Abstract
Aims
To characterize potential mechanism-based inactivation (MBI) of major human drug-metabolizing cytochromes P450 (CYP) by monoamine oxidase (MAO) inhibitors, including the antitubercular drug isoniazid.
Methods
Human liver microsomal CYP1A2, CYP2C9, CYP2C19, CYP2D6 and CYP3A activities were investigated following co- and preincubation with MAO inhibitors. Inactivation kinetic constants (KI and kinact) were determined where a significant preincubation effect was observed. Spectral studies were conducted to elucidate the mechanisms of inactivation.
Results
Hydrazine MAO inhibitors generally exhibited greater inhibition of CYP following preincubation, whereas this was less frequent for the propargylamines, and tranylcypromine and moclobemide. Phenelzine and isoniazid inactivated all CYP but were most potent toward CYP3A and CYP2C19. Respective inactivation kinetic constants (KI and kinact) for isoniazid were 48.6 µm and 0.042 min−1 and 79.3 µm and 0.039 min−1. Clorgyline was a selective inactivator of CYP1A2 (6.8 µm and 0.15 min−1). Inactivation of CYP was irreversible, consistent with metabolite-intermediate complexation for isoniazid and clorgyline, and haeme destruction for phenelzine. With the exception of phenelzine-mediated CYP3A inactivation, glutathione and superoxide dismutase failed to protect CYP from inactivation by isoniazid and phenelzine. Glutathione partially slowed (17%) the inactivation of CYP1A2 by clorgyline. Alternate substrates or inhibitors generally protected against CYP inactivation.
Conclusions
These data are consistent with mechanism-based inactivation of human drug-metabolizing CYP enzymes and suggest that impaired metabolic clearance may contribute to clinical drug–drug interactions with some MAO inhibitors.
Keywords: cytochrome P450, isoniazid, MAO inhibitors, mechanism-based inactivation
Introduction
The human monoamine oxidases (MAO-A and MAO-B) are flavin-containing enzymes that regulate the oxidative degradation of catecholamine neurotransmitters in the central nervous system. Drugs that inhibit MAOs have been extensively studied for the treatment of neuropathological disorders and are classified according to irreversible or reversible inhibition and selectivity between isoforms [1]. Nonselective irreversible inhibitors such as tranylcypromine, phenelzine and iproniazid have been utilized in the treatment of depression and panic disorders, but these can potentiate the sympathomimetic action of indirectly acting amines (e.g. food-derived tyramine) with resultant hypertensive crisis, the so-called ‘cheese effect’ [2]. The antitubercular drug isoniazid is also a nonselective irreversible MAO inhibitor, although hypertensive crisis is less frequent since inhibition is weak [3]. Development of selective inhibitors and reversible inhibitors regenerated interest in MAO as a therapeutic target when it was demonstrated that the ‘cheese effect’ could be avoided. The selective irreversible MAO-B inhibitor selegiline has found a role in the treatment of Parkinson's disease and possibly dementia [4]. Moreover, moclobemide is a selective and reversible MAO-A inhibitor efficacious in the treatment of major depression [5].
Drugs that behave as irreversible MAO inhibitors require metabolism to intermediate species that covalently modify the enzyme and cause impairment of function [6]. This type of inhibition is defined as mechanism-based inactivation (MBI). MBI is not limited to oxidations catalysed by MAOs [7]. Importantly, the reactive terminal acetylene, alkylamine and hydrazine functions of MAO inhibitors (Figure 1) are also associated with MBI of cytochromes P450 (CYP) [8]. This can have clinical consequences, since CYP inactivation often leads to severe and long-lasting impairment of metabolic clearance and clinically important drug–drug interactions [9]. In vitro data consistent with MBI have been reported for the inhibition of rodent CYP2B1 by selegiline and clorgyline [10] and the inhibition of rodent liver microsomal CYP-dependent activities by phenelzine [11], isoniazid [12], iproniazid and pargyline [13]. Furthermore, time-dependent inhibition of human CYP1A2, CYP2A6, CYP2C19 and CYP3A was recently demonstrated using isoniazid, but the mechanism was not elucidated [14]. In addition to previous work with cyclopropylamines [15], these studies together suggest that parallel MBI of MAO and CYP may be relatively common.
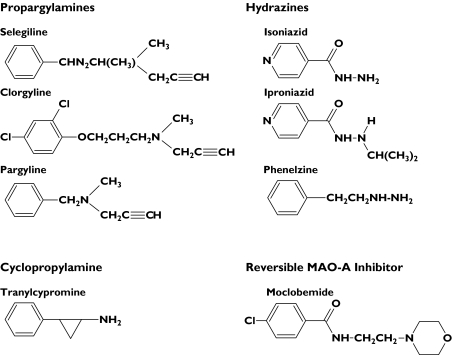
Figure 1.
Structures of monoamine oxidase inhibitors
Pharmacokinetic interaction studies have demonstrated that isoniazid decreases the metabolic clearance of numerous drugs eliminated by CYP [16–18] and moclobemide is known to inhibit the activity of CYP1A2, CYP2C19 and CYP2D6 in vivo[5]. However, there are no well-documented reports for other MAO inhibitors. Limited data suggest that tranylcypromine, phenelzine and moclobemide impair their own metabolism during chronic treatment [19]. Despite these observations, most clinical interactions involving MAO inhibitors (e.g. sympathomimetic agents, tricyclic antidepressants, selective serotonin reuptake inhibitors, carbamazepine, barbiturates and opioid analgesics) are attributed to the pharmacodynamic effects of biogenic amines. However, there is increasing awareness that impaired metabolic clearance is a pharmacokinetic factor that may contribute to many interactions with MAO inhibitors [20]. For example, the potentially fatal combination of meperidine with phenelzine and the toxicity of antiepileptic drugs when coadministered with isoniazid are drug–drug interactions attributed to this mechanism [16, 21].
A previous study in this laboratory described simultaneous inactivation of human liver microsomal CYP2C8 and CYP3A4 by isoniazid and phenelzine [22]. Despite many decades of clinical utilization, however, there have been no systematic investigations of potential MBI of major human drug-metabolizing CYPs by MAO inhibitors. Given the current deficits in understanding the pharmacokinetics of drug–drug interactions involving MAO inhibitors and the propensity for CYP inactivation, this work aimed to: evaluate a range of MAO inhibitors as MBIs of human liver microsomal CYP1A2, CYP2C9, CYP2C19, CYP2D6 and CYP3A, characterize the kinetics of inactivation, and determine the mechanism(s) of inactivation. Drugs investigated as potential MBIs included propargylamines (selegiline, clorgyline, pargyline), hydrazines (phenelzine, isoniazid, iproniazid), the cyclopropylamine tranylcypromine, and the reversible MAO-A inhibitor, moclobemide (Figure 1).
Methods
Chemicals
Phenacetin, paracetamol (S)-mephenytoin (S)-4′-hydroxymephenytoin, dextromethorphan, dextrorphan, testosterone, 6β-hydroxytestosterone, troleandomycin, selegiline, clorgyline, pargyline, tranylcypromine, isoniazid, iproniazid, phenelzine, caffeine, ciclosporin, quinidine, 4-methylumbelliferone, glucose 6-phosphate, glucose 6-phosphate dehydrogenase, β-nicotinamide adenine dinucleotide phosphate (NADP), reduced glutathione and superoxide dismutase were purchased from Sigma-Aldrich (Sydney, Australia). Other chemicals were kindly donated by the following sources: moclobemide, Dr J. Hoskins (University of Sydney, Australia); torsemide and tolyl methylhydroxytorsemide, Boehringer Mannheim International (Mannheim, Germany); tolbutamide, Hoechst Australia (Melbourne, Australia); and omeprazole, AstraZeneca (Molndal, Sweden).
Human liver microsomes and recombinant CYP
The Flinders Medical Centre Ethics Review Committee approved the use of human liver tissue obtained from the liver ‘bank’ of the Department of Clinical Pharmacology for in vitro drug metabolism studies. Microsomes from six livers (H6, H7, H10, H12, H13 and H40) were prepared by differential centrifugation and the protein concentrations determined as previously reported [22]. Inhibition and spectral studies were performed utilizing pooled human liver microsomes (HLM) with equal amounts of microsomal protein from each liver. Recombinant human CYP2C9, CYP2C19, CYP2D6 and CYP3A4 were coexpressed with either human or rat cytochrome P450 oxidoreductase (rOxR) in Escherichia coli and membrane fractions prepared according to published methods [22–25].
