Abstract
Aim
To develop a population pharmacokinetic (PK) model using sparse sampling of long-term treatment with paroxetine in elderly depressed subjects, incorporating CYP2D6 genotype as well as other covariates.
Methods
Elderly subjects (age ≥70 years) with nonpsychotic, nonbipolar major depressive disorder from the inpatient and outpatient clinic were treated with paroxetine in a 5-year clinical trial investigating ‘Maintenance Therapies in Late-Life Depression’ (MTLD-2). Plasma concentrations were collected during regular visits. CYP2D6 genotype was determined using polymerase chain reaction (PCR) for each individual. A nonlinear mixed-effects model was developed with NONMEM® for these subjects who received 10–40 mg day−1 of paroxetine during treatment. One- and two-compartment models with linear and nonlinear elimination (Michaelis–Menten) were evaluated. PK parameters as well as interindividual and residual variability were estimated. The effects of age, weight, sex, race and CYP2D6 genotypes on the pharmacokinetics of paroxetine were evaluated.
Results
One hundred and seventy-one subjects with a mean age of 77 years (range 69–95) and a mean weight of 72.0 kg (range 32.9–137.0) were enrolled in the MTLD-2 clinical trial. A total of 1970 paroxetine concentrations were available for population PK analyses. Approximately 10 samples were taken per subject. A two-compartment nonlinear PK model with additive and proportional error provided the best base model for description of the data. Weight and CYP2D6 polymorphisms were found to have a significant effect on maximal velocity (Vm), whereas sex had an effect on volume of distribution of the central compartment. The Vm estimates in each of the CYP2D6 phenotypic groups were: 125 µg h−1 in poor metabolizer (n = 1), 182 µg h−1 in intermediate metabolizers (n = 28), 454 µg h−1 in extensive metabolizers (n = 36) and 3670 µg h−1 in ultra-rapid metabolizers (n = 5).
Conclusions
The population PK model adequately described paroxetine data in this elderly depressed population. The data indicate that female and male subjects with different CYP2D6 polymorphisms have different elimination rates and therefore may need to be dosed differently based on metabolizer genotype.
Keywords: CYP2D6 polymorphism, geriatrics, NONMEM, paroxetine, pharmacokinetics
Introduction
Selective serotonin reuptake inhibitors (SSRIs) are the first-line antidepressants used in primary care and psychiatric practices. The response rate after the first drug administration can be as low as 60% in the general adult population and 39% in the geriatric population [1]. Large interindividual variability (IIV) has been found in pharmacokinetic (PK) parameters such as clearance (CL), half-life, area under the curve (AUC), and pharmacodynamic (PD) parameters such as time to response, recurrence, and side-effects [2–7]. This represents a significant clinical problem in the treatment of psychiatric illness in geriatric subjects. Some studies have suggested that the PK of SSRIs are associated with drug effect [8, 9]. Thus, understanding the IIV in PK is important for a PD study.
Paroxetine, one of the most potent SSRIs, is widely used in the treatment of depression and anxiety [1]. A wide range of IIVs was observed for the PK parameters of paroxetine [10–13]. Following single or multiple administration of paroxetine at doses of 20–50 mg, the mean elimination half-life for healthy subjects was approximately 24 h, with a range of 7–65 h having been reported [5]. Elderly subjects taking paroxetine have higher plasma concentrations and slower elimination than younger subjects [5, 13]. Although plasma concentrations have not yet been correlated with paroxetine response or adverse events, these results suggested the initial dosage in the elderly subjects should be reduced.
Paroxetine was mainly metabolized by CYP2D6 [5, 12]. More than 80 allelic variants have been identified for the CYP2D6 gene among different ethnic populations [14]. These polymorphisms result in variable enzymatic activity and drug-metabolizing phenotypes which can be classified as poor (PM), intermediate (IM), extensive (EM) and ultra-rapid (UM) metabolizers [15, 16]. A limited number of studies have reported an association between CYP2D6 polymorphism and paroxetine PK. Two studies have investigated the differences in paroxetine PK between EMs and PMs [11, 12]. A sevenfold difference in the median AUC0–∞ was found for healthy subjects receiving a single dose of 30 mg of paroxetine and a twofold difference in the median AUC at steady state (AUCss) after multiple 30-mg doses [11]. The other study found a threefold difference in median steady-state concentration (Css) after multiple 30-mg doses [12]. Some studies have suggested that the distribution of CYP2D6 activity in EMs also displays substantial IIV. Ozdemir [17] found a twofold higher median Css in healthy heterozygous EMs when compared with healthy homozygous EMs receiving paroxetine (20 mg day−1). No PMs were included in this study.
Factors that contribute to the variability of paroxetine PK parameters in the geriatric depressed population have not been reported. The association between CYP2D6 polymorphisms and paroxetine PK in the geriatric population after chronic paroxetine treatment is unknown. To investigate this association, we applied a nonlinear mixed-effect modelling approach to characterize paroxetine PK in a placebo-controlled study (the MTLD-2 trial) of the efficacy of paroxetine in preventing recurrence of major depressive episodes in people aged ≥70 years. The mixed effect population PK approach is the study of the sources and correlates of variability in plasma concentrations between individuals [18], which is currently widely used in evaluation of drug safety and efficacy. Compared with the traditional pharmacokinetic approach, population PK is more suitable for analysing large-scale clinical trials, where only a few samples are available per subject.
The purpose of this study was: (i) to apply a nonlinear mixed effect modelling approach to describe paroxetine PK parameters using limited sampling in a large number of geriatric subjects from the MTLD-2 clinical trial, and (ii) to evaluate the impact of covariates including age, weight, sex, race and CYP2D6 polymorphisms on the PK parameters.
Subjects and methods
The Maintenance Therapies in Late-Life Depression (MTLD-2) study [19] assessed paroxetine as a maintenance treatment for prevention of recurrent episodes of major depression in geriatric subjects. Subjects (aged ≥70 years) were included if they met the diagnostic criteria from the Structured Clinical Interview for DSM-IV Axis I disorders (SCID) for a current episode of major depressive disorder, nonbipolar, nondelusional and not actively suicidal. Subjects also were required to score ≥15 on the 17-item Hamilton Rating Scale for Depression (HAMD-17) and ≥18 on the Folstein Mini-Mental State Exam (MMSE). All the study sites were located in Pittsburgh. The study was approved by institutional review committee of the University of Pittsburgh and written informed consent to participate was obtained from each subject.
