Abstract
Aims
CYP2D6 protein expression is determined by the number of functional CYP2D6 alleles. It is also higher in individuals with at least one CYP2D6*2 allele. This study has investigated the effect of the number of functional CYP2D6 alleles and the influence of CYP2D6*2 alleles on plasma perhexiline concentrations in patients administered a standard loading regimen over 3 days.
Methods
Eighteen patients with myocardial ischaemia who were not taking any drugs known to inhibit CYP2D6 metabolism in vivo commenced treatment with 200 mg of perhexiline twice per day. On the fourth day, blood was drawn for genotyping and the measurement of trough plasma concentrations of perhexiline and its major metabolite, cis-OH-perhexiline.
Results
The only genotypic CYP2D6 poor metabolizer had a trough plasma perhexiline concentration of 2.70 mg l−1 and no detectable cis-OH-perhexiline. The mean ± SD trough plasma perhexiline concentration in patients with one functional allele was significantly higher (0.63 ± 0.31 mg l−1, n = 8, P = 0.05) than in patients with two functional alleles (0.37 ± 0.17 mg l−1, n = 9). Conversely, the mean metabolic ratio was significantly lower in patients with one functional allele (2.90 ± 1.76, P < 0.01) compared with patients with two functional alleles (6.52 ± 3.26). Patients with at least one CYP2D6*2 allele had a lower plasma perhexiline concentration (0.20 ± 0.09 mg l−1, n = 5, P < 0.001) and a higher metabolic ratio (7.86 ± 2.51, P < 0.01) than the non-poor metabolizer patients with no CYP2D6*2 alleles (0.62 ± 0.23 mg l−1 and 3.55 ± 2.54, respectively, n = 12).
Conclusion
Patients with only one functional allele and not CYP2D6*2 have diminished CYP2D6 metabolic capacity for perhexiline.
Keywords: CYP2D6, cytochrome P450, metabolic ratio, perhexiline, polymorphism, therapeutic drug monitoring
Introduction
The considerable efficacy of perhexiline as monotherapy for the treatment of exertional angina was established over 30 years ago [1], although its application has been limited following the association of persistently high plasma perhexiline concentrations with polymorphism of the cytochrome P450 2D6 (CYP2D6) isoform and eventual development of hepatic and neurological toxicity [2–4]. Perhexiline inhibits carnitine palmitoyltransferase -1 and -2 to enhance carbohydrate utilization by myocytes, increasing ATP synthesis per unit oxygen utilization during ischaemia [5, 6]. This metabolic method of action has proved incrementally effective beyond β-blockers, nitrates and calcium channel blockers in patients with severe stable angina unsuitable for coronary revascularization, provided plasma perhexiline concentrations are maintained within 0.15–0.60 mg l−1[7]. Furthermore, recent investigations have provided increasing evidence of the efficacy of perhexiline in acute coronary syndromes such as unstable angina pectoris [8], which may be associated with the recently described effect of perhexiline in sensitizing platelets to nitric oxide [9].
CYP2D6 poor metabolizers (PM) have a profoundly impaired capacity to form the primary metabolite cis-OH-perhexiline [4, 10]. The frequency distributions of the metabolic ratio of plasma concentrations of cis-OH-perhexiline to perhexiline and the apparent oral clearance (CL/F) at steady state are bimodal, with PM subjects forming a subpopulation that can be differentiated from phenotypic CYP2D6 extensive, intermediate and ultra-rapid metabolizers (EM, IM and UM, respectively) [10]. Because the metabolic ratio reflects oral clearance, it can be used to guide perhexiline dosage, which can vary from 70 mg week−1 for PM up to 500 mg day−1 for UM [10]. In addition, the metabolic ratio may also be used to identify IM subjects who are likely to be more susceptible to significant interactions with CYP2D6 inhibitors due to diminished CYP2D6 metabolic capacity [11].