Construction of CYP1A2 and rOxR expression plasmids
The N-terminal membrane anchor of wild-type CYP1A2 cDNA (accession NM_000761) was replaced with a modified sequence derived from CYP17A to promote high levels of bacterial expression. Generation of the 17α-hydroxylase leader sequence in CYP1A2 cDNA utilized polymerase chain reaction (PCR)-directed mutagenesis to delete codons 3–10 using the following primers: Sense, 5′-TACATATGGCTCTGTTATTAGCAGTTTTTCTGTTCTGCCTGG TATTCTGGGTGC-3′; antisense, 5′-ATAAGCTTTCA ATTGATGGAGAAGCGCCGC-3′. To facilitate directional ligation into the pCW plasmid, NdeI and HindIII restriction sites (bold text) were incorporated into the sense and antisense CYP1A2 oligonucleotides, respectively. The 1515-bp 17α-CYP1A2 PCR products were digested with NdeI and HindIII prior to ligation with the pCW ori(+) plasmid. The rOxR expression construct containing the OmpA signal sequence was generated using the bacterial plasmid pACYC184, as documented previously [22, 23].
Heterologous coexpression of pCW-17aCYP1A2 with pACYC OmpA-rOxR
Clonal pCW-17αCYP1A2 was cotransformed into DH5αE. coli cells with pACYC OmpA-rOxR. Ampicillin/chloramphenicol selected colonies were screened for the correct plasmid by restriction enzyme analysis. Plasmid DNA was purified with the QIAprep Spin Miniprep Kit (Qiagen Pty Ltd, Doncaster, Australia) and confirmed on both strands by sequencing (ABI Prism 3100). Cells were cultured and membrane fractions separated [23]. Total protein concentration of membrane fractions, holoenzyme quantification and measurement of rOxR activity were determined as described previously [22].
Probe substrates
Phenacetin, torsemide (S)-mephenytoin, dextromethorphan and testosterone were used as the respective ‘probe’ substrates to assess CYP1A2, CYP2C9, CYP2C19, CYP2D6 and CYP3A activities. Rates of formation of paracetamol, tolyl methylhydroxytorsemide (S)-4′-hydroxymephenytoin, dextrorphan and 6β-hydroxytestosterone were linear with respect to protein concentration and incubation time under the incubation conditions utilized. Probe substrates were added to incubations as aqueous solutions (torsemide and dextromethorphan) or following dissolution in acetonitrile ((S)-mephenytoin and testosterone) or methanol (phenacetin). Where included, the final concentration of solvent in incubations was 1% (v/v), a concentration which did not affect reaction rates. The kinetics of phenacetin O-deethylation, (S)-mephenytoin 4′-hydroxylation and dextromethrophan O-demethylation were determined using microsomes from six livers (H6, H7, H10, H12, H13 and H40). The kinetics of torsemide tolyl methylhydroxylation and testosterone 6β-hydroxylation have been documented in previous publications from this laboratory [22, 26].
Preliminary screening experiments to identify potential MBI
Coincubation
MAO inhibitors were incubated with HLM and NADPH-regenerating system in the presence of probe substrates at 37 °C using the incubation conditions shown in Table 1. Coincubation mixtures contained HLM, NADPH-regenerating system (1 mm NADP, 10 mm glucose 6-phosphate, 2 IU ml−1 glucose 6-phosphate dehydrogenase, 5 mm MgCl2), MAO inhibitor, the probe substrate (at a concentration corresponding to the approximate Km for metabolite formation) and phosphate buffer (0.1 m, pH 7.4). MAO inhibitors were added to incubations as aqueous solutions. With the exception of the phenacetin assay, which was terminated by the addition of 200 µl of ice-cold acetonitrile, reactions were terminated by adding 2 or 5 µl of 70% perchloric acid. 4-Methylumbelliferone was added as an internal standard for the torsemide assay and samples prepared for high-performance liquid chromatography (HPLC) analysis according to Boye et al. [23]. Incubation samples from other CYP assays were vortex mixed, chilled on ice, and then centrifuged (4000 g) before analysis of supernatant fractions by HPLC.
Table 1.
Incubation conditions utilized for preliminary mechanism-based inactivation screening and derived kinetic constants for human liver microsomal CYP activities
| Phenacetin O-deethylation (CYP1A2) | Torsemide tolyl methylhydroxylation (CYP2C9) | (S)-Mephenytoin 4′-hydroxylation (CYP2C19) | Dextromethorphan O-demethylation (CYP2D6) | Testosterone 6β-hydroxylation (CYP3A) | |
|---|---|---|---|---|---|
| Final incubation volume (µl) | 200 | 500 | 200 | 200 | 500 |
| Protein concentration (mg ml−1) | 0.2 | 0.35 | 0.675 | 0.5 | 0.1 |
| Incubation time (min) | 30 | 20 | 45 | 20 | 15 |
| Kinetic model | Michaelis–Menten (two enzyme) | Michaelis–Menten | Michaelis–Menten | Michaelis–Menten (two enzyme) | Hill Equation |
| Km or Km1* (µm) | 20 ± 6.1 | 11.2 ± 1.3† | 25.4 ± 5.8 | 4.7 ± 2.5 | |
| S50 (µm) | 35.6 ± 4.3‡ | ||||
| Vmax or Vmax1* (pmol min−1 mg−1) | 486 ± 348 | 530 ± 110† | 69.3 ± 35.2 | 348.3 ± 165.2 | 2413 ± 90‡ |
| n (Hill coefficient) | 1.39 ± 0.05‡ | ||||
| Substrate concentration for MBI screening (µm) | 20 | 10 | 25 | 5 | 40 |
Rates of metabolite formation (velocity) were fitted either to the Michaelis–Menten, two-enzyme Michaelis–Menten or Hill equations and the kinetic constants determined as described under Methods. Data represent the mean (±SD) of six separate determinations for microsomes from human livers.
Km and Vmax for high-affinity component.
Data from Miners et al. (1995).
Data from Polasek et al. (2004).
Preincubation
MAO inhibitors were preincubated with HLM and NADPH-regenerating system in the absence of probe substrates for 30 min at 37 °C. Preincubation mixtures contained HLM, NADPH-regenerating system, MAO inhibitor and phosphate buffer (0.1 m, pH 7.4). Following preincubation, probe substrates were added so that the concentration and final volume remained as described for the coincubations. Assays were subsequently performed as described above.
Kinetics of CYP inactivation
Time- and concentration-dependent inhibition of human liver microsomal CYP by selected MAO inhibitors was characterized by determining residual enzyme activity using a conventional experimental protocol involving two steps. Inactivation assays contained HLM (2.0 mg ml−1 microsomal protein for measurement of CYP1A2, CYP2C9, CYP2C19 and CYP2D6 activity, and 1.0 mg ml−1 microsomal protein for measurement of CYP3A activity), NADPH-regenerating system and MAO inhibitor (five different concentrations) in phosphate buffer (0.1 m, pH 7.4). Final incubation volumes were as shown in Table 1. At selected preincubation times, 20-µl or 50-µl aliquots were removed and diluted 10-fold to activity assays containing either phenacetin (120 µm), torsemide (200 µm) (S)-mephenytoin (250 µm), dextromethorphan (25 µm) or testosterone (250 µm) (saturating concentrations of probe substrates). The preincubation times for experiments with isoniazid were 0, 5, 10, 20 and 30 min (CYP1A2, CYP2C9, CYP2D6) or 0, 2.5, 5, 10 and 15 min (CYP2C19 and CYP3A). All preincubation sampling times in experiments with phenelzine, and those investigating the effects of clorgyline on CYP1A2, were 0, 2.5, 5, 10 and 15 min. Activity assay reactions were allowed to proceed for incubation times as specified in Table 1 before termination and preparation of samples for analysis by HPLC. Since phenacetin O-deethylation and dextromethorphan O-demethylation by HLM followed biphasic kinetics, the relative contribution of CYP1A2 to phenacetin metabolism and CYP2D6 to dextromethorphan metabolism was estimated (by substitution of derived kinetic parameters in the two-enzyme Michaelis–Menten equation) to exceed 90% of the total activity when 120 µm phenacetin and 25 µm dextromethorphan were utilized in activity assays.