There were three phases of paroxetine treatment. Subjects were treated during an initial acute phase (which could last up to 26 weeks) to assess for response, followed by a 16-week continuation phase for those subjects who responded. Subjects with a continued response (recovered based on the 16-week continuation phase) entered the 2-year maintenance phase and were randomly assigned to a placebo group or to paroxetine. A responder was defined by a HAMD-17 score of <10 for three consecutive weeks. Recovery was defined as being free of significant depressive symptoms for 16 weeks of continuation treatment. Recurrence in maintenance treatment was defined by a HAMD-17 score >15 for at least two consecutive weeks and meeting SCID criteria for syndromal major depressive episode, confirmed by an independent senior geriatric psychiatrist. The acute and continuation phases were open-label and the maintenance phase was double-blind. Subjects visited the clinic weekly during acute treatment, twice monthly during continuation treatment and monthly during maintenance treatment. Plasma paroxetine samples were taken at each visit for concentration measurement. No specific timing was scheduled for the paroxetine sampling. The dosage time was noted for inpatient subjects and was self-reported by outpatients.
Paroxetine was started at 10 mg daily and could be titrated to a higher dose based on response. Subjects received paroxetine doses ranging from 10 mg to 40 mg daily. De-identified data were applied in a population pharmacokinetic analysis, where the identification (ID) number for each subject was changed by replacing the original ID numbers by a randomly generated number.
Analytical procedures
Paroxetine plasma concentrations were determined by the high-performance liquid chromatography technique, as previous described [20]. Briefly, plasma was extracted using ethyl acetate and heptane (1 : 4, v/v) and back extracted into 0.025 m potassium phosphate, pH 2.4. Separation was achieved using a Beckman Column Ultrasphere C18 (150 × 2 mm; Rainin Instrument Co., Woburn, MA, USA). Detection wavelength was 205 nm and flow rate was 0.35 ml min−1. Fluoxetine hydrochloride was used as the internal standard. The limit of quantification was 5 ng ml−1. The linear range was 5–500 ng ml−1 with interassay variability ranging from 3.4 to 5.4% for spiked controls.
CYP2D6 genotyping
After separating lymphocytes from whole blood using BD Vacutainer CPTTM tubes (BD, Franklin Lakes, NJ, USA), DNA was extracted using the standard procedure [21, 22]. Genomic DNA fractions were stored at −20 °C.
A previously described [23] polymerase chain reaction (PCR)-based allele-specific analysis was used to determine whether individuals were carrying duplicated CYP2D6 genes (CYP2D6 *XN) and long PCR was used to amplify a fragment spanning the potential crossing-over sites [23, 24]. An allele-specific long PCR method developed by Steen et al. [24, 25] was used to detect CYP2D6 *5 (gene deletion). Nested PCR was performed to detect CYP2D6 *2, *4, *10 and *17 by amplifying the entire CYP2D6 gene (5 kb) [23, 26–28]. After amplification of the entire gene, subsequent internal PCR was performed to identify the presence of the CYP2D6*2 (C2938T), *4 (G1934A), *10 (G4286C) and *17 (C1111T) allele. When no mutations were found, the allele was defined as CYP2D6 *1. The specific primers, restriction enzyme, restriction pattern and agarose gel for these alleles are shown in Table 1.
Table 1A.
Conditions of the CYP2D6 genotyping study, including the specific primers, restriction enzyme, restriction pattern and agarose gel aLong PCR for CYP2D6 genotype determination
| CYP2D6 allele | Specific primers | PCR product (kb) | Agarose gels, % |
|---|---|---|---|
| 2D6 | F: 5′-CCAGAAGGCTTTGCAGGCTTCA-3′R: 5′-ACTGAGCCCTGGGAGGTAGGTA-3′ | 5.0 | 0.85 |
| 2D6-dup | F: 5′-CCTGGGAAGGCCCCA TGGAAG-3′R: 5′-CAGTTA CGGCAGTGGTCAGCT-3′ | 3.5 | 0.85 |
| *5 (deletion) | F: 5′-ACCAGGCACCTGTACTCCTCA-3′R: 5′-GCATGAGCTAAGGCACCCAGAC-3′ | 3.5 | 0.9 |
Table 1B.
Re-amplification reactions performed for CYP2D6 genotype determination
| CYP2D6 allele (mutation) | Specific primers | Restriction enzyme | Restriction pattern (bp) | Agarose gels, % |
|---|---|---|---|---|
| *2 (C2938T) | F: 5′-AGGCCTTCCTGGCAGAGATGGAG-3′ R: 5′-CCCCTGCACTGTTTCCCAGA-3′ | cfo I | wt:260, 126 mut: 386 | 2.0 |
| *4 (G1934A) | F: 5′-TGCCGCCTTCGCCAACCACT-3′ R: 5′-CTCGGTCTCTCGCTCCGCAC-3′ | Bst NI | wt:292 mut: 111, 181 | 1.5 |
| *10 (G4286C) | F: 5′-GAGACAAACCAGGACCTGCCA-3′ R: 5′-GCCTCAACGTACCCCTGTCTC-3′ | Bst EII | wt:860 mut: 240, 620 | 1.8 |
| *17 (1111C) (wt) | F: 5′-CCAAGGTTCAAATAGGACTA-3′ | wt: 237 | 1.5 | |
| *17 (1111T) (mut) | F: 5′-CCAAGGTTCAAATAGGACTA-3′ R: 5′-CCCGAAACCCAGGATCTGGA-3′ | mut: 237 | 1.5 |
wt, Wild type; mut, mutant; F, forward primer; R, reverse primer.
The allelic frequency was calculated using the equation:
CYP2D6 genotype was classified into one of four phenotype groups (Table 2) based on the phenotype–genotype relationship reported in the literature [29, 30]. Subjects carrying two nonfunctional alleles (*0/*0) were assigned to the PM group. Subjects carrying one normal or reduced functional allele and one reduced or nonfunctional allele were assigned to the IM group. Subjects carrying two normal functional alleles were assigned to the EM group and subjects carrying one *XN allele were assigned to the UM group.
Table 2.
