Abstract
Aim
Rabeprazole is metabolized to some extent by CYP2C19. The purpose of this study was to elucidate the pharmacokinetics of each rabeprazole enantiomer in three different CYP2C19 genotype groups.
Methods
Twenty-four healthy subjects, of whom each each were homozygous extensive metabolizers (homEMs), heterozygous extensive metabolizers (hetEMs) and poor metabolizers (PMs) for CYP2C19, participated in our study. After a single oral dose of 20 mg of racemic rabeprazole, the plasma concentrations of the rabeprazole enantiomers were measured over the course of 24 h.
Results
The area under the plasma concentration–time curves (AUC) of (R)-rabeprazole in homEMs, hetEMs and PMs were 1.8-, 2.2- and 2.4-fold, respectively, greater than those of (S)-rabeprazole; the relative AUC ratios of (R)- and (S)-rabeprazole in homEMs, hetEMs and PMs were 1 : 1.1 : 2.1 and 1 : 0.9 : 1.5, respectively. The mean maximum plasma concentrations (Cmax) of (R)-rabeprazole in homEMs, hetEMs and PMs were 1.7-, 1.9- and 1.8-fold higher, respectively, than those of the corresponding (S)-enantiomer (P < 0.05). There was no difference between homEMs and PMs in the elimination half-life of (S)-rabeprazole, whereas the elimination half-life of (R)-rabeprazole was significantly longer in PMs than in homEMs [1.7 h (1.4, 2.0) (mean (95% confidence interval)]vs. 0.8 h (0.6, 1.0), respectively, P < 0.0001).
Conclusions
(R)-Rabeprazole disposition was influenced to a greater degree by CYP2C19 genetic polymorphisms than was that of (S)-rabeprazole. The effect of CYP2C19 polymorphisms on the stereoselective disposition of rabeprazole was less than those of lansoprazole and omeprazole.
Keywords: CYP2C19, enantiomer, rabeprazole
Introduction
Rabeprazole [[(±)-sodium 2-[[4-methoxypropoxy]-3-me thylpyridin-2-yl]-methylsulfinyl]-1H-benzimidazole] is a proton pump inhibitor (PPI) that inhibits gastric acid secretion by interacting with (H+/K+)-ATPase in gastric parietal cells [1–3]. Rabeprazole is primarily converted non-enzymatically to rabeprazole-thioether, but some is oxidized to demethylated rabeprazole and rabeprazole sulphone by CYP2C19 and CYP3A4, respectively [4–7]. Hence, compared with omeprazole and lansoprazole, CYP2C19 contributes less to the overall metabolism of rabeprazole. Sakai et al. have reported that the relative area under the plasma concentration–time curve (AUC) ratios of omeprazole, lansoprazole and rabeprazole in homozygous extensive metabolizers (homEMs), heterozygous extensive metabolizers (hetEMs) and poor metabolizers (PMs) were 1 : 1.7 : 7.4, 1 : 1.4 : 3.7 and 1 : 1.1 : 1.2, respectively [8]. Similar to other PPIs, rabeprazole also possesses a chiral benzimidazole sulfoxide structure and it has been commercially marketed as a racemic mixture.
In general, the pharmacokinetics of each enantiomer of chiral compounds differs in humans. For lansoprazole, the plasma concentrations of (R)-lansoprazole in either healthy subjects or renal transplant recipients are considerably higher than those of the (S)-enantiomer after administration of an identical lansoprazole dose [9–11]. (R)-Lansoprazole is less influenced by CYP2C19 genetic polymorphisms because CYP2C19-catalysed metabolism is minimal compared with (S)-lansoprazole [10, 12]. (R)-Lansoprazole is the main active compound. In the case of omeprazole, the plasma concentrations of (S)-omeprazole are higher and less influenced by CYP2C19 genetic polymorphisms compared with those of (R)-omeprazole and racemic omeprazole [13–15]. This finding has led to the development of esomeprazole, the (S)-enantiomer of omeprazole, as the first single enantiomer PPI. We have previously described the kinetic disposition of rabeprazole enantiomers in renal transplant recipients who were CYP2C19 EMs [11]. In that report, the AUCs from 0 to 24 h (AUC0−24) and the elimination half-life of (R)-rabeprazole were 1.2-fold greater and 1.6-fold longer, respectively, than those of the (S)-enantiomer, whereas there was no difference in the maximum plasma concentration (Cmax) between the two rabeprazole enantiomers. However, no information is available on the effect of CYP2C19 genetic polymorphisms on the enantioselective disposition of rabeprazole.