CYP2D6 expression increases with the number of functional CYP2D6 gene copies per genome [12] and is responsible for the gene–dose effect seen with respect to perhexiline metabolism [11, 13]. Increased gene copy number as a consequence of alleles containing multiple gene copies predicts only about 20% of all individuals with UM phenotype [14]. The presence of a CYP2D6*2 allele results in increased CYP2D6 protein expression [12] and has been associated with the UM phenotype as measured by debrisoquine [15] and sparteine metabolic ratios [12]. In contrast, CYP2D6*41 codes for CYP2D6.2, but has profoundly reduced expression [12] and has been associated with the IM phenotype [12, 16]. Importantly, Zanger et al.[12] have reported that, irrespective of the number of functional alleles, the average expression of CYP2D6 protein in individuals possessing at least one CYP2D6*2 allele is at least twice as much as for all other non-PM. Therefore, the aim of this study was to investigate the relationship between CYP2D6 genotype and trough plasma perhexiline concentration and metabolic ratio after a standard loading regimen of perhexiline, with particular respect to the CYP2D6*2 allele and the number of functional alleles.
Methods
Patients
This study was approved by the Ethics of Human Research Committee of The Queen Elizabeth Hospital. Eighteen caucasian patients admitted to the Cardiology Unit of The Queen Elizabeth Hospital gave written informed consent prior to participating in the study. Thirteen of these patients had previously participated in a group of 24 to study the effect of perhexiline on CYP2D6 activity assessed by dextromethorphan [11]. The patients were between 48 and 87 years of age upon entry to the study, with a mean (± SD) age of 73.0 ± 11.6 years. Patients had a diagnosis of stable or unstable angina pectoris or acute myocardial infarction for which they commenced perhexiline treatment under the instruction of their attending cardiologist with a loading regimen of 200 mg twice daily for 3 days. Perhexiline was withheld on the fourth day and a blood sample was drawn for determination of CYP2D6 genotype [17] and trough plasma perhexiline and cis-OH-perhexiline concentrations by high-performance liquid chromatography [10]. Measurement of trough plasma perhexiline and cis-OH-perhexiline concentrations on the fourth day of treatment is standard clinical practice at The Queen Elizabeth Hospital. The plasma perhexiline concentration and the initial metabolic ratio are used to individualize patients’ maintenance dosage regimens [10]. None of these patients was taking any concomitant medications known to inhibit CYP2D6-catalysed metabolism in vivo.
Genotyping
Genomic DNA was isolated from whole blood samples using a QIAamp DNA mini kit, according to the manufacturer's protocol (Qiagen Pty Ltd, Clifton Hill, Australia). The alleles CYP2D6*1 to *4, *6 to *9 and *41 were determined from allele specific combinations of single nucleotide polymorphisms (SNPs) detected by sequencing bases 1556–1942 and 2379–3027 (based on translation start site) [17]. Samples assigned as CYP2D6*2 or *41 according to the 2988G→A SNP [18] were also analysed by a real-time polymerase chain reaction (PCR) assay (performed by the Dr Margarete Fischer-Bosch Institute of Clinical Pharmacology) to test for the −1584C→G SNP that is alternatively employed to discriminate between these two genotypes [12]. CYP2D6*10 alleles were detected using the method of Marez et al.[19]. Detection of alleles containing deleted genes (CYP2D6*5 and *16) or amplified genes (CYP2D6*1xN, *2xN and *4xN) was performed using PCR-based assays [20, 21]. The gene amplification assay included positive controls that had been previously confirmed by sequencing, but was not designed to determine which allele carried the increased gene copies due to clinical time constraints. Consequently, genotypes with amplified genes were assigned the nomenclature CYP2D6(*x/*y)xN, where x and y refer to the alleles and xN signifies amplification. In the clinical setting this genotyping assay is applicable to the routine diagnostic analysis of patients at the commencement of drug therapy due to its 24-h turnaround time.
Statistical analyses
Patients were segregated by their number of functional CYP2D6 alleles (i.e. 0, 1 or 2), irrespective of the total functional gene copy number contributed by both alleles, or by the presence of at least one CYP2D6*2 allele. Comparisons between groups were carried out by two-way anova (GraphPad Software Inc., San Diego, CA, USA). P-values ≤0.05 were considered to be statistically significant. Plasma perhexiline and cis-OH-perhexiline concentrations and the metabolic ratio of cis-OH-perhexiline to perhexiline are presented as mean ± SD.
Results
Six CYP2D6*41 alleles (−1584C) and six CYP2D6*2 alleles (−1584G) were detected (Table 1). All the CYP2D6*2 alleles were wild type at 2988 and all the CYP2D6*41 alleles had the 2988G→A SNP, confirming its suitability as a marker in the current genotyping assay for CYP2D6*41, in agreement with Raimundo et al.[18].
