present study we found similar expression of CYP3A4 and CYP3A5 mRNA in the duodenum. The expression of CYP3A5 protein is polymorphic, and individuals with at least one CYP3A5*1 allele express greater amounts of the enzyme. Based on our data, the expression of CYP3A5 mRNA is monomorphic, since the CYP3A5*3 allelic variant also expresses mRNA to a varying degree [33].
In the colon there was greater expression of CYP3A5 mRNA compared with that of CYP3A4, which is in agreement with findings of McKinnon et al. [25]. Other data also indicate that CYP3A5 is the predominant CYP3A enzyme in colonic mucosa [26, 32]. Furthermore, there appears to be greater expression of CYP3A5 in the stomach than CYP3A4 [26], which is consistent with the findings of the present study.
It seems that there is a relatively low CYP3A phenotypic variability among healthy adults compared with that seen in archived intestinal and liver tissue samples [34]. One possible explanation for this is the different rates of CYP3A4 and CYP3A5 degradation during the harvesting and cold storage of the tissues [35], which could explain the discrepancies between the relative proportion of CYP3A4 and CYP3A5 mRNA expression in our study and previous reports [29, 30]. It is unclear whether the rates of degradation of CYP3A4 and CYP3A5 are different in the GI tract compared with the liver.
Lown et al. [2] found that variation in intestinal P-gp accounted for about 30% of the variability in peak blood concentration of orally administered cyclosporine. We found a continuous increase in MDR1 expression along the GI tract to distal descending colon. Our findings are in agreement with those of Fojo et al. [36], although this report was based on colon samples from different patients and only one jejunum and rectum sample. Mouly and Paine [37] showed an increase in P-gp expression from proximal to distal small intestine. It has been suggested by Stephens et al. [38] that the ileum and distal colon are regions with high permeability for xenobiotics and that this is compensated for by enhanced expression of P-gp. The distribution pattern of MDR1 mRNA is opposite to that of CYP3A4 mRNA. These two proteins have overlapping substrate specificity and tissue distribution [24]. High expression of P-gp in the more distal part of the GI tract may be important in expelling xenobiotics not yet metabolized by CYP3A.
We found no influence of smoking, alcohol intake or sex on mRNA concentrations. However, the groups were quite small, and hence these findings must be interpreted with caution. There was no detectable relationship between the medication taken by patients and mRNA expression. However, the reported drug consumption was very heterogeneous among the patients.
We conclude that among the CYP enzymes investigated CYP2E1 mRNA expression was highest throughout the GI tract, with that of CYP3A being greatest in the duodenum. This finding is relevant because of the emerging role of the gut in the metabolism of a large number of clinically important drugs. MDR1 mRNA expression increased continuously from duodenum to colon, a finding that may be linked to a natural role for P-gp in the protection against xenobiotics produced by the intestinal microflora.
Acknowledgments
We thank Dr Roberto Canaparo for preparing some of the standards and probes, and all doctors and staff at the Endoscopy Unit at the University Hospital of Uppsala for help with collection of the samples. We also thank Professor Magnus Ingelman-Sundberg for help in the validation of the data. This work was supported by the Swedish Science Council (Medicine 04496), the Mary Eriksson foundation at Uppsala University, and grants from the research foundation, county of Västmanland and the Karolinska Institute.