Effect of trapping agents and alternate substrates or inhibitors on CYP inactivation
Inactivation assays were additionally conducted in the presence of glutathione, superoxide dismutase or alternate CYP substrates/inhibitors. For these experiments, inactivation assays (as described under the Kinetics of CYP inactivation section) contained HLM, NADPH-regenerating system, MAO inhibitor, trapping agent (2 mm glutathione or 1000 U ml−1 superoxide dismutase) or alternate substrate/inhibitor and phosphate buffer (0.1 m, pH 7.4). Isoniazid and phenelzine were included in incubations at various concentrations (see Table 3), whereas 5 µm clorgyline was used to study the inactivation of CYP1A2. The respective alternate substrates or inhibitors for CYP1A2, CYP2C9, CYP2C19, CYP2D6 and CYP3A assays were caffeine (500 µm), tolbutamide (500 µm), omeprazole (100 µm), quinidine (0.1 µm) and ciclosporin (10 µm). Inactivation assays were conducted for 15 min at 37 °C prior to 10-fold dilution and determination of residual activity. Control samples were prepared in the absence of MAO inhibitors.
Table 3.
Effect of trapping agents and alternate substrates or inhibitors on the inactivation of human liver microsomal CYP by isoniazid and phenelzine
| CYP probe substrate activity (percent of control) | ||||||||
|---|---|---|---|---|---|---|---|---|
| Isoniazid | Phenelzine | |||||||
| NADPH | NADPH + GSH (2 mm) | NADPH + SOD (1000 U ml−1) | NADPH + alternate substrate/ inhibitor | NADPH | NADPH + GSH (2 mm) | NADPH + SOD (1000 U ml−1) | NADPH + alternate substrate/ inhibitor | |
| CYP1A2 (400 µm ISO; 50 µm PHE) | 40.4 | 41.8 | 36.5 | 85.1 | 25.5 | 30.8 | 30.6 | 62.5 |
| CYP2C9 (1000 µm ISO; 100 µm PHE) | 75.0 | 70.7 | 74.0 | 101.6 | 60.5 | 64.3 | 69.1 | 37.4 |
| CYP2C19 (200 µm ISO; 40 µm PHE) | 61.8 | 56.7 | 58.7 | 91.0 | 56.7 | 46.1 | 45.1 | 70.3 |
| CYP2D6 (1000 µm ISO; 200 µm PHE) | 84.3 | 84.9 | 87.3 | 102.6 | 63.7 | 69.7 | 70.4 | 91.6 |
| CYP3A (100 µm ISO; 20 µm PHE) | 56.5 | 56.9 | 56.5 | 79.8 | 42.6 | 69.9 | 103.2 | 76.1 |
Human liver microsomes were preincubated with monoamine oxidase inhibitor for 15 min at 37°C. Control preparations were preincubated with NADPH-regenerating system alone and either glutathione (GSH), superoxide dismutase (SOD), or alternate substrate or inhibitor, but lacking test drug. Directly following preincubation, aliquots were removed and diluted 10-fold to determine remaining CYP activities. Data represent the mean of duplicate determinations. ISO, isoniazid; PHE, phenelzine.
Effect of ultrafiltration on CYP inactivation
Ultrafiltration experiments with recombinant CYP enzymes were conducted to determine whether catalytic function could be restored following preincubation with isoniazid, phenelzine and clorgyline. Recombinant enzymes rather than HLM were employed since the microsomal protein concentrations required for inactivation studies (i.e. 1.0 or 2.0 mg ml−1 of microsomal protein, see Kinetics of CYP inactivation section) block membrane filters. Inactivation assays, containing 250 pmol ml−1 of recombinant CYP1A2, CYP2C9, CYP2C19, CYP2D6 or CYP3A4, were performed for 15 min at 37 °C using isoniazid (100 µm), phenelzine (10 µm) or clorgyline (1 µm) (CYP1A2 only). Aliquots were removed and diluted 10-fold to activity assays containing saturating concentrations or probe substrates (200 µm phenacetin, 200 µm torsemide, 250 µm (S)-mephenytoin, 100 µm dextromethorphan or 250 µm testosterone). In comparison with the HLM studies (see the Kinetics of CYP inactivation section), higher concentrations of phenacetin and dextromethorphan were utilized to ensure full saturation during activity assays. Metabolite formation was quantified prior to and following ultrafiltration, as described by Polasek et al. [22]. Control measurements were performed in the absence of MAO inhibitors and without ultrafiltration.
Spectral difference scanning
Incubations contained HLM (1300 µg ml−1) in phosphate buffer (0.1 m, pH 7.4), with or without NADPH-regenerating system (final volume 500 µl). Mixtures were divided between two cuvettes and a baseline of equal light absorbance was established using a Cary 300 double-beam UV-visible spectrophotometer (Varian Inc. Melbourne, Australia). Isoniazid, phenelzine or clorgyline (200 µm) was added to the sample cuvette and an equivalent volume of solvent was added to the reference cuvette. Both were placed in a shaking water bath at 37 °C and the difference spectra recorded after 2.5, 7.5 and 15 min. In the experiments that included NADPH-regenerating system, potassium ferricyanide (200 µm) was added to both cuvettes following the 15-min scan and the difference spectra re-recorded. Clorgyline (10, 50 and 200 µm) was also investigated by the same method using recombinant CYP1A2 (500 pmol ml−1).
Cytochrome P450 reduced CO-difference spectroscopy
Incubations (500 µl in 0.1 m phosphate buffer, pH 7.4) containing HLM (1300 µg ml−1), 1 mm EDTA, isoniazid, phenelzine or clorgyline (200 µm) were performed for 15 min at 37 °C in the presence or absence of NADPH-regenerating system. The reactions were terminated by the addition of 50 µl of ice-cold phosphate buffer containing 10% glycerol and 1% Emulgen 913. Sodium dithionite was added and dissolved thoroughly. The mixtures were divided between two cuvettes and a baseline of equal light absorbance was established. CO gas was gently bubbled through the sample cuvette for 1 min and the spectrum of the reduced CO complex recorded between 500 and 400 nm. Clorgyline (10 µm) was also incubated with recombinant CYP1A2 (500 pmol ml−1) and the P450 content determined as for the studies with HLM.
HPLC assays
Metabolites of CYP probe substrates were quantified by reversed-phase HPLC using an Agilent 1100 series HPLC system (Agilent Technologies, Sydney, Australia) unless otherwise stated. Concentrations of paracetamol, tolyl methylhydroxytorsemide, (S)-4′-hydroxymephenytoin, dextrorphan and 6β-hydroxytestosterone in incubations were determined by comparison of the peak area ratios (or metabolite to internal standard ratio in the case of tolyl methylhydroxytorsemide) with those of standard curves generated using authentic standards. Standard curves were constructed in the concentration range of 0.5–5 µm with paracetamol, dextrorphan and 6β-hydroxytestosterone, and in the concentration range of 0.25–2.5 µm with tolyl methylhydroxytorsemide and (S)-4′-hydroxymephenytoin. The overall reproducibility of CYP assays was assessed by measuring metabolite formation for replicate incubations (n = 10) using HLM at low and high CYP probe substrate concentrations. The within-day and between-day coefficients of variation for phenacetin O-deethylation (10 µm and 2000 µm), torsemide tolyl methylhydroxylation (10 µm and 50 µm) (S)-mephenytoin 4′-hydroxylation (20 µm and 100 µm), dextromethorphan O-demethylation (5 µm and 50 µm) and testosterone 6β-hydroxylation (25 µm and 100 µm), were <10%.
Phenacetin O-deethylation (CYP1A2)
Chromatography was performed using a Beckman Ultrasphere ODS C18 column (25 cm × 4.6 mm inner diameter, 5 µm particle size; Beckman Coulter Inc., Fullerton, CA, USA). The mobile phase, which consisted of sodium acetate (5 mm, adjusted to pH 4.3 with glacial acetic acid) plus acetonitrile (87.5 : 12.5) (A) and acetonitrile (B), was delivered at a flow rate of 1.5 ml min−1 according to the following gradient: initial conditions 100% A held for 2.5 min, then changed to 55% A/45% B over 0.1 min, which was held for 1 min before returning to initial conditions. The retention time of paracetamol, detected by UV absorbance at 254 nm, was 3.1 min.