The aim of this investigation was to elucidate the pharmacokinetics of each enantiomer of rabeprazole among three different CYP2C19 genotype groups (homozygous EMs, heterozygous EMs and PMs) in healthy Japanese subjects.
Materials and methods
Subjects
Healthy Japanese subjects consisted of eight of each CYP2C19 genotype: homEMs, hetEMs and PMs. There were no differences among the three CYP2C19 genotypes in terms of subject profiles, including age (23.4 ± 0.7, 24.1 ± 4.2 and 24.0 ± 1.3 years, respectively), body weight (51.6 ± 7.8, 56.9 ± 9.0 and 55.6 ± 6.5 kg, respectively) and male/female ratios (2 : 6, 6 : 2 and 3 : 5, respectively). None of the subjects had a history of significant medical illness or hypersensitivity to any drug and none was a smoker. Furthermore, they were not allowed to take drugs or medications during the study periods. The study protocol was approved by the Ethics Committee of Akita University Hospital and Hirosaki University Hospital, and all subjects gave their written informed consent before participating.
Study protocols
Each subject received a single oral dose of 20 mg of rabeprazole sodium (Pariet®; Eisai, Tokyo, Japan) with a glass of tap water at 09.00 h. Venous blood samples used to determine the plasma concentration of rabeprazole enantiomers were taken before and 1, 2, 3, 4, 5, 6, 8, 10, 12 and 24 h after dosing. The samples were centrifuged at 3000 g immediately after collection and stored at −80 °C until analysis. All subjects fasted for 10 h before the administration of rabeprazole and had a standard meal 4 h later. Alcohol and caffeinated beverages were forbidden during the test period.
CYP2C19 genotyping
The genotyping procedure used to identify the CYP2C19 wild-type gene and its two mutant alleles, CYP2C19*2 in exon 5 and CYP2C19*3 in exon 4, was a polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) method [16]. CYP2C19 genotype analysis revealed four different patterns as follows: *1/*1 in eight, *1/*2 in five, *1/*3 in three and *2/*2 in eight subjects. Individuals with these genotype patterns were divided into three groups; homEMs (*1/*1, n = 8); hetEMs (*1/*2 and *1/*3, n = 8); and PMs (*2/*2, n = 8).
Reagents and chemicals
Rabeprazole enantiomers and omeprazole-thioether were donated from Eisai Co. Ltd.. An Oasis HLB extraction cartridge was purchased from Waters (Milford, MA, USA). All solvents used were of high-performance liquid chromatogrphy (HPLC) grade (Wako Pure Chemical Industries, Osaka, Japan) and all other reagents and chemicals were purchased from either Wako Chemical Industries or Nacalai Tesque (Kyoto, Japan).
Analysis of rabeprazole enantiomers in plasma
The plasma concentrations of rabeprazole enantiomers were determined according to the HPLC method of Miura et al.[11]. Briefly, omeprazole-thioether (20 ng) in methanol (10 µl) was added to the samples (100 µl) as an internal standard, then the samples were diluted with water (1.0 ml) and the solutions were mixed for a short time. Each mixture was applied to an Oasis HLB extraction cartridge that had been previously activated with methanol and water (1.0 ml each). The cartridges were then washed with 60% methanol in water (1.0 ml), followed by elution with 100% methanol (1.0 ml). The eluates were evaporated to dryness in a vacuum at 60°C by a rotary evaporator (Iwaki, Tokyo, Japan). The residues were dissolved in 50 µl of methanol and 50 µl of the mobile phase, and each aliquot (50 µl) was injected into the HPLC apparatus. The HPLC column used was a Chiral CD-Ph (250 × 4.6 mm i.d.; Shiseido Co. Ltd, Tokyo, Japan). The mobile phase consisted of 0.5 m NaClO4-acetonitrile (60 : 40, v/v), which was degassed in an ultrasonic bath prior to use. A flow rate of 0.5 ml min−1 was used at ambient temperature, with the wavelength set at 285 nm. The lower limit of quantification for this assay was 5.0 ng ml−1 for each rabeprazole enantiomer. The coefficient of variation of inter- and intraday assays was <7.8% and the accuracy was within 4.7% for both analytes at concentrations of 5, 50, 250, 500 and 1000 ng ml−1.