Genotype/phenotype frequencies in caucasian (CA) and African-American (AA) subjects in the MTLD-2 trial
| Allele status | Assigned phenotype | Genotype | CA Frequency (%) | n | AA Frequency (%) | n |
|---|---|---|---|---|---|---|
| 0 | PM | *0/*0 | 1.6 | 1 | 0 | 0 |
| 1 | IM | IM/*0, IM/IM, EM/*0, EM/IM | 39 | 25 | 75 | 1 |
| 2 | EM | EM/EM | 52 | 33 | 25 | 3 |
| 3 | UM | UM/*X | 7.8 | 5 | 0 | 0 |
0, Nonfunctional alleles (e.g. *4, *5); 1, one normal functional (EM: *1, *2) or reduced-functional allele (IM: *10, *17), plus a reduced or nonfunctional allele; 2, two functional alleles or *XN allele plus other allele (UM).
Population pharmacokinetic analysis
The population PK analysis includes the base model and final (covariate) model development. The base model defines the PK parameters and describes the plasma concentration–time profile. The final model describes the influence of fixed effects (i.e. demographic factors) on the PK parameters. Analysis platform, minimization methods, model building criteria and model validation are described below.
Analysis platform
Nonlinear mixed effects modelling was used for the population PK analysis using the NONMEM computer program (Version 5, level 1.1; University of California at San Francisco, CA, USA) [31, 32]. The models consisted of a structural model that described the disposition of the drug following oral administration, and a pharmacostatistical model that described the inter- and intraindividual variability. Diagnostic graphics, exploratory analyses and postprocessing of NONMEM outputs were performed using S-PLUS (Version 6.2; Insightful, Seattle, WA, USA).
Minimization methods and model building criteria
The first order estimation method (FO) was used for model building. The adequacy of the developed structural models was evaluated using both statistical and graphical methods. The likelihood ratio test was used to discriminate between alternative models. The likelihood ratio test was based on the property that the ratios of the NONMEM objective function values (OFV) (−2 log-likelihood) were asymptotically χ2 distributed. An objective function decrease of 3.84 units was considered significant (χ2P < 0.05 d.f. = 1). Standard errors for all parameters were obtained using the covariance option in NONMEM.
Base model development
Structure PK model
The structure PK model represents the best description of the data without considering the effect of subject-specific covariates. The population PK analysis was performed using NONMEM®[31, 32] with the subroutine ADVAN9, ADVAN2 TRANS2 and ADVAN4 TRANS4. Various structure models were tested, including one- and two-compartment models, model with linear, nonlinear elimination (Michaelis–Menten) and combination nonlinear with linear elimination.
Inter-individual variability
It was assumed that the IIV of the PK parameters was log-normally distributed. The relationship between a PK parameter (P) and its variance could therefore be expressed as shown below:
where, Pj is the value of the PK parameter for the jth individual, PTV is the typical value of P for the population, and ηP denotes the difference between Pj and PTV, independently, which was identically distributed with a mean of zero and variance of ωP2.
Intra-individual variability
The residual variability, which was comprised of, but not limited to, intra-individual variability, experimental errors, process noise and/or model misspecifications, was modelled using additive, proportional and combined error structures as described below:
where yij is the jth observation in the ith individual, yij is the corresponding model prediction, and ɛij (or ɛij′) is a normally distributed random error with a mean of zero and a variance of σ2.
Final model development
The final model was developed by testing the effect of subject specific covariates, including age, weight, sex, race and CYP2D6 polymorphisms on PK parameter estimates. The two types of covariates, including continuous covariates (e.g. age and weight) and discrete covariates (e.g. sex, race and CYP2D6 polymorphisms) were introduced into each parameter in a stepwise fashion. The following example showed the effect of a continuous covariate on Vm (maximal rate):
where TVVm is the typical value for the population and ηi is the random effect representing the difference of the ith subject from the population mean. The random effects of between-subject variability were assumed to be log-normally distributed, with a mean of zero and SD of ω. Cov was the continuous covariate that was affecting Vm and MedCov was the median Cov.
The following example shows the effect of a discrete covariate (sex) on Vm:
When sex was female (male = 0, female = 1), TVVm =θVm since numeric value for (1-female) = 0 resulting in a zero multiplier for the covariate effect. For male subjects, the θSex term was added to the population estimate of Vm to modify it.
Categorical variables were assigned to each of the four CYP2D6 phenotype groups and for the subjects without CYP2D6 phenotype information (i.e. PMs = 1, IMs = 2, EMs = 3, UMs = 4, Missing = 0). The incorporation of this covariate is shown here for the parameter Vm below:
The graphical assessment of POSTHOC parameter estimates vs. covariates was evaluated to help identify possible covariate relationships using S-PLUS 6.2. In addition, goodness of fit plots were utilized to assess model robustness [33]. The covariate was retained in the model if it decreased the objective function value (OFV) by 3.84 (χ2P < 0.05, d.f. = 1). Covariate influence on interindividual variability and goodness of fit was also examined. In cases where the covariate value was not recorded at any time during the study for the subject, the median value calculated from the population dataset was used.
Results
Patient characteristics
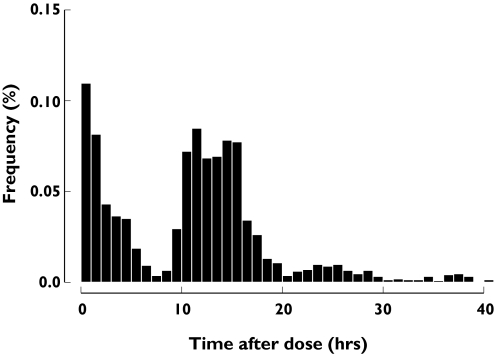
The MTLD-2 clinical trial included 171 elderly subjects (58 males) who provided 1970 paroxetine concentrations. Subjects had an average (mean ± SD) age and weight of 77.1 ± 5.7 years and 72.0 ± 16.4 kg, respectively (Table 3). With the exception of three subjects who were 69 years of age, subjects aged ≥70 years were included in the MTLD-2 study. The majority of the subjects were caucasian (CA) (n = 156) and only 15 subjects were African-American (AA). The distribution of paroxetine sampling time (time after dose) is shown in Figure 1.
Table 3.
Patient characteristics for the MTLD-2 study
| Demographics | Mean ± SD (range) |
|---|---|
| Sample size | 171 |
| No. of observations | 1970 |
| Age (years) | 77.1 ± 5.8 (69–95) |
| Weight (kg) | 72.0 ± 16.4 (32.9–137.0) |
| Gender: Male | 58 |
| Female | 113 |
| Race: Caucasian | 156 |
| African-American | 15 |
Figure 1.