Torsemide tolyl methylhydroxylation (CYP2C9)
Measurement of tolyl methylhydroxytorsemide in incubations was performed as described previously [23], with minor modifications. Briefly, a Waters Nova-Pak C18 column (15 cm × 3.9 mm inner diameter, 4 µm particle size; Waters, Milford, MA, USA) was eluted with mobile phase, consisting of sodium acetate buffer (10 mm, adjusted to pH 4.3 by glacial acetic acid) plus acetonitrile (95 : 5) (A) and acetonitrile (B), at a flow rate of 2 ml min−1. Gradient conditions were initially 95% A/5% B changed to 87.5% A/12.5% B over 9 min, then 25% A/75% B over 0.1 min, which was held for 0.3 min before returning to the initial conditions. Peaks were monitored using UV detection at 290 nm. The retention times of tolyl methylhydroxytorsemide and 4-methylumbelliferone (internal standard) were 3.6 and 8.4 min, respectively.
(S)-Mephenytoin 4′-hydroxylation (CYP2C19)
Separation of (S)-4′-hydroxymephenytoin was achieved using a Waters Nova-Pak C18 column (15 cm × 3.9 mm inner diameter, 4 µm particle size). A mobile phase of 5% acetonitrile in water (A) and acetonitrile (B) was delivered to the column at 1 ml min−1 according to the following gradient: initial conditions 87.5% A/12.5% B held for 4 min, then changed to 30% A/70% B over 0.1 min, which was held for 1 min before returning to the initial conditions. The retention time of (S)-4′-hydroxymephenytoin, which was detected by UV absorbance at 223 nm, was 4.6 min.
Dextromethorphan O-demethylation (CYP2D6)
The system to quantify dextrorphan formation consisted of a Shimadzu SIL-9 A autoinjector, a Shimadzu LC-6A pump (Shimadzu, Kyoto, Japan) and a Perkin Elmer LC 240 fluorescence detector (Perkin Elmer UK, Beaconsfield, UK). The stationary phase was a radial compression module containing a Waters Nova-Pak CN cartridge (10 cm × 8 mm inner diameter, 4 µm particle size; Waters) and a CN precolumn. The mobile phase consisted of a mixture of 10% acetonitrile and 0.2% triethylamine in water adjusted to pH 3.0 with orthophosphoric acid, which was delivered at 1 ml min−1. The excitation and emission wavelengths, the response and the attentuation factor were set at 224 and 320 nm, 5 and 32, respectively. The retention times of dextrorphan and 3-methoxymorphinan were 6.9 and 10.4 min, respectively.
Testosterone 6b-hydroxylation (CYP3A)
The concentration of 6β-hydroxytestosterone was determined by HPLC with UV detection at 241 nm according to the method previously reported by Polasek et al. [22].
Data analysis
The kinetic constants for the CYP probe substrates were derived (EnzFitter; Biosoft, Cambridge, UK) by fitting to either the Michaelis–Menten equation, the two-enzyme Michaelis–Menten equation or the Hill equation [27].
Inhibition difference (‘preincubation effect’) in the preliminary screen to identify potential MBI was calculated by subtracting the remaining CYP activity (as percentage of control) after preincubation from the remaining CYP activity following coincubation at the corresponding MAO inhibitor concentrations, i.e. a positive number indicated greater inhibition following preincubation compared with coincubation.
The mean value of remaining CYP activity expressed as a percentage of control was used to estimate the kinetic constants of inactivation according to Silverman [7]. Briefly, the observed inactivation rate constant (kobs) at each MAO inhibitor concentration was calculated from the initial slopes of the remaining enzyme activity, plotted semilogarithmically against the preincubation time. The following equation was fitted to inactivation data and the values of KI and kinact derived using nonlinear least-squares regression (EnzFitter; Biosoft).
| (1) |
where [I] is the initial inhibitor concentration, kinact is the maximal inactivation rate constant, and KI is the inhibitor concentration required for half the maximal rate of inactivation.
Results
In vitro kinetics of P450 probe substrates
The kinetics of metabolite formation for each of the CYP probe substrates were characterized in this laboratory prior to the preliminary MBI screening experiments (Table 1). Data for torsemide tolyl methylhydroxylation and testosterone 6β-hydroxylation have been reported elsewhere [22, 26]. (S)-Mephenytoin 4′-hydroxylation exhibited hyperbolic (Michaelis–Menten) kinetics, whereas biphasic kinetics consistent with the two-enzyme Michaelis–Menten model were observed for phenacetin O-deethylation and dextrorphan formation (Table 1). Derived kinetic parameters for all CYP probe substrate reactions were consistent with literature data.
Preliminary screening experiments to identify potential MBI
Since catalytic turnover is required for inactivation, greater inhibition following a preincubation step (i.e. a positive inhibition difference) may be used to identify potential MBI. Table 2 shows the inhibition difference between co- and preincubation conditions for the eight MAO inhibitors investigated here. Incubations were also conducted at one other MAO inhibitor concentration (5 or 100 µm) and consistency between the two concentrations was noted for both positive and negative inhibition differences in all cases (data not shown), with the exception of phenelzine (see Discussion below). Excluding clorgyline inhibition of CYP1A2 and selegiline inhibition of CYP2D6, the inhibitory potency of propargylamine MAO inhibitors toward CYP enzymes was not increased following preincubation. Negative inhibition differences (i.e. coincubation inhibition >preincubation inhibition) were observed using tranylcypromine. In contrast, the hydrazine MAO inhibitors generally exhibited greater inhibition following preincubation, although iproniazid did not consistently mimic isoniazid. Indeed, only inhibition of CYP2C9 and CYP2D6 were greater following preincubation with iproniazid. The reversible MAO-A inhibitor moclobemide demonstrated minor positive inhibition differences toward CYP1A2 and CYP2D6 (<10%).
Table 2.
Preliminary screening for potential mechanism-based inactivation of human liver microsomal CYP enzymes
| Inhibition difference (percentage of control) | |||||
|---|---|---|---|---|---|
| Phenacetin O-deethylation (CYP1A2) | Torsemide tolyl methylhydroxylation (CYP2C9) | (S)-Mephenytoin 4′-hydroxylation (CYP2C19) | Dextromethorphan O-demethylation (CYP2D6) | Testosterone 6β-hydroxylation (CYP3A) | |
| Selegiline | 0.5 | N.O. | −10.7 | 6.0 | 0.5* |
| Clorgyline | 21.6† | −14.3 | −16.6 | −4.8 | 0.8 |
| Pargyline | −1.9 | N.O. | N.O. | −8.8 | 1.3 |
| Tranylcypromine | −7.4 | −0.9 | −12.1 | −1.3 | −6.7* |
| Phenelzine | 25.4 | 46.8 | 24.2 | 11.9 | 12.5* |
| Isoniazid | 11.4 | 14.8 | 18.0 | 4.0 | 15.9* |
| Iproniazid | −4.0 | 15.7 | −0.7 | 4.0 | −1.6 |
| Moclobemide | 3.1 | N.O. | −3.0 | 9.9 | −3.2 |
Monoamine oxidase inhibitors (20 µm) were co- and preincubated as described under Methods and the inhibition difference between co- and preincubation conditions determined. Data represent the mean of at least duplicate determinations.
Data from Polasek et al. (2004).
Results using 5 µm clorgyline. N.O., Co- and preincubation inhibition not observed.
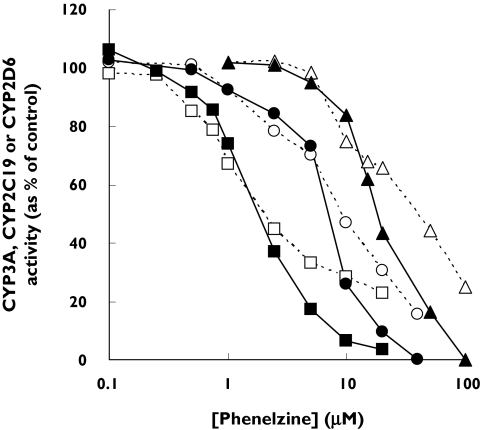
Unexpectedly, CYP3A, CYP2C19 and CYP2D6 activities were not further reduced by preincubation at the lower phenelzine concentrations corresponding to <40% inhibition in the coincubation (i.e. there was no preincubation effect) (Figure 2). In contrast, the activities of CYP1A2 and CYP2C9 were further reduced by preincubation at all phenelzine concentrations in the range of 5–100 µm, including those that caused minor (<10%) inhibition during coincubation (data not shown).