Pharmacokinetic analysis
Pharmacokinetic analysis of the rabeprazole enantiomers was carried out by a standard noncompartmental method using WinNonlin (Pharsight Co., Mountain View, CA, USA; version 4.0.1). The elimination half-life was obtained by log-linear regression of the terminal phase of the concentration–time data with at least three sampling points (elimination half-life = ln2/ke; ke = elimination rate constant). The total area under the observed plasma concentration–time curve (AUC) was calculated using the linear trapezoidal rule. Extrapolation of AUC from the last measurable concentration (Ct) to infinity (AUCt–∝) was performed by adding the value Ct/ke (where Ct = plasma concentration for t hours after rabeprazole administration). The maximum plasma concentration (Cmax) and time required to reach the Cmax (tmax) were obtained directly from the profile.
Statistical analysis
All results are expressed as mean values ± standard deviation and 95% confidence intervals (CI). Statistical comparisons of the parameters were supplemented with the multiple comparison procedure of Scheffe using StatView software (SAS Institute, Cary, NC, USA; version 5.0). A P-value of <0.05 was considered to be statistically significant.
The necessary sample size was calculated using the following assumption. The AUC, Cmax and elimination half-life for rabeprazole are of the same proportion between the EMs and the PMs, as reported in the literature (n = 6 each) [17], and so six subjects are necessary for each genotype with α = 0.05 and β = 0.2 (power of 80%). To allow for stratification by genotype, eight subjects for each group were targeted for enrolment in this study. This analysis was performed with S-PLUS (Mathematical System Inc, Tokyo, Japan; version 6.0).
Results
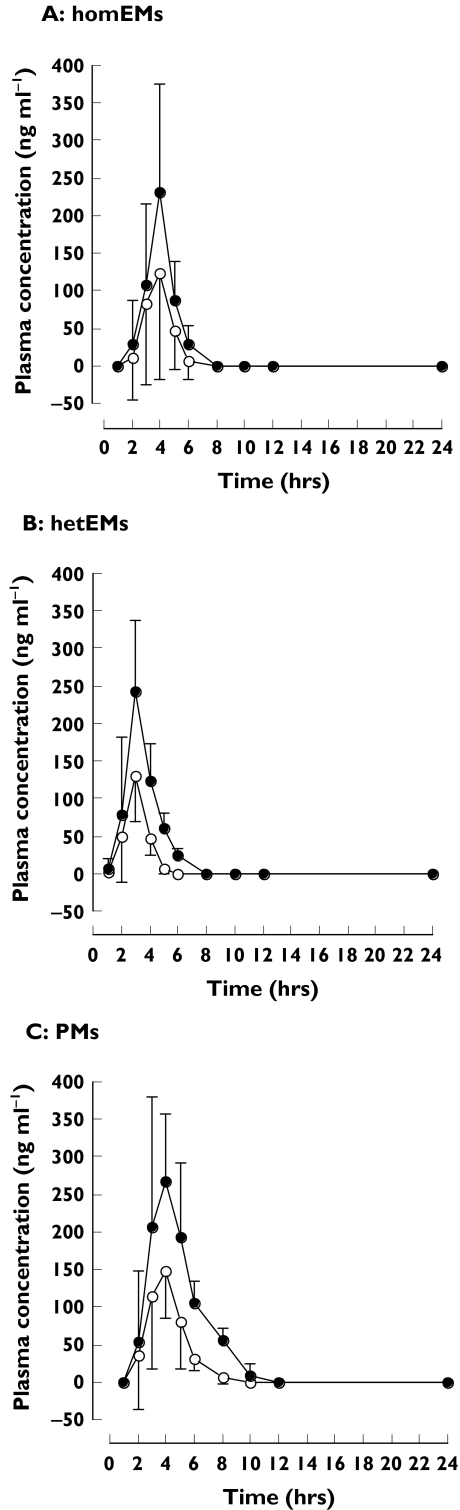
The mean plasma concentrations of (R)-rabeprazole were much higher than those of (S)-rabeprazole in all three CYP2C19 genotype groups (Figure 1, Table 1). The AUCs of (R)-rabeprazole in homEMs, hetEMs and PMs were 1.8- (1.4, 2.1) [mean (95% CI)] (P < 0.05), 2.2- (2.0, 2.4) (P < 0.005) and 2.4-fold (2.1, 2.7) (P < 0.0001) higher, respectively, than those of the corresponding (S)-enantiomer. The relative AUC ratios of (R)- and (S)-enantiomer in homEMs, hetEMs and PMs were 1 : 1.1 : 2.1 and 1 : 0.9 : 1.5, respectively. The mean maximum plasma concentrations (Cmax) of (R)-rabeprazole in homEMs, hetEMs and PMs were 1.7- (1.5, 2.0) (P < 0.05), 1.9- (1.7, 2.0) (P < 0.05) and 1.8-fold (1.3, 2.3) (P < 0.005) higher, respectively, than those of the corresponding (S)-enantiomer. However, there were no significant differences for the mean Cmax values of (R)- or (S)-rabeprazole across the three CYP2C19 genotype groups. In contrast, although there was no difference in the elimination half-life of (S)-rabeprazole across the three different CYP2C19 genotype groups, the elimination half-life of (R)-rabeprazole was significantly longer in PMs than in homEMs (P < 0.0001).