Table 1.
Trough plasma perhexiline concentrations (mg l−1) in ascending order for 18 patients following a standard perhexiline loading regimen and their corresponding plasma cis-OH-perhexiline concentration (mg l−1) and CYP2D6 genotype
| Plasma perhexiline (mg l−1) | Plasma Cis-OH- perhexiline (mg l−1) | CYP2D6 genotype |
|---|---|---|
| 0.04 | 0.36 | *1/*2 |
| 0.20 | 2.24 | *2/*2 |
| 0.21 | 1.28 | *2/*4 |
| 0.26 | 1.24 | *2/*4 |
| 0.28 | 2.30 | (*2/*41)xN |
| 0.30 | 1.93 | *1/*41 |
| 0.42 | 3.27 | (*1/*1)xN |
| 0.44 | 0.81 | (*4/*10)xN |
| 0.48 | 4.01 | *1/*41 |
| 0.52 | 1.87 | *1/*1 |
| 0.53 | 1.10 | (*1/*41)xN |
| 0.54 | 1.07 | *1/*41 |
| 0.63 | 1.16 | *3/*41 |
| 0.72 | 0.41 | *4/*9 |
| 0.80 | 1.97 | *1/*4 |
| 0.97 | 2.76 | *1/*5 |
| 1.04 | 2.90 | (*1/*4)xN |
| 2.70 | 0.00 | *4/*4 |
The mean trough plasma perhexiline and cis-OH-perhexiline concentrations of the 18 patients who underwent identical loading regimens were 0.62 ± 0.58 and 1.70 ± 1.08 mg l−1, respectively. One patient was a genotypic PM (CYP2D6*4/*4) and had the highest plasma perhexiline concentration (2.70 mg l−1) and the lowest cis-OH-perhexiline/perhexiline metabolic ratio of zero (Table 1).
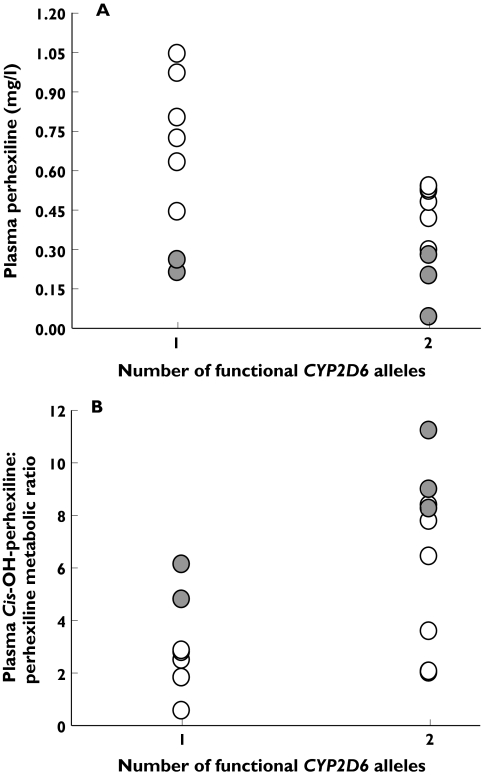
For the remaining 17 non-PM patients, both the number of functional CYP2D6 alleles and the presence of at least one CYP2D6*2 allele appeared to influence plasma perhexiline concentrations. Patients with one functional allele had a mean plasma perhexiline concentration of 0.63 ± 0.31 mg l−1 that exceeded the therapeutic range in five of the eight patients. It was significantly higher (P = 0.05, two-way anova) than the mean plasma perhexiline concentration of 0.37 ± 0.17 mg l−1 for patients with two functional alleles (n = 9). Similarly, patients without a CYP2D6*2 allele (n = 12) had a mean plasma perhexiline concentration of 0.62 ± 0.23 mg l−1 that was significantly higher (P < 0.001, two-way anova) than the mean plasma perhexiline concentration of 0.20 ± 0.09 mg l−1 for patients with at least one CYP2D6*2 allele (n = 5) (Figure 1).
Figure 1.