References
- 1.Wu C-Y, Benet LZ, Hebert MF, Gupta SK, Rowland M, Gomez DY, Wacher VJ. Differentiation of absorption and first-pass gut and hepatic metabolism in humans: studies with cyclosporine. Clin Pharm Ther. 1995;58:492–7. doi: 10.1016/0009-9236(95)90168-X. [DOI] [PubMed] [Google Scholar]
- 2.Lown KS, Mayo RR, Leichtman AB, Hsaio H, Turgeon K, Schmiedlin-Ren P, Brown MB, Guo W, Rossi SJ, Benet LZ, Watkins PB. Role of intestinal P-glycoprotein (mdr1) in interpatient variation in the oral bioavailability of cyclosporine. Clin Pharm Ther. 1997;62:248–60. doi: 10.1016/S0009-9236(97)90027-8. [DOI] [PubMed] [Google Scholar]
- 3.Greiner B, Eichelbaum M, Fritz P, Kreichgauer H-P, Von Richter O, Zundler J, Kroemer HK. The role of intestinal P-glycoprotein in the interaction of digoxin and rifampin. J Clin Invest. 1999;104:147–53. doi: 10.1172/JCI6663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Robin DW, Lee M, Hasan SS, Wood AJ. Triazolam in cirrhosis: pharmacokinetics and pharmacodynamics. Clin Pharmacol Ther. 1993;54:630–7. doi: 10.1038/clpt.1993.200. [DOI] [PubMed] [Google Scholar]
- 5.Kolars JC, Awni WM, Merion RM, Watkins PB. First-pass metabolism by the gut. Lancet. 1991;338:1448–90. doi: 10.1016/0140-6736(91)92302-i. [DOI] [PubMed] [Google Scholar]
- 6.Paine MF, Shen DD, Kunze KL, Perkins JD, Marsh CL, McVicar JP, Barr DM, Gillies BS, Thummel KE. First-pass metabolism of midazolam by the human intestine. Clin Pharmacol Ther. 1996;60:14–24. doi: 10.1016/S0009-9236(96)90162-9. [DOI] [PubMed] [Google Scholar]
- 7.Gonzales FJ. Molecular genetics of the P-450 superfamily. Pharmac Ther. 1990;45:1–38. doi: 10.1016/0163-7258(90)90006-n. [DOI] [PubMed] [Google Scholar]
- 8.Nebert DW, Russel DW. Clinical importance of the cytochromes P450. Lancet. 2002;360:1155–62. doi: 10.1016/S0140-6736(02)11203-7. [DOI] [PubMed] [Google Scholar]
- 9.Wrighton SA, Vandenbranden M, Ring BJ. The human drug metabolising cytochromes P450. J Pharmacokin Biopharm. 1996;24:461–73. doi: 10.1007/BF02353474. [DOI] [PubMed] [Google Scholar]
- 10.Ueda K, Cornwell MM, Pastan I, Roninson IB, Ling V, Riordan JR. The mdr1 gene, responsible for multidrug-resistance, codes for P-glycoprotein. Biochem Biophys Res Commun. 1986;141:956–62. doi: 10.1016/s0006-291x(86)80136-x. [DOI] [PubMed] [Google Scholar]
- 11.Willingham MC, Cornwell MM, Cardarelli CO, Gottesman MM, Pastan I. Single cell analysis of daunomycin uptake and efflux in multidrug-resistant and -sensitive KB cells: effects of verapamil and other drugs. Cancer Res. 1986;46:5941–6. [PubMed] [Google Scholar]
- 12.Thiebaut F, Tsuruo T, Hamada H, Gottesman MM, Pastan I, Willingham MC. Cellular localisation of the multidrug-resistance gene product P-glycoprotein in normal human tissues. Proc Natl Acad Sci USA. 1987;84:7735–8. doi: 10.1073/pnas.84.21.7735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schwab M, Schaeffler E, Marx C, Fromm MF, Kaskas B, Metzler J, Stange E, Herfarth H, Schoelmerich J, Gregor M, Walker S, Cascorbi I, Roots I, Brinkmann U, Zanger U, Eichelbaum M. Association between the C3435T MDR1 gene polymorphism and susceptibility for ulcerative colitis. Gastroenterology. 2003;124:26–33. doi: 10.1053/gast.2003.50010. [DOI] [PubMed] [Google Scholar]