Figure 1.
Mean ± SD plasma concentration–time profiles of (R)-rabeprazole (•) and (S)-rabeprazole (○) for A: homozygous extensive metabolizers (EMs); B: heterozygous EMs; and C: poor metabolizers, after a 20-mg oral dose of racemic rabeprazole
Table 1.
Pharmacokinetic parameters of (R)- and (S)-rabeprazole in three CYP2C19 genotype groups
| Study group | Homozygous EMs | Heterozygous EMs | PMs |
|---|---|---|---|
| (R)-Rabeprazole | |||
| Cmax (ng ml−1) | 257 ± 116 | 279 ± 76 | 330 ± 97 |
| (95% CI) | (160, 354) | (200, 332) | (282, 425) |
| tmax (h) | 3.6 ± 0.7 | 2.9 ± 0.4 | 3.5 ± 0.9 |
| (95% CI) | (3.0, 4.3) | (2.6, 3.2) | (2.7, 4.3) |
| Half-life (h) | 0.8 ± 0.2*** | 0.9 ± 0.2*** | 1.7 ± 0.4 |
| (95% CI) | (0.6, 1.0) | (0.8, 1.0) | (1.4, 2.0) |
| AUC0–∝ (ng h−1 ml−1) | 514 ± 258*** | 573 ± 75*** | 1068 ± 212 |
| (95% CI) | (299, 730) | (484, 627) | (891, 1245) |
| (S)-Rabeprazole | |||
| Cmax (ng ml−1) | 145 ± 50† | 153 ± 51† | 201 ± 44†† |
| (95% CI) | (103, 186) | (109, 197) | (164, 237) |
| tmax (h) | 3.5 ± 0.8 | 2.8 ± 0.5 | 3.5 ± 0.8 |
| (95% CI) | (2.9, 4.1) | (2.4, 3.1) | (2.9, 4.1) |
| Half-life (h) | 0.8 ± 0.3 | 0.7 ± 0.2 | 1.0 ± 0.1††† |
| (95% CI) | (0.6, 1.1) | (0.5, 0.9) | (0.9, 1.1) |
| AUC0–∝ (ng h−1 ml−1) | 294 ± 154† | 260 ± 49*†† | 445 ± 88††† |
| (95% CI) | (166, 422) | (213, 307) | (372, 519) |
| R/S | |||
| Cmax | 1.7 ± 0.3 | 1.9 ± 0.2 | 1.8 ± 0.2 |
| (95% CI) | (1.5, 2.0) | (1.7, 2.0) | (1.3, 2.3) |
| AUC0–∝ | 1.8 ± 0.4* | 2.2 ± 0.3 | 2.4 ± 0.4 |
| (95% CI) | (1.4 ± 2.1) | (2.0 ± 2.4) | (2.1 ± 2.7) |
The values are shown as the mean ± SD. Cmax, Maximum plasma concentration; tmax, time to reach Cmax; AUC0–∝, area under the plasma concentration–time curve from 0 to infinity; EM, extensive metabolizer; PM, poor metabolizer.