Trough plasma perhexiline concentration (mg l−1) (A) and plasma cis-OH-perhexiline to perhexiline metabolic ratio (B) in patients with either one or two functional CYP2D6 alleles following a standard perhexiline loading regime of 200 mg twice per day for 3 days. Patients without a CYP2D6*2 allele are represented with open symbols and those possessing at least one CYP2D6*2 allele are represented with closed symbols
In addition, the number of functional CYP2D6 alleles and the presence of at least one CYP2D6*2 allele influenced the cis-OH-perhexiline/perhexiline metabolic ratio. Patients with one functional allele had a mean metabolic ratio of 2.90 ± 1.76, which was significantly lower (P < 0.01, two-way anova) than the mean metabolic ratio of 6.52 ± 3.26 for patients with two functional alleles. Likewise, patients with no CYP2D6*2 alleles had a mean metabolic ratio of 3.55 ± 2.54, which was significantly lower (P < 0.01, two-way anova) than the mean metabolic ratio of 7.86 ± 2.51 for patients with at least one CYP2D6*2 allele (Figure 1).
There was no significant overall interaction between the number of functional alleles and the presence of a CYP2D6*2 allele with respect to plasma perhexiline concentration (P = 0.18, two-way anova) or metabolic ratio (P = 0.63, two-way anova). However, the possibility that this may have been due to the relatively small sample size (Figure 1) cannot be excluded.
Discussion
The higher mean plasma perhexiline concentration and lower mean metabolic ratio in patients with one functional CYP2D6 allele when compared with patients with two functional CYP2D6 alleles confirmed a gene–dose effect that has previously been demonstrated for perhexiline metabolism [11, 13]. In addition, an effect of the CYP2D6*2 allele on the metabolism of perhexiline was also demonstrated.
Three recent studies have described no difference between CYP2D6*2 and wild type with respect to in vivo function as measured by urinary metabolic ratios following single doses of sparteine or dextromethorphan [18, 22] or by the steady-state perhexiline plasma metabolic ratio [13]. However, the mean expression of CYP2D6 by non-PM individuals with at least one CYP2D6*2 allele has been shown to be more than twice that of other non-PM individuals without a CYP2D6*2 allele [12] and two studies have associated the CYP2D6*2 allele with the UM phenotype [12, 15], regardless of genotype or the number of functional alleles. Consistent with these observations, this study has demonstrated that the presence of a CYP2D6*2 allele significantly enhances in vivo perhexiline metabolism. Importantly, the results remained significant even when subjects positive for gene amplification were excluded from the analysis. The standard multiple dose perhexiline loading regimen permits very sensitive CYP2D6 phenotyping because the range of plasma perhexiline concentrations achieved are near the Km for the CYP2D6 dependent cis-hydroxylation of perhexiline [23] and consequently reflect differences in metabolic capacity.
Barclay et al. (2003) [13] have similarly demonstrated a relationship between CYP2D6 genotype and phenotype as determined by the steady-state perhexiline plasma metabolic ratio, although they did not specifically investigate the effect of the CYP2D6*2 allele. In patients receiving different perhexiline doses the steady-state metabolic ratio is unlikely to separate adequately UM from EM. The metabolic ratio of UM decreases from its initial measurement to steady state as a result of increased plasma perhexiline concentrations and saturated CYP2D6 metabolism from the increased perhexiline dosage necessary to achieve the therapeutic range [10].
Although our results should be confirmed with a larger clinical study, the current pilot study suggests that patients possessing only one functional allele are likely to be classified as IM, have a diminished CYP2D6 metabolic capacity and be more likely to experience clinically significant inhibition of CYP2D6-catalysed metabolism [11]. In addition, patients possessing only one functional allele that is not CYP2D6*2 are likely to have the lowest CYP2D6 expression and may be most at risk from excessive plasma perhexiline concentrations following a standard perhexiline loading regimen. We have previously reported that the cis-OH-perhexiline/perhexiline metabolic ratio measured within the first week of initiating treatment with perhexiline may be useful for assessing the relative CYP2D6 metabolic capacity of patients and thus for individualizing their long-term maintenance dosage regimens [10]. The current data further support the use of the metabolic ratio in dosage individualization. Importantly, the data also suggest that CYP2D6 genotyping for clinical applications should be optimized to identify IM as well as PM.