- 14.Panwala C, Jones J, Viney J. A novel model of inflammatory bowel disease: mice deficient for the multiple drug resistance gene, mdrla, spontaneously develop colitis. J Imunnol. 1998;161:5733–44. [PubMed] [Google Scholar]
- 15.Bustin SA. Absolute quantification of mRNA using real-time reverse transcription polymerase chain reaction assays. J Mol Endocrinol. 2000;25:169–93. doi: 10.1677/jme.0.0250169. [DOI] [PubMed] [Google Scholar]
- 16.Finnström N, Thörn M, Lööf L, Rane A. Independent patterns of cytochrome P450 gene expression in liver and blood in patients with suspected liver disease. Eur J Clin Pharmacol. 2001;57:403–9. doi: 10.1007/s002280100318. [DOI] [PubMed] [Google Scholar]
- 17.Thörn M, Lundgren S, Herlenius G, Ericzon B-G, Lööf L, Rane A. Gene expression of cytochrome P450 in liver transplants over time. Eur J Clin Pharmacol. 2004;60:413–20. doi: 10.1007/s00228-004-0786-4. [DOI] [PubMed] [Google Scholar]
- 18.Bowen WP, Carey JE, Miah A, McMurray HF, Munday PW, James RS, Coleman RA, Brown AM. Measurement of cytochrome P450 gene induction in human hepatocytes using quantitative real-time reverse transcriptase-polymerase chain reaction. Drug Metab Disp. 2000;28:781–8. [PubMed] [Google Scholar]
- 19.Klotz U, Hoensch H, Schütz T, Beaune P, Zanger U, Bode JC, Fritz P. Expression of intestinal drug-metabolising enzymes in patients with chronic inflammatory bowel disease. Curr Therapeut Res. 1998;59:556–63. [Google Scholar]
- 20.George J, Liddle C, Murray M, Byth K, Farrell GC. Pre-translational regulation of cytochrome P450 genes is responsible for disease-specific changes of individual P450 enzymes among patients with cirrhosis. Biochem Pharmacol. 1995;49:873–81. doi: 10.1016/0006-2952(94)00515-n. [DOI] [PubMed] [Google Scholar]
- 21.Sumida A, Kinoshita K, Fukada T, Matsuda H, Yamamoto I, Inaba T, Azuma J. Relationship between mRNA levels quantified by reverse transcription-competitive PCR and metabolic activity of CYP3A4 and CYP2E1 in human liver. Biochem Biophys Res Commun. 1999;262:499–503. doi: 10.1006/bbrc.1999.1233. [DOI] [PubMed] [Google Scholar]
- 22.Rodrigez-Antona C, Donato MT, Pareja E, Gómez-Léchon M-J, Castell JV. Cytochrome P-450 mRNA expression in human liver and its relationship with enzyme activity. Arch Biochem Biophys. 2001;393:308–15. doi: 10.1006/abbi.2001.2499. [DOI] [PubMed] [Google Scholar]
- 23.Watkins PB, Wrighton SA, Schuetz EG, Molowa DT, Guzelian PS. Identification of a glucocorticoid-inducible cytochromes P-450 in the intestinal mucosa of rats and man. J Clin Invest. 1987;80:1029–36. doi: 10.1172/JCI113156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wacher VJ, Wu CY, Benet LZ. Overlapping substrate specificity and tissue distribution of cytochrome P450 3A and P-glycoprotein: implications for drug delivery and activity in cancer chemotherapy. Mol Carcin. 1995;13:129–34. doi: 10.1002/mc.2940130302. [DOI] [PubMed] [Google Scholar]
- 25.McKinnon RA, Burgess WM, de la Hall PM, Roberts-Thompson SJ, Gonzalez FJ, McManus ME. Characterisation of CYP3A gene subfamily expression in human gastro-intestinal tissues. Gut. 1995;36:259–67. doi: 10.1136/gut.36.2.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kolars JC, Lown KS, Schmiedlin-Ren P, Ghosh M, Fang C, Wrighton SA, Merion RM, Watkins PB. CYP3A gene expression in human gut epithelium. Pharmacogenetics. 1994;4:247–59. doi: 10.1097/00008571-199410000-00003. [DOI] [PubMed] [Google Scholar]