Frequency histogram showing the sampling distribution for paroxetine sampling measurements. The abscissa is broken into 1-h bins. The ordinate is the proportion of samples taken in each interval
CYP2D6 genotyping
CYP2D6 genotype was classified into one of four phenotype groups (Table 2) based on phenotype–genotype relationship reported in the literature, as described in Subjects and methods [29, 30]. Of the 171 subjects, whole blood was available from 68 subjects for CYP2D6 genotyping analysis. Of these 68 subjects, four were AA and 64 were CA. Five subjects were identified as UMs, 36 as EMs, 26 as IMs and one subject as a PM.
The frequency of each CYP2D6 allele is summarized in Table 4. The CYP2D6 *17 allele, an African and African-American specific allele found in previous studies [16, 34], was found in only 25% of the AAs in this study. CYP2D6 *2N was found only in CA subjects.
Table 4.
CYP2D6 allele frequency in caucasian (CA) and African-American (AA) patients
| CYP2D6 allele | CA, n | Allele frequency in CA | AA, n | Allele frequency in AA |
|---|---|---|---|---|
| *1 | 48 | 0.38 | 1.00 | 0.13 |
| *2 | 45 | 0.35 | 4.00 | 0.50 |
| *4 | 22 | 0.17 | 1.00 | 0.13 |
| *5 | 4 | 0.03 | 0.00 | 0.00 |
| *17 | 0 | 0.00 | 2.00 | 0.25 |
| *10 | 4 | 0.03 | 0.00 | 0.00 |
| *2 × 2 | 5 | 0.04 | 0.00 | 0.00 |
Population PK modelling
Base model
The population PK analysis was performed by using NONMEM® (version V; GloboMax, Hanover, MD, USA) [31] with the subroutine ADVAN9. A two-compartment nonlinear model with exponential interindividual variability on Vm, the Michaelis–Menten constant (Km) and volume of distribution of the central compartment (V2) adequately described the data. The best residual error model was a combined additive and proportional model. The basic PK parameters of Vm, Km, V2, volume of distribution of the peripheral compartment (V3) and absorption rate constant Ka were shown in Table 5.
Table 5.
Pharmacokinetic parameter estimates for the two-compartment model
| Parameters | Base model estimates | SE % | Parameters | Final model estimates | SE % |
|---|---|---|---|---|---|
| Vm (µg h−1) | 208 | 32.5 | Vm (µg h−1) | 474 | 19.3 |
| Km (µg l−1) | 157 | 83.4 | Km (µg l−1) | 205 | 24.67 |
| V2 (l) | 254 | 6.70 | V2 (l per 75 kg wt) | 230 | 8.60 |
| V3 (l) | 2350 | 102.1 | V3 (l) | 900 | 81.1 |
| Q (l h−1) | 1.33 | 12.3 | Q (l h−1) | 1.05 | 25.3 |
| Ka (h−1) | 9.24 | 8.8 | Ka (h−1) | 9.81 | 28.0 |
| PM (θVm) | N/A | N/A | PM (θVm) | 125 | 48.8 |
| IM (θVm) | N/A | N/A | IM (θVm) | 182 | 19.4 |
| EM (θVm) | N/A | N/A | EM (θVm) | 454 | 49.5 |
| UM (θVm) | N/A | N/A | UM (θVm) | 3670 | 34.6 |
| Wt (θV2) | N/A | N/A | Wt (θV2) | 1.83 | 50.1 |
| Sex (θV2) | N/A | N/A | Sex (θV2) | 99.3 | 42.9 |
| ωVm% | 155 | 168.8 | ωVm% | 90.0 | 57.3 |
| ωKm% | 79 | 19.4 | ωKm% | 109 | 56.1 |
| ωv2% | 134 | 245.3 | ωv2% | 77.8 | 18.2 |
| σ1% | 40 | 14.0 | σ1% | 35.1 | 14.4 |
| σ2 (µg l−1) | 10.8 | 90.6 | σ2 (µg l−1) | 8.60 | 103 |
Vm, Maximal rate; KmMichaelisvMenten constant (concentration at half Vm); SE, standard error; Wt, total body weight; V2, volume of distribution of central compartment; V3volume of distribution of peripheral compartment;ωcoefficient of variation of interindividual variability;σ, coefficient of variation of residual error; N/A, not available; unit of weight = kg.
A two-compartment model was determined to be the most robust, based on the Akaike information criterion (AIC) [35] (AIC = 17265.0, one-compartment; AIC = 17149.9, two-compartment). Moreover, the decrease in residual error (50% decrease in additive residual error) and bias of data fitting were also observed. The nonlinear elimination model improved OFV by 114.0 units (P < 0.001) compared with the linear elimination model. A combination of linear and nonlinear elimination models did not further improve model fitness or reduce the OFV value (ΔOFV = − 3.2, P >0.05).
Final model
CYP2D6 phenotype was the covariate on Vm that resulted in the largest reduction in objective function value (ΔOFV = −137.9; P < 0.005). Weight was a significant covariate on V2 (ΔOFV = −69.64; P < 0.005). After incorporating the CYP2D6 phenotypic effect on Vm, sex was a significant covariate on V2 (ΔOFV = −107.1; P < 0.005). After incorporating the CYP2D6 phenotype on Vm and sex on V2, the incorporation of weight on Vm further improved model fitness by reducing OFV by 62.66 units (P < 0.005). The detailed covariate selection during model development is shown in Table 6. The final model Vm and V2 was:
Table 6.