P < 0.0001 compared with the PM group;
P < 0.05;
P < 0.005;
P < 0.0001 compared with (R)-rabeprazole.
P < 0.05;
Discussion
In the present study, we examined the pharmacokinetics of rabeprazole enantiomers in relation to CYP2C19 genotype status by administering 20 mg of racemic rabeprazole to healthy Japanese subjects. We found no statistically significant differences in the pharmacokinetics of (S)-rabeprazole between homEMs and PMs. However, our results showed that the pharmacokinetics of (R)-rabeprazole were more intensely affected by CYP2C19 polymorphisms than those of the (S)-enantiomer. The elimination half-life of (R)-rabeprazole in PMs was significantly longer than in homEMs (P < 0.0001), whereas there was no significant difference in Cmax value between homEMs and PMs (P = 0.0721). A power analysis based on the observed differences revealed that more than six subjects of each genotype would have been necessary to demonstrate statistical significance with a power of 0.8. The power between homEMs and PMs calculated from our present data, which consisted of eight of each genotype, was 0.478 for the Cmax value of (R)-rabeprazole and 0.677 and 0.665, respectively, for the AUC and Cmax of (S)-rabeprazole. In the present study, there was no CYP2C19-specific genetic effect on the first-pass metabolism of either rabeprazole enantiomer.
In CYP2C19 EMs, the Cmax of (R)-rabeprazole was significantly higher than that of the (S)-enantiomer (P < 0.05), but the elimination half-life did not differ for the two enantiomers. On the other hand, in CYP2C19 PMs, the Cmax and the elimination half-life of (R)-rabeprazole were significantly higher and longer, respectively, than those of the (S)-enantiomer (P < 0.005 and P < 0.0001, respectively). Racemic rabeprazole has a low oral bioavailability of about 52% because of extensive first-pass metabolism [18]. Instability of (S)-rabeprazole in the human body seems to cause the low bioavailability of racemic rabeprazole. The significant difference in Cmax between the rabeprazole enantiomers may be due to the stereoselective reduction of (S)-rabeprazole into rabeprazole thioether by non-enzymatic means, or stereoselective oxidation into rabeprazole sulphone by CYP3A4. However, no information is available about the stereoselective properties of rabeprazole metabolism from in vitro studies with human liver microsomes. The AUC ratios for (R)-rabeprazole to (S)-rabeprazole in homEMs, hetEMs and PMs were 1.8, 2.2 and 2.4, respectively. Hence, the R/S ratio for the AUC in PMs was 1.3-fold higher than in homEMs (P < 0.05).
The degree to which CYP2C19 participates in the overall metabolism of rabeprazole has been reported to be much less compared with that of lansoprazole and omeprazole [5, 8, 19, 20]. The R/S ratios for the AUC of lansoprazole in homEMs, hetEMs and PMs are 12.7, 8.5 and 5.8, respectively [10], and the R/S ratios for the AUC of omeprazole in EMs and PMs are 0.5 and 1.7, respectively [13]. Thus, in comparison with lansoprazole and omeprazole, the enantioselective disposition of rabeprazole is less affected by CYP2C19 genetic polymorphisms.
The pharmacokinetics of rabeprazole enantiomers in the present study using healthy CYP2C19 EM subjects differed from those reported in CYP2C19 EM renal transplant recipients receiving tacrolimus [11]. The mean Cmax values of (S)-rabeprazole are higher and the elimination half-life of (R)-rabeprazole is longer in renal transplant recipients on tacrolimus, a substrate of CYP3A4 [21, 22], than in our present study's healthy subjects. CYP3A4 is also the high-affinity enzyme responsible for rabeprazole sulphone formation. Further studies may be needed to elucidate the interactions between rabeprazole and tacrolimus.
The difference in the pharmacological activity between (R)- and (S)-rabeprazole has not yet been established. Therefore, it is difficult to evaluate clinical outcomes based on our results. Even if the pharmacological activity of racemic rabeprazole is predominantly mediated by one enantiomer, the pharmacodynamics of each rabeprazole enantiomer would be less affected by CYP2C19-related genetic differences compared with omeprazole and lansoprazole. Especially in the case of (S)-rabeprazole, the influence of CYP2C19 polymorphisms is likely to be small.