Figure 2.
Inhibition of human liver microsomal CYP3A (squares), CYP2C19 (circles) and CYP2D6 (triangles) by phenelzine. Phenelzine was coincubated (□, ○ and ▵) and preincubated (▪, • and ▴) as described in Methods and the CYP activities determined. Each data point represents the mean of duplicate measurements
Inactivation kinetics for isoniazid, phenelzine and clorgyline
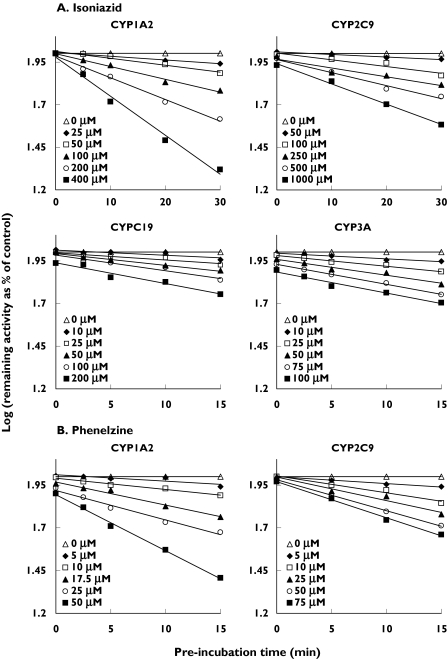
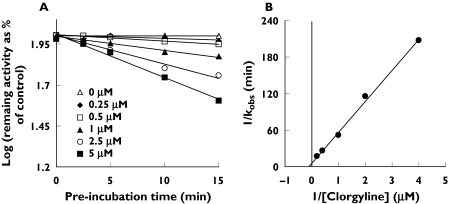
MAO inhibitors that exhibited positive inhibition differences during the preliminary screening experiments were considered for further investigation. Isoniazid and phenelzine were considered as potential nonselective MBIs and clorgyline as a potential selective MBI of CYP1A2. Given that the preincubation effects observed for selegiline and moclobemide toward CYP2D6 were minor (<10%) (Table 2), potential MBI was not further investigated. Figure 3A shows the inactivation of CYP1A2, CYP2C9, CYP2C19 and CYP3A by isoniazid, characterized by respective inactivation kinetic constants (KI and kinact) of 364 µm and 0.081 min−1, 530 µm and 0.042 min−1, 79.3 µm and 0.039 min−1 and 48.6 µm and 0.042 min−1. Inactivation was saturable in all cases and pseudo first-order inactivation kinetics were observed over the preincubation times and concentrations studied. Time-dependent inhibition of CYP2D6 by isoniazid was too weak for accurate kinetic analysis (data not shown). Furthermore, inactivation kinetic constants could not be determined for phenelzine-mediated inactivation of CYP2C19, CYP2D6 and CYP3A using the conventional experimental protocol because time-dependent inhibition occurred only at concentrations above approximately 50 µm, 200 µm and 15 µm, respectively (data not shown). However, studies were possible with CYP1A2 (KI = 317 µm and kinact = 0.54 min−1) and CYP2C9 (KI = 14.3 µm and kinact = 0.052 min−1) (Figure 3B). Clorgyline was a selective MBI of CYP1A2 (KI = 6.8 µm and kinact = 0.15 min−1) that exhibited saturable pseudo first-order inactivation kinetics over the preincubation times and concentrations studied (Figure 4). Inhibition of other CYP enzymes by clorgyline was not time-dependent (data not shown), consistent with preliminary screening experiments (Table 2).
Figure 3.
Time- and concentration-dependent inhibition of human liver microsomal CYP1A2, CYP2C9, CYP2C19 and CYP3A by isoniazid (A) and human liver microsomal CYP1A2 and CYP2C9 by phenelzine (B). Aliquots were removed from the primary reaction mixtures at the indicated time points and assayed for residual CYP activity. Each data point represents the mean of at least triplicate measurements
Figure 4.
(A)Time- and concentration-dependent inhibition of human liver microsomal CYP1A2 by clorgyline and (B) the corresponding double reciprocal plot of the rate of inactivation (kobs). Each data point represents the mean of triplicate measurements
Effect of trapping agents and alternate substrates or inhibitors
With the exception of phenelzine-mediated inactivation of CYP3A, glutathione and superoxide dismutase failed to protect CYP from inactivation by isoniazid and phenelzine (Table 3). Human liver microsomal CYP1A2 activity was reduced to 38% of control following inactivation assays with 5 µm clorgyline. This was not altered in the presence of superoxide dismutase, but was partially restored by glutathione (55% of control). The addition of alternate substrates or inhibitors to incubations slowed or prevented inactivation in almost all cases, including the protection of clorgyline-mediated CYP1A2 inactivation by caffeine (data not shown). The exception was phenelzine inactivation of CYP2C9, which appeared to be enhanced by the alternate substrate tolbutamide (Table 3).
Effect of ultrafiltration on recombinant CYP inactivation
Since HLM could not be used for ultrafiltration experiments, recombinant CYP preparations were employed to determine whether CYP inhibition by isoniazid, phenelzine and clorgyline was reversible or irreversible. Recombinant CYP activities were not restored by ultrafiltration following incubations with isoniazid and phenelzine (data not shown). Similarly, recombinant CYP1A2 activity remained at approximately 50% of control following inactivation assays with 1 µm clorgyline, irrespective of ultrafiltration. Loss of recombinant CYP activities attributed to ultrafiltration alone was <25% (e.g. comparison of control sample activities in the absence of MAO inhibitors prior to and following ultrafiltration).
Spectral difference scanning and reduced CO-difference spectra
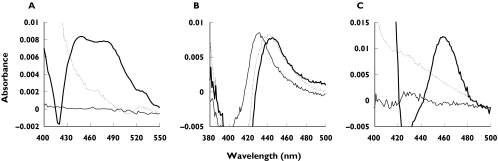
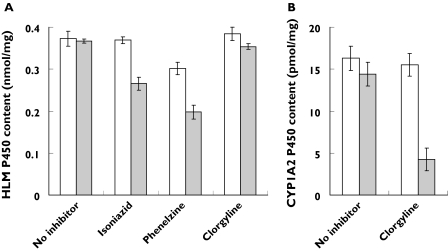
Spectral studies were conducted to investigate whether the inactivation of CYP enzymes by isoniazid, phenelzine and clorgyline occurred via the formation of metabolite–intermediate complexes (MICs). Incubations with isoniazid and an NADPH-regenerating system resulted in a broad increase in absorbance between 510 and 420 nm that was sensitive to ferricyanide (Figure 5A) and a corresponding reduction in spectrally detectable P450 (Figure 6A). This is consistent with MIC formation by isoniazid previously observed using rat liver microsomes [12]. Phenelzine bound to microsomal CYP in the absence of NADPH-regenerating system to give a typical type II binding spectrum (Figure 5B). For incubations including NADPH-regenerating system, the absorbance maximum was 445 nm and the apparent P450 content was reduced by approximately 45% (Figure 6A). Since incubations with clorgyline and HLM produced no spectral changes (scans not shown) and P450 content was reduced by <5% (Figure 6A), recombinant CYP1A2 was substituted as the enzyme source. When incubated in the presence of NADPH-regenerating system, clorgyline (10 µm) showed time-dependent and ferricyanide-sensitive changes in absorbance with a maximum at 458–460 nm (Figure 5C). There was an approximate 70% loss in detectable recombinant CYP1A2 P450 (Figure 6B). This observation resembles MIC formation by alkylamine drugs [28]. Interestingly, the MIC was not observed when clorgyline concentrations >50 µm were employed. Rather, time- and NADPH-dependent increases in absorbance maximum at 424 nm were recorded that were insensitive to ferricyanide (scans not shown).
Figure 5.