Population pharmacokinetic model development (two-compartment with nonlinear elimination)
| Covariate | Model | −2LL | Δ-2LL | P-value |
|---|---|---|---|---|
| 1 | Base model | 17 137.900 | ||
| 2–1 | ||||
| Vm | ||||
| CYP2D6 | M1 | 16 977.144 | −160.76 | <0.005 |
| Wt | M2 | 17 079.98 | −57.92 | <0.005 |
| Age | M3 | 17 143.06 | 5.155 | >0.05 |
| Race | M4 | 17 133.65 | −4.25 | <0.05 |
| Sex | M5 | 17 070.42 | −67.48 | <0.005 |
| 2–2 | ||||
| V2 | ||||
| CYP2D6 | M6 | 17 071.83 | −66.07 | <0.005 |
| Wt | M7 | 17 068.262 | −69.64 | <0.005 |
| Age | M8 | 17 134.346 | −3.55 | >0.05 |
| Race | M9 | 17 142.224 | 4.32 | >0.05 |
| Sex | M10 | 17 080.828 | −57.07 | <0.005 |
| 3–1 | ||||
| Vm, V2 | ||||
| Vm(CYP2D6, Wt) | M11 | 16 876.32 | −100.82 | <0.005 |
| Vm(CYP2D6), V2(Wt) | M12 | 16 900.59 | −76.55 | <0.005 |
| Vm(CYP2D6), V2(sex) | M13 | 16 892.90 | −84.24 | <0.005 |
| Vm(CYP2D6, race) | M14 | 16 977.165 | 0.021 | >0.05 |
| 3–2 | ||||
| Vm and V2 | ||||
| Vm(CYP2D6), V2(sex, Wt) | M15 | 16 861.53 | −31.37 | <0.005 |
| Vm(CYP2D6, Wt), V2(sex) | M16 | 16 830.24 | −62.66 | <0.005 |
Wt, Weight; V2, volume of distribution of central compartment;Δ-2LL was objective function value (OFV) from covariate model minus base model; −2LL values in 2–1 and 2–2 were compared with base model; −2LL values in 3–1 were compared with model M1; while values in 3–2 were compared with 3–1 M13 and M11. The incorporation of covariates is described in Subjects and methods.
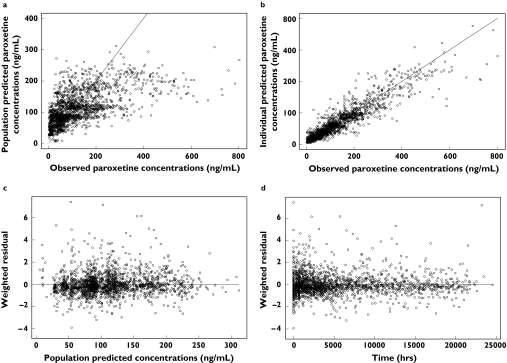
The final PK parameter estimates are shown in Table 5. Diagnostic plots are shown in Figure 2, including observed paroxetine concentrations vs. population predicted paroxetine concentrations (Figure 2a); observed paroxetine concentrations vs. individual predicted paroxetine concentrations (Figure 2b); weighted residual error (WRES) vs. population predicted concentrations (Figure 2c); and WRES vs. time (Figure 2d). Compared with the base model, proportional and additive residual error was reduced by 12.5% and 20.4%, respectively, in the final model. The interindividual variability in Vm and V2 both decreased by 41.9%. The standard error (SE) of IIV estimation of Vm was reduced by 66.1% and SE of IIV estimation of V2 was reduced by 92.6%. The SE of Vm, Km and V3 estimates were also decreased in the final model. However, the estimation of IIV on Km and the SE of IIV of Km increased.
Figure 2.
Diagnostic plots of final pharmacokinetic model. (a) Plot of population predicted paroxetine concentrations vs. observed paroxetine concentrations. Individual data points are shown as dots and the unity line is shown as a solid line. (b) Plot of individual population predicted paroxetine concentrations vs. observed paroxetine concentrations. Individual data points are shown as dots and the unity line is shown as a solid line. (c) Plot of weighted residual error (WRES) vs. population predicted concentrations. (d) Plot of WRES vs. time
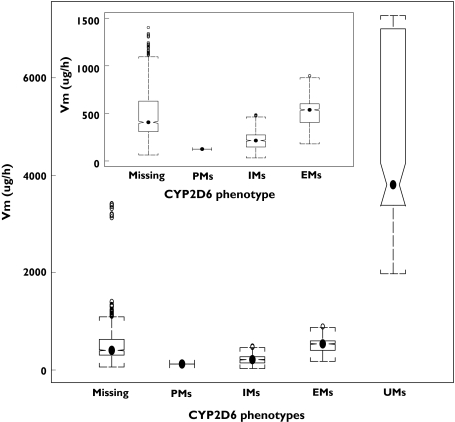
The order of magnitude for the Vm estimates by CYP2D6 phenotype was: UMs > EMs > IMs > PMs (Figure 3), which corresponded to the functional allele of the CYP2D6 gene. The population mean (%SE) of Vm estimates in the final model for each CYP2D6 phenotype group were: 125 µg h−1 (48.8%) in PM, 182 µg h−1 (19.4%) in IMs, 454 µg h−1 (49.5%) in EMs and 3670 µg h−1 (34.6%) in UMs. The 95% confidence intervals (CI) of Vm in each phenotype group were: 64.41, 444.09 µg h−1 in IMs, 191.35, 895.96 µg h−1 in EMs and 2073.70, 7006.30 µg h−1 in UMs (95% CI is unavailable for PM group with n = 1). The estimates of V2 in male subjects were 461.30 ± 259.75 l and in female subjects were 346.41 ± 255.81 l.
Figure 3.
Box plot of Vm estimates for each CYP2D6 phenotype group. Dots in each group were median values. Notches show approximate 95% confidence limits for the median. CYP2D6 genotype was classified into one of the four CYP2D6 phenotype groups based on the phenotype–genotype relationship. In this plot, PMs = poor metabolizers, IMs = Intermediate metabolizers, EMs = extensive metabolizers, UMs = ultra-rapid metabolizers. Missing = Subject was missing CYP2D6 phenotype information
Age did not affect paroxetine disposition in this study, although the Vm estimates in subjects aged ≥80 years appeared to be lower than in subjects <80 years. The median (25th and 75th percentile) of Vm estimates in subjects aged ≥80 years was 275 µg h−1 (198 µg h−1 and 468 µg h−1); in subjects aged <80 years was 419 µg h−1 (291 µg h−1 and 620 µg h−1).
Race was a significant covariate on paroxetine PK if CYP2D6 phenotype was not included in the PK model (Table 3). However, once the CYP2D6 phenotype was included in the model, race did not significantly impact on paroxetine PK parameters.