In conclusion, the present study indicates that the plasma concentrations and the degree of CYP2C19-mediated metabolism of (R)-rabeprazole are higher and greater, respectively, than those of the (S)-enantiomer. However, the R/S ratios for the AUC of rabeprazole in homEMs, hetEMs and PMs are 1.8, 2.2 and 2.4, respectively, suggesting a lesser effect of CYP2C19 polymorphisms on the stereoselective disposition of rabeprazole compared with lansoprazole and omeprazole.
Acknowledgments
We thank Eisai Co. Ltd. (Tokyo, Japan) for providing the rabeprazole enantiomers. This work was supported by a grant (no.17923061) from the Japan Society for the Promotion of Science, Tokyo, Japan.
References
- 1.Prakash A, Faulds D. Rabeprazole. Drugs. 1998;55:261–7. doi: 10.2165/00003495-199855020-00009. [DOI] [PubMed] [Google Scholar]
- 2.Morii M, Takata H, Fujisaki H, Takeguchi N. The potency of substituted benzimidazoles such as E3810, omeprazole, Ro 18-5364 to inhibit gastric H+, K(+)-ATPase is correlated with the rate of acid-activation of the inhibitor. Biochem Pharmacol. 1990;39:661–7. doi: 10.1016/0006-2952(90)90143-9. [DOI] [PubMed] [Google Scholar]
- 3.Fujisaki H, Shibata H, Oketani K, Murakami M, Fujimoto M, Wakabayashi T, Yamatsu I, Yamaguchi M, Sakai H, Takeguchi N. Inhibitions of acid secretion by E3810 and omeprazole, and their reversal by glutathione. Biochem Pharmacol. 1991;42:321–8. doi: 10.1016/0006-2952(91)90719-l. [DOI] [PubMed] [Google Scholar]
- 4.Ishizaki T, Horai Y. Review article: cytochrome P450 and the metabolism of proton pump inhibitors — emphasis on rabeprazole. Aliment Pharmacol Ther. 1999;13:27–36. doi: 10.1046/j.1365-2036.1999.00022.x. [DOI] [PubMed] [Google Scholar]
- 5.Yasuda S, Horai Y, Tomono Y, Nakai H, Yamato C, Manabe K, Kobayashi K, Chiba K, Ishizaki T. Comparison of the kinetic disposition and metabolism of E3810, a new proton pump inhibitor, and omeprazole in relation to S-mephenytoin 4′-hydroxylation status. Clin Pharmacol Ther. 1995;58:143–54. doi: 10.1016/0009-9236(95)90192-2. [DOI] [PubMed] [Google Scholar]
- 6.VandenBranden M, Ring BJ, Binkley SN, Wrighton SA. Interaction of human liver cytochromes P450 in vitro with LY307640, a gastric proton pump inhibitor. Pharmacogenetics. 1996;6:81–91. doi: 10.1097/00008571-199602000-00007. [DOI] [PubMed] [Google Scholar]
- 7.Klotz U. Pharmacokinetic considerations in the eradication of Helicobacter pylori. Clin Pharmacokinet. 2000;38:243–70. doi: 10.2165/00003088-200038030-00004. [DOI] [PubMed] [Google Scholar]
- 8.Sakai T, Aoyama N, Kita T, Sakaeda T, Nishiguchi K, Nishitora Y, Hohda T, Sirasaka D, Tamura T, Tanigawara Y, Kasuga M, Okumura K. CYP2C19 genotype and pharmacokinetics of three proton pump inhibitors in healthy subjects. Pharm Res. 2001;18:721–7. doi: 10.1023/a:1011035007591. [DOI] [PubMed] [Google Scholar]
- 9.Kim KA, Shon JH, Park JY, Yoon YR, Kim MJ, Yun DH, Kim MK, Cha IJ, Hyun MH, Shin JG. Enantioselective disposition of lansoprazole in extensive and poor metabolizers of CYP2C19. Clin Pharmacol Ther. 2002;72:90–9. doi: 10.1067/mcp.2002.126176. [DOI] [PubMed] [Google Scholar]
- 10.Miura M, Tada H, Yasui-Furukori N, Uno T, Sugawara K, Tateishi T, Suzuki T. Pharmacokinetic differences between the enantiomers of lansoprazole and its metabolite, 5-hydroxylansoprazole, in relation to CYP2C19 genotypes. Eur J Clin Pharmacol. 2004;60:623–8. doi: 10.1007/s00228-004-0809-1. [DOI] [PubMed] [Google Scholar]