- 27.Hebert MF. Contributions of hepatic and P-glycoprotein to cyclosporine and tacrolimus oral drug delivery. Adv Drug Deliv Rev. 1997;27:201–4. doi: 10.1016/s0169-409x(97)00043-4. [DOI] [PubMed] [Google Scholar]
- 28.Paine MF, Khalighi M, Fisher JM, Shen DD, Kunze KL, Marsh CL, Perkins JD, Thummel KE. Characterization of interintestinal and intraintestinal variations in human CYP3A-dependent metabolism. J Pharmacol Exp Ther. 1997;283:1552–62. [PubMed] [Google Scholar]
- 29.Zhang Q-Y, Dunbar D, Ostrowska A, Zeisloft S, Yang J, Kaminsky LS. Characterization of human small intestinal cytochromes P-450. Drug Metab Dispos. 1999;27:804–9. [PubMed] [Google Scholar]
- 30.Kivistö KT, Bookjans G, Fromm MF, Griese EU, Munzel P, Kroemer HK. Expression of CYP3A4, CYP3A5 and CYP3A7 in human duodenal tissue. Br J Clin Pharmacol. 1996;42:387–9. doi: 10.1046/j.1365-2125.1996.42615.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lown KS, Kolars JC, Thummel KE, Barnett JL, Kunze KL, Wrighton SA, Watkins PB. Interpatient heterogeniety in expression of CYP3A4 and CYP3A5 in small bowel. Lack of prediction by the erythromycin breath test. Drug Metab Dispos. 1994;22:947–55. [PubMed] [Google Scholar]
- 32.Lown KS, Bailey DG, Fontana RJ, Janardan SK, Adair CH, Fortlage LA, Brown MB, Guo W, Watkins PB. Grape-fruit juice increases felodipine oral availability in humans by decreasing intestinal CYP3A protein expression. J Clin Invest. 1997;99:2545–53. doi: 10.1172/JCI119439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kuehl P, Zhang J, Lin Y, Lamba J, Assem M, Schuetz J, Watkins PB, Daly A, Wrighton SA, Hall SD, Maurel P, Relling M, Brimer C, Yasuda K, Venkataramanan R, Strom S, Thummel K, Boguski MS, Schuetz E. Sequence diversity in CYP3A promoters and characterization of the genetic basis of polymorphic CYP3A5 expression. Nature Genet. 2001;27:383–91. doi: 10.1038/86882. [DOI] [PubMed] [Google Scholar]
- 34.Thummel KE. Does the CYP3A5*3 polymorphism affect in vivo drug elimination? Pharmacogenetics. 2003;13:585–7. doi: 10.1097/00008571-200310000-00001. [DOI] [PubMed] [Google Scholar]
- 35.Floyd MD, Gervasini G, Masica AL, Mayo G, George AL, Jr, Bhat K, Kim RB, Wilkinson GR. Genotype-phenotype associations for common CYP3A4 and CYP3A5 variants in the basal and induced metabolism of midazolam in European- and African-American men and women. Pharmacogenetics. 2003;13:595–606. doi: 10.1097/00008571-200310000-00003. [DOI] [PubMed] [Google Scholar]
- 36.Fojo AT, Ueda K, Slamon DJ, Poplack DG, Gottesman MM, Pastan I. Expression of a multidrug-resistance gene in human tumors and tissues. Proc Natl Acad Sci USA. 1987;84:265–9. doi: 10.1073/pnas.84.1.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mouly S, Paine MF. P-glycoprotein increases from proximal to distal regions of human small intestine. Pharm Res. 2003;20:1595–9. doi: 10.1023/a:1026183200740. [DOI] [PubMed] [Google Scholar]
- 38.Stephens RH, Tanianis-Hughes J, Higg NB, Humphrey M, Warhurst G. Region-dependent modulation of intestinal permeability by drug efflux transporters: in vitro studies in mdra (-/-) mouse intestine. J Pharmacol Exp Ther. 2002;303:1095–101. doi: 10.1124/jpet.102.041236. [DOI] [PubMed] [Google Scholar]