Discussion
The molecular basis of the CYP2D6 polymorphism has been intensively studied and more than 80 allelic variants, including nonfunctional, normal, reduced or increased functional alleles, have been identified for the CYP2D6 gene among different ethnic populations [14, 16]. These polymorphisms result in variable enzymatic activity and drug-metabolizing phenotypes, which can be classified as PMs, IMs, EMs and UMs metabolizers [15, 16]. Paroxetine is mainly metabolized by the CYP2D6 enzyme. However, the relationship between CYP2D6 genotype and paroxetine PKs in the geriatric population has not been reported. This is the first study to assess the impact of CYP2D6 genotype as well as other factors (e.g. weight, sex, age and race) on paroxetine PK by using a population modelling approach in a geriatric depressed population with small a number of samples per subject. In addition, we have captured the individual specific drug exposure magnitude over time. This provides a basis where the magnitude of exposure can be examined in conjunction with the maintenance response of subjects in this study in a future study as response data become available.
Both one- and two-compartment nonlinear PK models demonstrated that CYP2D6 polymorphisms and weight were significantly related to paroxetine Vm and sex significantly impacted on V2. The two-compartment PK model was a better description of the data than the one-compartment model, based on the significantly reduced OFV value and a better goodness of fit. The Vm estimates in each CYP2D6 phenotype group showed that UMs had a higher Vm than other CYP2D6 phenotype groups, while PMs had the lowest Vm population estimate. The order of magnitude for Vm estimates by CYP2D6 phenotype was: UMs > EMs > IMs > PMs and the Vm estimates were: 125 µg h−1 in PMs, 182 µg h−1 in IMs, 454 µg h−1 in EMs and 3670 µg h−1 in UMs. The order of magnitude of the Vm estimate was consistent with the CYP2D6 functional alleles, where the UM phenotype could be caused by alleles carrying multiple 2D6 gene copies [16, 26] and the PM phenotype was the result of inheriting any two nonfunctional (null) alleles (genotype *0/*0). The IM phenotype was the result of both heterozygosity for a null allele and homozygosity for two alleles with impaired function (e.g. *9, *10, *17). Moreover, the model was able to differentiate IM from PM groups. Comparing the differences of Vm estimates between CYP2D6 genotype groups, Vm estimates between PMs (125 µg h−1) and IMs (182 µg h−1) were similar, which agrees with the suggestion from several studies that the IM phenotype is of clinical importance because drug PK in IMs could be more similar to the PMs than to the normal EMs, especially after chronic treatment [30]. The Vm for subjects without having CYP2D6 genotype information was 474 µg h−1, which is similar to the estimate of Vm in the EMs (454 µg h−1), since EM is the most frequent genotype in both the CA and AA populations.
Nonlinear PK of paroxetine was found in EMs in the study of Sindrup et al. [11, 12]. One explanation for this finding is saturation of CYP2D6 metabolic capacity, where CYP2D6 enzyme activity seems readily to saturate as the paroxetine dose increases, as demonstrated by Sindrup et al. [12] and Preskorn [36]. Another possibility relates to the self-inhibition of paroxetine metabolism, since paroxetine itself can inhibit CYP2D6 enzyme activity [12, 37]. Without considering self-inhibition, the model with nonlinear elimination (Michaelis–Menten) could underpredict paroxetine concentrations. However the result of MTLD-2 data analysis did not support the self-inhibition mechanism, as no bias or underprediction was found in the diagnostic plots of the weighted residual vs. predicted paroxetine concentrations and the weighted residual vs. time. Moreover, the model was tested with different elimination mechanisms (e.g. simple noncompetitive inhibition and uncompetitive inhibition [38, 39]; data not shown). Model fitness was not significantly improved based on OFV values and diagnostic plots.
Previous studies have reported that paroxetine was mainly metabolized by the high-affinity enzyme CYP2D6 [5, 12] and the low-affinity enzyme, e.g. CYP3A4 [5, 11]. Accordingly, models with linear, nonlinear and combined linear and nonlinear elimination were evaluated. Results showed that the nonlinear elimination model improved the model fitness and significantly decreased the OFV compared with the linear elimination model. However, the combined model with linear and nonlinear elimination did not provide further improvement in model fitness.
Elderly subjects taking oral paroxetine had higher plasma concentrations than younger subjects [5, 13]. Age was identified as a significant covariate on the PK of another SSRI, citalopram [6]. Age was not a significant covariate on paroxetine PK in this study, although the Vm estimates in subjects aged ≥80 years appeared to be lower than in subjects <80 years. The small sample size in the MTLD-2 study may lead to a decreased ability to detect an age effect in this study.
Race was determined to be a significant covariate in both the one- and two-compartment PK models when CYP2D6 genotype information was not incorporated [40]. One possible explanation was related to the correlation between race and genotype. The frequency of *4, the most frequent null allele in CAs, was about threefold higher than in AAs [16, 26]. The CYP2D6 *17 allele was an African and AA-specific allele found in this and previous studies [16, 41]. CYP2D6 *2N was found only in CA subjects in this study. When the CYP 2D6 genotype was incorporated, this race effect was no longer significant. The frequency of UMs in CA was 8%, which may be more reflective of the Pittsburgh CA population, since the frequency of UMs is higher in Southern Europe (10%) than in North Europe (1–2%) [16].
Conclusion
The population PK model adequately described paroxetine PK parameters in subjects with late-life depression. The results suggest that weight, sex and genotype contribute to the variability in PK parameters and that individuals of different sex or with a different genotype may therefore need to be dosed differently from one another.
Conflict of interest
Y.F., R.E.F. and M.A.K.: none to declare. R.R.B. has received funding from the National Institute of Mental Health, National Cancer Institute and the National Institute for Bioimaging and Bioengineering. B.G.P. has received grant/research support from the National Institute of Mental Health, Janssen Pharmaceutica, Forest Pharmaceuticals and GlaxoSmithKline. He is a consultant for Forest Pharmaceuticals, Janssen Pharmaceuticals and GlaxoSmithKline. He is also a member of the speakers’ bureau of Forest Pharmaceuticals and Glaxo Smith Kline & Sepracor. C.F.R. has received grant/research support from the National Institute of Mental Health, GlaxoSmithKline, Forest, Pfizer and Lilly.
Acknowledgments
The authors would like to acknowledge Kristin Bigos for her assistance in preparing this manuscript. Advanced Center for Interventions and Services Research for the Study of Late Life Mood Disorders: R37 MH43832 (the MTLD-2 study), P30 MH71944 (the late-life ACISR), P30 MH52247, R01 MH37869, MH65416, MH30915, MH55756 and NIH MH64173; National Institute for Biomedical Imaging and Bioengineering (NIBIB) Grant no. P41 EB001975-06.