- 11.Miura M, Kagaya H, Tada H, Sagae Y, Satoh S, Habuchi T, Suzuki T. Comparison of enantioselective disposition of rabeprazole versus lansoprazole in renal transplant recipients who are CYP2C19 extensive metabolizers. Xenobiotica. 2005;35:479–86. doi: 10.1080/00498250500111562. [DOI] [PubMed] [Google Scholar]
- 12.Miura M, Tada H, Yasui-Furukori N, Uno T, Sugawara K, Tateishi T, Suzuki T. Enantioselective disposition of lansoprazole in relation to CYP2C19 genotypes in the presence of fluvoxamine. Br J Clin Pharmacol. 2005;60:61–8. doi: 10.1111/j.1365-2125.2005.02381.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Andersson T, Hassan-Alin M, Hasselgren G, Rohss K, Weidolf L. Pharmacokinetic studies with esomeprazole, the (S)-isomer of omeprazole. Clin Pharmacokinet. 2001;40:411–26. doi: 10.2165/00003088-200140060-00003. [DOI] [PubMed] [Google Scholar]
- 14.Kanazawa H, Okada A, Higaki M, Yokota H, Mashige F, Nakahara K. Stereospecific analysis of omeprazole in human plasma as a probe for CYP2C19 phenotype. J Pharm Biomed Anal. 2003;30:1817–24. doi: 10.1016/s0731-7085(02)00524-1. [DOI] [PubMed] [Google Scholar]
- 15.Tybring G, Bottiger Y, Widen J, Bertilsson L. Enantioselective hydroxylation of omeprazole catalyzed by CYP2C19 in Swedish white subjects. Clin Pharmacol Ther. 1997;62:129–37. doi: 10.1016/S0009-9236(97)90060-6. [DOI] [PubMed] [Google Scholar]
- 16.De Morais SM, Wilkinson GR, Blaisdell J, Meyer UA, Nakamura K, Goldstein JA. Identification of a new genetic defect responsible for the polymorphism of (S)-mephenytoin metabolism in Japanese. Mol Pharmacol. 1994;46:594–8. [PubMed] [Google Scholar]
- 17.Horai Y, Kimura M, Furuie H, Matsuguma K, Irie S, Koga Y, Nagahama T, Murakami M, Matsui T, Yao T, Urae A, Ishizaki T. Pharmacodynamic effects and kinetic disposition of rabeprazole in relation to CYP2C19 genotypes. Aliment Pharmacol Ther. 2001;15:793–803. doi: 10.1046/j.1365-2036.2001.00980.x. [DOI] [PubMed] [Google Scholar]
- 18.Sclar DA, Tartaglione TA, Fine MJ. Overview of issues related to medical compliance with implications for the outpatient management of infectious diseases. Infect Agents Dis. 1994;3:266–73. [PubMed] [Google Scholar]
- 19.Ishizaki T, Horai Y. Review article: cytochrome P450 and the metabolism of proton pump inhibitors — emphasis on rabeprazole. Aliment Pharmacol Ther. 1999;3:27–36. doi: 10.1046/j.1365-2036.1999.00022.x. [DOI] [PubMed] [Google Scholar]
- 20.Desta Z, Zhao X, Shin JG, Flockhart DA. Clinical significance of the cytochrome P450 2C19 genetic polymorphism. Clin Pharmacokinet. 2002;41:913–58. doi: 10.2165/00003088-200241120-00002. [DOI] [PubMed] [Google Scholar]
- 21.Sattler M, Guengerich FP, Yun CH, Christians U, Sewing KF. Cytochrome P-450 3A enzymes are responsible for biotransformation of FK506 and rapamycin in man and rat. Drug Metab Dispos. 1992;20:753–61. [PubMed] [Google Scholar]
- 22.Shiraga T, Matsuda H, Nagase K, Iwasaki K, Noda K, Yamazaki H, Shimada T, Funae Y. Metabolism of FK506, a potent immunosuppressive agent, by cytochrome P450 3A enzymes in rat, dog and human liver microsomes. Biochem Pharmacol. 1994;47:727–35. doi: 10.1016/0006-2952(94)90136-8. [DOI] [PubMed] [Google Scholar]