Representative 15-min difference spectra for incubations of human liver microsomes with isoniazid (A) and phenelzine (200 µm) (B), and an incubation of recombinant CYP1A2 with clorgyline (10 µm) (C). Incubations were performed in the absence (thin solid line) or presence of NADPH-regenerating system (thick solid line). Potassium ferricyanide (dashed line) was added following incubations in which NADPH-regenerating system was present
Figure 6.
Apparent human liver microsomal P450 content following 15-min incubations with isoniazid, phenelzine and clorgyline (200 µm) (A) and recombinant CYP1A2 P450 content following 15-min incubations with clorgyline (10 µm) (B). Incubations were performed in the absence (clear bars) or presence (grey bars) of NADPH-regenerating system. Data represent the mean of six separate determinations with 95% confidence intervals
Discussion
The present in vitro studies were conducted to investigate potential MBI of major human drug-metabolizing CYP enzymes by MAO inhibitors, including the antitubercular drug isoniazid. Since time-dependent inhibition is characteristic of MBI, preliminary experiments compared inhibition following co- and preincubation [7, 22, 29]. The hydrazine MAO inhibitors generally demonstrated preincubation effects, whereas, with the exception of clorgyline inhibition of CYP1A2, time-dependent inhibition of CYP enzymes by the proparglyamines and moclobemide was absent or minor (Table 2). Inhibition of CYP by tranylcypromine was reduced following preincubation, consistent with previous rodent studies [13, 30]. This may be explained by consumption of the reversible inhibitor over time [30].
Although they are useful as a screen, caution is necessary when interpreting data from the preliminary experiments (Table 2). Metabolite(s) generated during the preincubation may also produce time-dependent inhibition by way of potent competitive inhibition [22]. In the absence of a dilution step and using a probe substrate concentration near Km, an observation of time-dependent inhibition is an indicator of possible MBI rather than verification [22]. For example, selegiline and moclobemide demonstrated minor preincubation effects on CYP2D6 (<10%). Both are converted to multiple metabolites [5, 31], some of which (e.g. formation of amphetamine from selegiline) are associated with MBI of CYP [32]. However, whether such minor time-dependent inhibition represents weak MBI or simply greater competition for the active site over time remains to be investigated. Indeed, the clinical relevance of the current data with CYP2D6 is unclear; metabolism-related drug–drug interactions involving selegiline are not reported, whereas interactions between moclobemide and CYP2D6 substrates (e.g. tricyclic antidepressants and metoprolol) are attributed to a pharmacodynamic mechanism only, despite evidence of effective CYP2D6 inhibition in vivo[5].
To confirm data from preincubation screening, both isoniazid and phenelzine were shown to be inactivators of all CYP enzymes studied, whereas clorgyline was a selective inactivator of CYP1A2. The in vitro MBI criteria (described in detail elsewhere [7, 22, 33]) were generally fulfilled despite minor inconsistencies; time-dependent inhibition of human liver microsomal CYP2D6 by isoniazid was too weak for kinetic analysis despite irreversible inhibition of recombinant CYP2D6; time-dependent inhibition of CYP2C19, CYP2D6 and CYP3A by phenelzine was unusual (see below); the alternate CYP2C9 substrate tolbutamide stimulated CYP2C9 inactivation by phenelzine, possible due to putative multiple CYP2C9 substrate binding sites; and finally, glutathione partially slowed the inactivation of CYP1A2 by clorgyline. With respect to the penultimate point, it should be noted that dapsone also activates the metabolism of numerous CYP2C9 substrates [34]. Spectral studies to investigate the mechanisms of inactivation by isoniazid, phenelzine and clorgyline were consistent with previous rodent studies. Here, isoniazid formed a hydrazide MIC with a corresponding reduction in spectrally detectable P450 (Figures 5A and 6A) as shown in rat [12]. The metabolism of phenelzine reduced P450 content via haeme destruction (Figure 6A) [11], whereas the inactivation of CYP1A2 by clorgyline occurred by alkylamine MIC formation [28] (Figures 5C and 6B) similar to previous work with rat CYP2B1 [35].
Recently, kinetic constants for the inactivation of human liver microsomal CYP1A2, CYP2A6, CYP2C19 and CYP3A by isoniazid were reported [14]. In the present study, isoniazid was a more potent inactivator of CYP3A and CYP2C19, but a less potent inactivator of CYP1A2 than reported by those authors. Furthermore, the derived maximal inactivation rate constants (kinact) calculated by us were lower by approximately half. Here, isoniazid was diluted 10-fold to the activity assay, where residual CYP activity was determined at a saturating concentration of the probe substrate. Therefore, any inhibitory effect of reversible metabolite(s) generated during the inactivation assay was minimized. Interestingly, more potent inhibition of CYP2C19 compared with CYP3A (midazolam 4-hydroxylation) has been reported [14, 16]. The greater potency toward CYP3A-catalysed testosterone 6β-hydroxylation in the present study suggests that inhibition by isoniazid may be substrate dependent, as described for numerous other CYP3A inhibitors [36]. Despite this, the classification and relative potency of isoniazid as an MBI of major drug-metabolizing CYP enzymes support the selectivity and magnitude of reported pharmacokinetic interaction studies. Interactions with CYP3A and CYP2C19 substrates such as carbamazepine and the benzodiazepines are common, whereas drugs eliminated predominantly by CYP1A2 (theophylline) and CY2C9 (warfarin) are less likely to be affected [16]. Additionally, there are no reports of isoniazid altering the metabolic clearance of drugs principally eliminated via CYP2D6.
The significant protection against phenelzine-mediated CYP3A inactivation offered by ‘trapping agents’ (Table 3) supports a major contribution from phenelzine-derived reactive intermediates that do not remain within the active site prior to inactivation. Hence, phenelzine is considered a metabolically activated inactivator of CYP3A [7]. Interestingly, the time-dependent inhibition of CYP2C19, CYP2D6 and CYP3A by phenelzine was unusual, with preincubation effects observed only above separate ‘threshold’ concentrations during screening (Figure 2), and in the experiments designed to estimate inactivation kinetic constants (data not shown). Radicals are formed during the oxidation of phenelzine [37] and delayed time-dependent inhibition may be explained if radical-mediated inactivation occurred under conditions of endogenous glutathione and oxygen scavenger exhaustion, presumably at the higher phenelzine concentrations. However, trapping agents did not significantly slow (defined as <10%) the inactivation of CYP2D6 or affect the inactivation rate of CYP2C19, despite similar concentration-dependent preincubation effects to CYP3A (i.e. time dependence occurred commonly at approximately 40% inhibition; Figure 2). The mechanistic basis for these observations remains unclear. Nevertheless, the metabolism of phenelzine by HLM gave rise to nonselective CYP inactivation, with CYP3A and CYP2C19 the most susceptible. CYP2B6, CYP3A and CYP2C19 were recently identified as the CYP enzymes responsible for meperidine N-demethylation [38]. Current data support the inhibition of CYP3A and CYP2C19 as the rationale for the meperidine and phenelzine interaction.
Clorgyline was shown here to be a selective MBI of human liver microsomal CYP1A2. In comparison with other drugs that are putatively selective CYP1A2 MBIs, clorgyline is more potent than zileuton [39] but less potent than furafylline [40]. Interestingly, the difference spectra recorded with recombinant CYP1A2 were dependent upon the clorgyline concentration. MIC formation was observed at concentrations <50 µm (Figure 5C), whereas increasing concentration resulted in type II binding spectra (scans not shown). Conceivably, the metabolite required for MIC formation might be a secondary metabolite that cannot compete with the generation of the primary metabolite at higher concentrations. This mechanism has been proposed previously for the concentration-dependent MIC formation rate observed with CYP2B1 and clorgyline [35]. Sequential N-demethylation and N-depropargylation of clorgyline yields an amphetamine-like compound that may be responsible for MIC formation [32, 35]. Binding of the primary metabolite desmethylclorgyline may explain the time- and NADPH-dependent but ferricyanide-insensitive spectra recorded here at greater clorgyline concentrations. Hence, the balance between reversible and irreversible inhibition by propargylamines may be complex.