References
- 1.Wagstaff AJ, Cheer SM, Matheson AJ, Ormrod D, Goa KL. Paroxetine: an update of its use in psychiatric disorders in adults. Drugs. 2002;62:655–703. doi: 10.2165/00003495-200262040-00010. [DOI] [PubMed] [Google Scholar]
- 2.Pollock BG, Ferrell RE, Mulsant BH, Mazumdar S, Miller M, Sweet RA, Davis S, Kirshner MA, Houck PR, Stack JA, Reynolds CF, Kupfer DJ. Allelic variation in the serotonin transporter promoter affects onset of paroxetine treatment response in late-life depression. Neuropsychopharmacology. 2000;23:587–90. doi: 10.1016/S0893-133X(00)00132-9. [DOI] [PubMed] [Google Scholar]
- 3.Reis M, Lundmark J, Bengtsson F. Therapeutic drug monitoring of racemic citalopram: a 5-year experience in Sweden, 1992–1997. Ther Drug Monit. 2003;25:183–91. doi: 10.1097/00007691-200304000-00007. [DOI] [PubMed] [Google Scholar]
- 4.Feng Y, Pollock B, Roose S, Kirshner M, Lotrich F, Kupfer D, Bies R. Mixed effects pharmacokinetic modeling of an age effect on citalopram disposition. AAPS Pharmsci. 2003;5 Abstract M1063. [Google Scholar]
- 5.Kaye CM, Haddock RE, Langley PF, Mellows G, Tasker TC, Zussman BD, Greb WH. A review of the metabolism and pharmacokinetics of paroxetine in man. Acta Psychiatr Scand Suppl. 1989;350:60–75. doi: 10.1111/j.1600-0447.1989.tb07176.x. [DOI] [PubMed] [Google Scholar]
- 6.Bies RR, Feng Y, Lotrich FE, Kirshner MA, Roose S, Kupfer DJ, Pollock BG. Utility of sparse concentration sampling for citalopram in elderly clinical trial subjects. J Clin Pharmacol. 2004;44:1352–9. doi: 10.1177/0091270004269647. [DOI] [PubMed] [Google Scholar]
- 7.Walters G, Reynolds CF3rd, Mulsant BH, Pollock BG. Continuation and maintenance pharmacotherapy in geriatric depression: an open-trial comparison of paroxetine and nortriptyline in patients older than 70 years. J Clin Psychiatry. 1999;60(Suppl. 20):21–5. [PubMed] [Google Scholar]
- 8.Meijer WE, Bouvy ML, Heerdink ER, Urquhart J, Leufkens HG. Spontaneous lapses in dosing during chronic treatment with selective serotonin reuptake inhibitors. Br J Psychiatry. 2001;179:519–22. doi: 10.1192/bjp.179.6.519. [DOI] [PubMed] [Google Scholar]
- 9.Henry ME, Moore CM, Kaufman MJ, Michelson D, Schmidt ME, Stoddard E, Vuckevic AJ, Berreira PJ, Cohen BM, Renshaw PF. Brain kinetics of paroxetine and fluoxetine on the third day of placebo substitution: a fluorine MRS study. Am J Psychiatry. 2000;157:1506–8. doi: 10.1176/appi.ajp.157.9.1506. [DOI] [PubMed] [Google Scholar]
- 10.Findling RL, Reed MD, Myers C, O'Riordan MA, Fiala S, Branicky L, Waldorf B, Blumer JL. Paroxetine pharmacokinetics in depressed children and adolescents. J Am Acad Child Adolesc Psychiatry. 1999;38:952–9. doi: 10.1097/00004583-199908000-00010. [DOI] [PubMed] [Google Scholar]
- 11.Sindrup SH, Brosen K, Gram LF, Hallas J, Skjelbo E, Allen A, Allen GD, Cooper SM, Mellows G, Tasker TC. The relationship between paroxetine and the sparteine oxidation polymorphism. Clin Pharmacol Ther. 1992;51:278–87. doi: 10.1038/clpt.1992.23. [DOI] [PubMed] [Google Scholar]
- 12.Sindrup SH, Brosen K, Gram LF. Pharmacokinetics of the selective serotonin reuptake inhibitor paroxetine: nonlinearity and relation to the sparteine oxidation polymorphism. Clin Pharmacol Ther. 1992;51:288–95. doi: 10.1038/clpt.1992.24. [DOI] [PubMed] [Google Scholar]
- 13.Sawamura K, Suzuki Y, Someya T. Effects of dosage and CYP2D6-mutated allele on plasma concentration of paroxetine. Eur J Clin Pharmacol. 2004;60:553–7. doi: 10.1007/s00228-004-0792-6. [DOI] [PubMed] [Google Scholar]
- 14.Human Cytochrome P450 (CYP) Allele Nomenclature Committee. [30 September 2005]; http://www.imm.ki.se/CYPalleles/ (last accessed:).