Acknowledgments
Determination of the −1584C/G polymorphism was kindly performed by the Dr Margarete Fischer-Bosch Institute of Clinical Pharmacology, Stuttgart, Germany. B.J.D. is the recipient of the MF and MH Joyner Scholarship in Medicine and the Freemasons Medical Research Scholarship. J.K.C. is the recipient of the CJ Martin Training Fellowship from the National Health and Medical Research Council of Australia.
References
- 1.Burns-Cox CJ, Chandrasekhar KP, Ikram H, Peirce TH, Pilcher J, Quinlan CD, Rees JR. Clinical evaluation of perhexiline maleate in patients with angina pectoris. BMJ. 1971;4:586–8. doi: 10.1136/bmj.4.5787.586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Singlas E, Goujet MA, Simon P. Pharmacokinetics of perhexiline maleate in anginal patients with and without peripheral neuropathy. Eur J Clin Pharmacol. 1978;14:195–201. doi: 10.1007/BF02089960. [DOI] [PubMed] [Google Scholar]
- 3.Morgan MY, Reshef R, Shah RR, Oates NS, Smith RL, Sherlock S. Impaired oxidation of debrisoquine in patients with perhexiline liver injury. Gut. 1984;25:1057–64. doi: 10.1136/gut.25.10.1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cooper RG, Evans DAP, Whibley EJ. Polymorphic hydroxylation of perhexiline maleate in man. J Med Genet. 1984;21:27–33. doi: 10.1136/jmg.21.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kennedy JA, Unger SA, Horowitz JD. Inhibition of carnitine palmitoyltransferase-1 in rat heart and liver by perhexiline and amiodarone. Biochem Pharmacol. 1996;52:273–80. doi: 10.1016/0006-2952(96)00204-3. [DOI] [PubMed] [Google Scholar]
- 6.Kennedy JA, Kiosoglous AJ, Murphy GA, Pelle MA, Horowitz JD. Effect of perhexiline and oxfenicine on myocardial function and metabolism during low-flow ischemia/reperfusion in the isolated rat heart. J Cardiovasc Pharmacol. 2000;36:794–801. doi: 10.1097/00005344-200012000-00016. [DOI] [PubMed] [Google Scholar]
- 7.Cole PL, Beamer AD, McGowan N, Cantillon CO, Benfell K, Kelly RA, Hartley LH, Smith TW, Antman EM. Efficacy and safety of perhexiline maleate in refractory angina. A double-blind placebo-controlled clinical trial of a novel antianginal agent. Circulation. 1990;81:1260–70. doi: 10.1161/01.cir.81.4.1260. [DOI] [PubMed] [Google Scholar]
- 8.Stewart S, Voss DW, Northley DL, Horowitz JD. Relationship between plasma perhexiline concentration and symptomatic status during short-term perhexiline therapy. Ther Drug Monit. 1996;18:635–9. doi: 10.1097/00007691-199612000-00002. [DOI] [PubMed] [Google Scholar]
- 9.Willoughby SR, Stewart S, Chirkov YY, Kennedy JA, Holmes AS, Horowitz JD. Beneficial clinical effects of perhexiline in patients with stable angina pectoris and acute coronary syndromes are associated with potentiation of platelet responsiveness to nitric oxide. Eur Heart J. 2002;23:1946–54. doi: 10.1053/euhj.2002.3296. [DOI] [PubMed] [Google Scholar]
- 10.Sallustio BC, Westley IS, Morris RG. Pharmacokinetics of the antianginal agent perhexiline: relationship between metabolic ratio and steady-state dose. Br J Clin Pharmacol. 2002;54:107–14. doi: 10.1046/j.1365-2125.2002.01618.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Davies BJL, Coller JK, James HM, Somogyi AA, Horowitz JD, Sallustio BC. Clinical inhibition of CYP2D6-catalysed metabolism by the antianginal agent perhexiline. Br J Clin Pharmacol. 2004;57:456–63. doi: 10.1046/j.1365-2125.2003.02033.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zanger UM, Fischer J, Raimundo S, Stüven T, Evert BO, Schwab M, Eichelbaum M. Comprehensive analysis of the genetic factors determining expression and function of hepatic CYP2D6. Pharmacogenetics. 2001;11:573–85. doi: 10.1097/00008571-200110000-00004. [DOI] [PubMed] [Google Scholar]