The clinical impact of MBI on CYP-mediated clearance may be simulated from in vitro inactivation data using the mechanism-based pharmacokinetic model developed by Hall and colleagues. The theoretical background of this model has been described previously [29] and its predictive utility demonstrated with other mechanism-based inactivators [29, 41–43]. Although details of simulations are beyond the scope of this study, if we assume no nonspecific binding during incubations [44], full inhibition of intestinal CYP3A [29] and a hepatic CYP3A degradation half-life of 64 h [45], use of the data presented here predicts that isoniazid will increase the area under the concentration–time curve (AUC) of oral midazolam by two- to eightfold at hepatic concentrations up to 1 µm. Similarly, the AUC of a coadministered drug that is exclusively eliminated by hepatic CYP2C19 metabolism is predicted to increase approximately twofold (using a hepatic CYP2C19 degradation half-life of 26 h). Lack of clinical drug–drug interaction studies with oral midazolam or a suitable CYP2C19 probe substrate precludes a direct comparison of predicted vs. actual AUC results. However, given that peak unbound plasma concentrations of isoniazid range between 20 and 60 µm following oral administration (300 mg) [46], we expect that clinically significant CYP3A and CYP2C19 inactivation is likely, especially considering its widespread use as monotherapy in the treatment and prevention of tuberculosis [16]. In comparison with isoniazid, the mean peak plasma concentration following an oral dose of phenelzine (30 mg) is low (0.085 µm) [47]. Despite this, extensive hepatic metabolism of phenelzine by free radical pathways impairs CYP-mediated function during chronic treatment [20, 37]. Clearly, quantitative predictions with phenelzine are likely to be complex, emphasized here by the difficulty in obtaining meaningful in vitro inactivation data for CYP2C19, CYP2D6 and CYP3A.
In conclusion, we have demonstrated that MAO inhibitors have the capacity to behave as MBIs of major human drug-metabolizing CYP enzymes. Minor time-dependent inhibition of CYP2D6 is described for selegiline and moclobemide, the significance of which remains to be investigated. Data are consistent with MBI of CYP1A2, CYP2C9, CYP2C19, CYP2D6 and CYP3A by isoniazid, MBI of CYP1A2, CYP2C9, CYP2C19 and CYP2D6 by phenelzine and selective MBI of CYP1A2 by clorgyline. Phenelzine acts as a metabolically activated inactivator of CYP3A. These studies confirm that parallel MBI of human MAOs and CYP enzymes occurs in some cases and suggest that impaired metabolic clearance may contribute to clinical drug–drug interactions with MAO inhibitors.
Acknowledgments
We thank Dr Janelle Hoskins for the gift of moclobemide and Dr Mark R. Hutchinson for the development of the dextromethorphan O-demethylation assay. This work was supported by a grant from the National Health and Medical Research Council of Australia. T.M.P. is the recipient of an Australian Post-Graduate Award.
References
- 1.Youdim MBH, Finberg JPM. New directions in monoamine oxidase A and B. Selective inhibitors and substrates. Biochem Pharmacol. 1991;41:155–62. doi: 10.1016/0006-2952(91)90471-g. [DOI] [PubMed] [Google Scholar]
- 2.Yamada M, Yasuhara H. Clinical pharmacology of MAO inhibitors: safety and future. Neurotoxicology. 2004;25:215–21. doi: 10.1016/S0161-813X(03)00097-4. [DOI] [PubMed] [Google Scholar]
- 3.DiMartini A. Isoniazid, tricyclics and the ‘cheese reaction’. Int Clin Psychopharm. 1995;10:197–8. doi: 10.1097/00004850-199510030-00009. [DOI] [PubMed] [Google Scholar]
- 4.Anttila M, Sotaniemi EA, Pelkonen O, Rautio A. Marked effect of liver and kidney function on the pharmacokinetics of selegiline. Clin Pharmacol Ther. 2005;77:54–62. doi: 10.1016/j.clpt.2004.09.004. [DOI] [PubMed] [Google Scholar]
- 5.Gram LF, Guentert TW, Grange S, Vistisen K, Brosen K. Moclobemide, a substrate of CYP2C19 and an inhibitor of CYP2C19, CYP2D6, and CYP1A2: a panel study. Clin Pharmacol Ther. 1995;57:670–7. doi: 10.1016/0009-9236(95)90230-9. [DOI] [PubMed] [Google Scholar]
- 6.Sablin SO, Yankovskaya V, Bernard S, Cronin CN, Singer TP. Isolation and characterization of an evolutionary precursor of human monoamine oxidases A and B. Eur J Biochem. 1998;253:270–9. doi: 10.1046/j.1432-1327.1998.2530270.x. [DOI] [PubMed] [Google Scholar]
- 7.Silverman RB. Mechanism-Based Enzyme InactivationChemistry and Enzymology. Boca Raton, FL: CRC Press; 1988. [Google Scholar]
- 8.Murray M. Drug-mediated inactivation of cytochrome P450. Clin Exp Pharmacol Physiol. 1997;24:465–70. doi: 10.1111/j.1440-1681.1997.tb01228.x. [DOI] [PubMed] [Google Scholar]
- 9.Lin JH, Lu AY. Inhibition and induction of cytochrome P450 and the clinical implications. Clin Pharmacokinet. 1998;35:361–90. doi: 10.2165/00003088-199835050-00003. [DOI] [PubMed] [Google Scholar]
- 10.Sharma U, Roberts ES, Hollenberg PF. Inactivation of cytochrome P4502B1 by the monoamine oxidase inhibitors R-(-)-deprenyl and clorgyline. Drug Metab Dispos. 1996;24:669–75. [PubMed] [Google Scholar]
- 11.Muakkassah SF, Yang WC. Mechanism of the inhibitory action of phenelzine on microsomal drug metabolism. J Pharmacol Exp Ther. 1981;219:147–55. [PubMed] [Google Scholar]
- 12.Muakkassah SF, Bidlack WR, Yang WC. Mechanism of the inhibitory action of isoniazid on microsomal drug metabolism. Biochem Pharmacol. 1981;30:1651–8. doi: 10.1016/0006-2952(81)90393-2. [DOI] [PubMed] [Google Scholar]
- 13.Dupont H, Davies DS, Strolin-Benedetti M. Inhibition of cytochrome P-450-dependent oxidation reactions by MAO inhibitors in rat liver microsomes. Biochem Pharmacol. 1987;36:1651–7. doi: 10.1016/0006-2952(87)90050-5. [erratum appears in Biochem Pharmacol 1987; 36: 3093]. [DOI] [PubMed] [Google Scholar]
- 14.Wen X, Wang JS, Neuvonen PJ, Backman JT. Isoniazid is a mechanism-based inhibitor of cytochrome P450 1A2, 2A6, 2C19 and 3A4 isoforms in human liver microsomes. Eur J Clin Pharmacol. 2002;57:799–804. doi: 10.1007/s00228-001-0396-3. [DOI] [PubMed] [Google Scholar]
- 15.Tullman RH, Hanzlik RP. Inactivation of cytochrome P-450 and monoamine oxidase by cyclopropylamines. Drug Metab Rev. 1984;15:1163–82. doi: 10.3109/03602538409033560. [DOI] [PubMed] [Google Scholar]
- 16.Desta Z, Soukhova NV, Flockhart DA. Inhibition of cytochrome P450 (CYP450) isoforms by isoniazid: potent inhibition of CYP2C19 and CYP3A. Antimicrob Agents Chemother. 2001;45:382–92. doi: 10.1128/AAC.45.2.382-392.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ochs HR, Greenblatt DJ, Knuchel M. Differential effect of isoniazid on triazolam oxidation and oxazepam conjugation. Br J Clin Pharmacol. 1983;16:743–6. doi: 10.1111/j.1365-2125.1983.tb02256.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ochs HR, Greenblatt DJ, Roberts GM, Dengler HJ. Diazepam interaction with antituberculosis drugs. Clin Pharmacol Ther. 1981;29:671–8. doi: 10.1038/clpt.1981.94. [DOI] [PubMed] [Google Scholar]