- 15.Kagimoto M, Heim M, Kagimoto K, Zeugin T, Meyer UA. Multiple mutations of the human cytochrome P450IID6 gene (CYP2D6) in poor metabolizers of debrisoquine. Study of the functional significance of individual mutations by expression of chimeric genes. J Biol Chem. 1990;265:17209–14. [PubMed] [Google Scholar]
- 16.Bradford LD. CYP2D6 allele frequency in European Caucasians, Asians, Africans and their descendants. Pharmacogenomics. 2002;3:229–43. doi: 10.1517/14622416.3.2.229. [DOI] [PubMed] [Google Scholar]
- 17.Ozdemir V, Tyndale RF, Reed K, Herrmann N, Sellers EM, Kalow W, Naranjo CA. Paroxetine steady-state plasma concentration in relation to CYP2D6 genotype in extensive metabolizers. J Clin Psychopharmacol. 1999;19:472–5. doi: 10.1097/00004714-199910000-00014. [DOI] [PubMed] [Google Scholar]
- 18.Aarons L. Population pharmacokinetics: theory and practice. Br J Clin Pharmacol. 1991;32:669–70. [PMC free article] [PubMed] [Google Scholar]
- 19.Thomas L, Mulsant BH, Solano FX, Black AM, Bensasi S, Flynn T, Harman JS, Rollman BL, Post EP, Pollock BJ, Reynolds CF., III Response speed and rate of remission in primary and specialty care of elderly patients with depression. Am J Geriatr Psychiatry. 2002;10:583–91. [PubMed] [Google Scholar]
- 20.Foglia JP, Sorisio D, Kirshner M, Pollock BG. Quantitative determination of paroxetine in plasma by high-performance liquid chromatography and ultraviolet detection. J Chromatogr B Biomed Sci Appl. 1997;693:147–51. doi: 10.1016/s0378-4347(97)00010-8. [DOI] [PubMed] [Google Scholar]
- 21.Bartlett J, White A. PCR Protocols. 2. Totowa, NJ: Humana Press; 2003. [Google Scholar]
- 22.Higuchi R. PCR TechnologyPrinciples and Applications for DNA Amplification. 2. New York: Oxford University Press; 1992. [Google Scholar]
- 23.Lundqvist E, Johansson I, Ingelman-Sundberg M. Genetic mechanisms for duplication and multiduplication of the human CYP2D6 gene and methods for detection of duplicated CYP2D6 genes. Gene. 1999;226:327–38. doi: 10.1016/s0378-1119(98)00567-8. [DOI] [PubMed] [Google Scholar]
- 24.Cheng S, Fockler C, Barnes WM, Higuchi R. Effective amplification of long targets from cloned inserts and human genomic DNA. Proc Natl Acad Sci USA. 1994;91:5695–9. doi: 10.1073/pnas.91.12.5695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Steen VM, Andreassen OA, Daly AK, Tefre T, Borresen AL, Idle JR, Gulbrandsen AK. Detection of the poor metabolizer-associated CYP2D6(D) gene deletion allele by long-PCR technology. Pharmacogenetics. 1995;5:215–23. doi: 10.1097/00008571-199508000-00005. [DOI] [PubMed] [Google Scholar]
- 26.Wan YJ, Poland RE, Han G, Konishi T, Zheng YP, Berman N, Lin KM. Analysis of the CYP2D6 gene polymorphism and enzyme activity in African-Americans in southern California. Pharmacogenetics. 2001;11:489–99. doi: 10.1097/00008571-200108000-00004. [DOI] [PubMed] [Google Scholar]
- 27.Mendoza R, Wan YJ, Poland RE, Smith M, Zheng Y, Berman N, Lin KM. CYP2D6 polymorphism in a Mexican American population. Clin Pharmacol Ther. 2001;70:552–60. doi: 10.1067/mcp.2001.120675. [DOI] [PubMed] [Google Scholar]
- 28.Masimirembwa C, Persson I, Bertilsson L, Hasler J, Ingelman-Sundberg M. A novel mutant variant of the CYP2D6 gene (CYP2D6*17) common in a black African population: association with diminished debrisoquine hydroxylase activity. Br J Clin Pharmacol. 1996;42:713–9. doi: 10.1046/j.1365-2125.1996.00489.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Raimundo S, Toscano C, Klein K, Fischer J, Griese EU, Eichelbaum M, Schwab M, Zanger UM. A novel intronic mutation, 2988G>A, with high predictivity for impaired function of cytochrome P450 2D6 in white subjects. Clin Pharmacol Ther. 2004;76:128–38. doi: 10.1016/j.clpt.2004.04.009. [DOI] [PubMed] [Google Scholar]
- 30.Rau T, Heide R, Bergmann K, Wuttke H, Werner U, Feifel N, Eschenhagen T. Effect of the CYP2D6 genotype on metoprolol metabolism persists during long-term treatment. Pharmacogenetics. 2002;12:465–72. doi: 10.1097/00008571-200208000-00007. [DOI] [PubMed] [Google Scholar]
- 31.Sheiner LB, Rosenberg B, Marathe VV. Estimation of population characteristics of pharmacokinetic parameters from routine clinical data. J Pharmacokinet Biopharm. 1977;5:445–79. doi: 10.1007/BF01061728. [DOI] [PubMed] [Google Scholar]
- 32.Beal B, Sheiner L. NONMEM User's Guide, Part I. San Francisco: University of California at San Francisco; 1992. [Google Scholar]
- 33.Ette EI, Ludden TM. Population pharmacokinetic modeling: the importance of informative graphics. Pharm Res. 1995;12:1845–55. doi: 10.1023/a:1016215116835. [DOI] [PubMed] [Google Scholar]
- 34.Wennerholm A, Johansson I, Massele AY, Lande M, Alm C, Aden-Abdi Y, Dahl ML, Ingelman-Sundberg M, Bertilsson L, Gustafsson LL. Decreased capacity for debrisoquine metabolism among black Tanzanians: analyses of the CYP2D6 genotype and phenotype. Pharmacogenetics. 1999;9:707–14. [PubMed] [Google Scholar]
- 35.Akaike H. A new look at the statistical model indentification. IEEE Trans Automat Control. 1974;AC-19:716–23. [Google Scholar]
- 36.Preskorn S. Targeted pharmacotherapy in depression management: comparative pharmacokinetics of fluoxetine, paroxetine and sertraline. Int Clin Psychopharmacol Supplement. 1994;3:13–9. doi: 10.1142/9789814440912_0082. [DOI] [PubMed] [Google Scholar]
- 37.Bertilsson L, Dahl M. Polymorphic drug oxidation. Relevance to the treatment of psychiatric disorders. CNS Drugs. 1996;5:200–23. [Google Scholar]
- 38.Cornish-Bowden A. Fundamentals of Enzyme Kinetics. London: Portland Press; 1995. [Google Scholar]
- 39.Cornish-Bowden A. Analysis of Enzyme Kinetic Data. Oxford, New York: Oxford University Press; 1995. [Google Scholar]
- 40.Feng Y, Pollock B, Reynolds C, Bies R. Paroxetine pharmacokinetics in geriatric patients. AAPS PharmSci. 2004 Abstract M1116. [Google Scholar]
- 41.Wennerholm A, Dandara C, Sayi J, Svensson JO, Abdi YA, Ingelman-Sundberg M, Bertilsson L, Hasler J, Gustafsson LL. The African-specific CYP2D617 allele encodes an enzyme with changed substrate specificity. Clin Pharmacol Ther. 2002;71:77–88. doi: 10.1067/mcp.2002.120239. [DOI] [PubMed] [Google Scholar]