- 13.Barclay ML, Sawyers SM, Begg EJ, Zhang M, Roberts RL, Kennedy MA, Elliott JM. Correlation of CYP2D6 genotype with perhexiline phenotypic metabolizer status. Pharmacogenetics. 2003;13:627–32. doi: 10.1097/00008571-200310000-00006. [DOI] [PubMed] [Google Scholar]
- 14.Zanger UM, Raimundo S, Eichelbaum M. Cytochrome P450 2D6: overview and update on pharmacology, genetics, biochemistry. Naunyn Schmiedebergs Arch Pharmacol. 2004;369:23–37. doi: 10.1007/s00210-003-0832-2. [DOI] [PubMed] [Google Scholar]
- 15.Løvlie R, Daly AK, Matre GE, Molven A, Steen VM. Polymorphisms in CYP2D6 duplication-negative individuals with the ultrarapid metabolizer phenotype: a role for the CYP2D6*35 allele in ultrarapid metabolism? Pharmacogenetics. 2001;11:45–55. doi: 10.1097/00008571-200102000-00006. [DOI] [PubMed] [Google Scholar]
- 16.Raimundo S, Fischer J, Eichelbaum M, Griese EU, Schwab M, Zanger UM. Elucidation of the genetic basis of the common ‘intermediate metabolizer’ phenotype for drug oxidation by CYP2D6. Pharmacogenetics. 2000;10:577–81. doi: 10.1097/00008571-200010000-00001. [DOI] [PubMed] [Google Scholar]
- 17.James HM, Coller JK, Gillis D, Bahnisch J, Sallustio BC, Somogyi AA. A new simple diagnostic assay for the identification of the major CYP2D6 genotypes by DNA sequencing analysis. Int J Clin Pharmacol Ther. 2004;42:719–23. doi: 10.5414/cpp42719. [DOI] [PubMed] [Google Scholar]
- 18.Raimundo S, Toscano C, Klein K, Fischer J, Griese EU, Eichelbaum M, Schwab M, Zanger UM. A novel intronic mutation, 2988G>A, with high predictivity for impaired function of cytochrome P4502D6 in white people. Clin Pharmacol Ther. 2004;76:128–38. doi: 10.1016/j.clpt.2004.04.009. [DOI] [PubMed] [Google Scholar]
- 19.Marez D, Sabbagh N, Legrand M, Lo-Guidice JM, Boone P, Broly F. A novel CYP2D6 allele with an abolished splice recognition site associated with the poor metabolizer phenotype. Pharmacogenetics. 1995;5:305–11. doi: 10.1097/00008571-199510000-00006. [DOI] [PubMed] [Google Scholar]
- 20.Griese EU, Zanger UM, Brudermanns U, Gaedigk A, Mikus G, Mörike K, Stüven T, Eichelbaum M. Assessment of the predictive power of genotypes for the in vivo catalytic function of CYP2D6 in a German population. Pharmacogenetics. 1998;8:15–26. doi: 10.1097/00008571-199802000-00003. [DOI] [PubMed] [Google Scholar]
- 21.Lundqvist E, Johansson I, Ingelman-Sundberg M. Genetic mechanisms for duplication and multiplication of the human CYP2D6 gene and methods for detection of duplicated CYP2D6 genes. Gene. 1999;226:327–38. doi: 10.1016/s0378-1119(98)00567-8. [DOI] [PubMed] [Google Scholar]
- 22.Chou WH, Yan FX, Robbins-Weilert DK, Ryder TB, Liu WW, Perbost C, Fairchild M, de Leon J, Koch WM, Wedlund PJ. Comparison of two CYP2D6 genotyping methods and assessment of genotype–phenotype relationships. Clin Chem. 2003;49:542–51. doi: 10.1373/49.4.542. [DOI] [PubMed] [Google Scholar]
- 23.Sorensen LB, Sorensen RN, Miners JO, Somogyi AA, Grgurinovich N, Birkett DJ. Polymorphic hydroxylation of perhexiline in vitro. Br J Clin Pharmacol. 2003;55:635–8. doi: 10.1046/j.1365-2125.2003.01805.x. [DOI] [PMC free article] [PubMed] [Google Scholar]