- 19.Mallinger AG, Smith E. Pharmacokinetics of monoamine oxidase inhibitors. Psychopharmacol Bull. 1991;27:493–502. [PubMed] [Google Scholar]
- 20.Baker GB, Urichuk LJ, McKenna KF, Kennedy SH. Metabolism of monoamine oxidase inhibitors. Cell Mol Neurobiol. 1999;19:411–26. doi: 10.1023/A:1006901900106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Clark B. The in vitro inhibition of the N-demethylation of pethidine by phenelzine (Phenethylhydrazine) Biochem Pharmacol. 1967;16:2369–85. doi: 10.1016/0006-2952(67)90222-5. [DOI] [PubMed] [Google Scholar]
- 22.Polasek TM, Elliot DJ, Lewis BC, Miners JO. Mechanism-based inactivation of human cytochrome P4502C8 by drugs in vitro. J Pharmacol Exp Ther. 2004;311:996–1007. doi: 10.1124/jpet.104.071803. [DOI] [PubMed] [Google Scholar]
- 23.Boye SL, Kerdpin O, Elliot DJ, Miners JO, Kelly L, Mckinnon RA, Bhasker CR, Yoovathaworn K, Birkett DJ. Optimizing bacterial expression of catalytically active human cytochromes P450: comparison of CYP2C8 and CYP2C9. Xenobiotica. 2004;34:49–60. doi: 10.1080/00498250310001636868. [DOI] [PubMed] [Google Scholar]
- 24.Cuttle L, Munns AJ, Hogg NA, Scott JR, Hooper WD, Dickinson RG, Gillam EM. Phenytoin metabolism by human cytochrome P450: involvement of P450 3A and 2C forms in secondary metabolism and drug–protein adduct formation. Drug Metab Dispos. 2000;28:945–50. [PubMed] [Google Scholar]
- 25.Parikh A, Gillam EM, Guengerich FP. Drug metabolism by Escherichia coli expressing human cytochromes P450. Nat Biotechnol. 1997;15:784–8. doi: 10.1038/nbt0897-784. [DOI] [PubMed] [Google Scholar]
- 26.Miners JO, Rees DL, Valente L, Veronese ME, Birkett DJ. Human hepatic cytochrome P450 2C9 catalyzes the rate-limiting pathway of torsemide metabolism. J Pharmacol Exp Ther. 1995;272:1076–81. [PubMed] [Google Scholar]
- 27.Segel IH. Enzyme Kinetics. Behavior and Analysis of Rapid Equilibrium and Steady State Enzyme Systems. New York: Wiley; 1993. [Google Scholar]
- 28.Bensoussan C, Delaforge M, Mansuy D. Particular ability of cytochromes P450 3A to form inhibitory P450–iron–metabolite complexes upon metabolic oxidation of aminodrugs. Biochem Pharmacol. 1995;49:591–602. doi: 10.1016/0006-2952(94)00477-4. [DOI] [PubMed] [Google Scholar]
- 29.Wang Y, Jones DR, Hall SD. Prediction of cytochrome P450 3A inhibition by verapamil enantiomers and their metabolites. Drug Metab Dispos. 2004;32:259–66. doi: 10.1124/dmd.32.2.259. [DOI] [PubMed] [Google Scholar]
- 30.Belanger PM, Atitse-Gbeassor A. Inhibitory effect of tranylcypromine on hepatic drug metabolism in the rat. Biochem Pharmacol. 1982;31:2679–83. doi: 10.1016/0006-2952(82)90719-5. [DOI] [PubMed] [Google Scholar]
- 31.Taavitsainen P, Anttila M, Nyman L, Karnani H, Salonen JS, Pelkonen O. Selegiline metabolism and cytochrome P450 enzymes: in vitro study in human liver microsomes. Pharmacol Toxicol. 2000;86:215–21. doi: 10.1034/j.1600-0773.2000.d01-38.x. [DOI] [PubMed] [Google Scholar]
- 32.Franklin MR. Inhibition of mixed-function oxidations by substrates forming reduced cytochrome P-450 metabolic intermediate complexes. Pharmacol Ther. 1977;2:227–45. [Google Scholar]
- 33.Palamanda JR, Casciano CN, Norton LA, Clement RP, Favreau LV, Lin C, Nomeir AA. Mechanism-based inactivation of CYP2D6 by 5-fluoro-2-[4-[(2-phenyl-1H-imidazol-5-yl)methyl]-1-piperazinyl]pyrimidine. Drug Metab Dispos. 2001;29:863–7. [PubMed] [Google Scholar]
- 34.Hutzler JM, Hauer MJ, Tracy TS. Dapsone activation of CYP2C9-mediated metabolism: evidence for activation of multiple substrates and a two-site model. Drug Metab Dispos. 2001;29:1029–34. [PubMed] [Google Scholar]
- 35.Sharma U, Roberts ES, Hollenberg PF. Formation of a metabolic intermediate complex of cytochrome P4502B1 by clorgyline. Drug Metab Dispos. 1996;24:1247–53. [PubMed] [Google Scholar]
- 36.Kenworthy KE, Bloomer JC, Clarke SE, Houston JB. CYP3A4 drug interactions: correlation of 10 in vitro probe substrates. Br J Clin Pharmacol. 1999;48:716–27. doi: 10.1046/j.1365-2125.1999.00073.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ortiz de Montellano PR, Watanabe MD. Free radical pathways in the in vitro hepatic metabolism of phenelzine. Mol Pharmacol. 1987;31:213–9. [PubMed] [Google Scholar]
- 38.Ramirez J, Innocenti F, Schuetz EG, Flockhart DA, Relling MV, Santucci R, Ratain MJ. CYP2B6, CYP3A4, and CYP2C19 are responsible for the in vitro N-demethylation of meperidine in human liver microsomes. Drug Metab Dispos. 2004;32:930–6. [PubMed] [Google Scholar]
- 39.Lu P, Schrag ML, Slaughter DE, Raab CE, Shou M, Rodrigues AD. Mechanism-based inhibition of human liver microsomal cytochrome P4501A2 by zileuton, a 5-lipoxygenase inhibitor. Drug Metab Dispos. 2003;31:1352–60. doi: 10.1124/dmd.31.11.1352. [DOI] [PubMed] [Google Scholar]
- 40.Racha JK, Rettie AE, Kunze KL. Mechanism-based inactivation of human cytochrome P450 1A2 by furafylline: detection of a 1 : 1 adduct to protein and evidence for the formation of a novel imidazomethide intermediate. Biochemistry. 1998;37:7407–19. doi: 10.1021/bi973011m. [DOI] [PubMed] [Google Scholar]
- 41.Mayhew BS, Jones DR, Hall SD. An in vitro model for predicting in vivo inhibition of cytochrome P450 3A4 by metabolic intermediate complex formation. Drug Metab Dispos. 2000;28:1031–7. [PubMed] [Google Scholar]
- 42.Ernest CS, 2nd, Hall SD, Jones DR. Mechanism-based inactivation of CYP3A by HIV protease inhibitors. J Pharmacol Exp Ther. 2005;312:583–91. doi: 10.1124/jpet.104.075416. [DOI] [PubMed] [Google Scholar]
- 43.Venkatakrishnan K, Obach RS. In vitro-in vivo extrapolation of CYP2D6 inactivation by paroxetine: prediction of nonstationary pharmacokinetics and drug interaction magnitude. Drug Metab Dispos. 2005;33:845–52. doi: 10.1124/dmd.105.004077. [DOI] [PubMed] [Google Scholar]
- 44.Austin RP, Barton P, Cockroft SL, Wenlock MC, Riley RJ. The influence of nonspecific microsomal binding on apparent intrinsic clearance, and its prediction from physicochemical properties. Drug Metab Dispos. 2002;30:1497–503. doi: 10.1124/dmd.30.12.1497. [DOI] [PubMed] [Google Scholar]
- 45.Galetin A, Burt H, Houston JB. Prediction of time-dependent drug interactions—impact of parallel pathways, enzyme degradation and intestinal inhibition. Drug Metab Rev. 2005;37:38. doi: 10.1124/dmd.105.006874. [DOI] [PubMed] [Google Scholar]
- 46.Sweetman SC. Martindale Complete Drug Reference. 32. London: Pharmaceutical Press; 1999. Isoniazid; p. 218.1. [Google Scholar]
- 47.Robinson DS, Nies A, Cooper TB. Relationships of plasma phenelzine levels to platelet MAO inhibition, acetylator phenotype, and clinical outcome in depressed outpatients. Clin Pharmacol Ther. 1980;27:280. [Google Scholar]