Abstract
Aims
The aims of this observational study were to assess the variability in imatinib pharmacokinetics and to explore the relationship between its disposition and various biological covariates, especially plasma α1-acid glycoprotein concentrations.
Methods
A population pharmacokinetic analysis was performed using NONMEM based on 321 plasma samples from 59 patients with either chronic myeloid leukaemia or gastrointestinal stromal tumours. The influence of covariates on oral clearance and volume of distribution was examined. Furthermore, the in vivo intracellular pharmacokinetics of imatinib was explored in five patients.
Results
A one-compartment model with first-order absorption appropriately described the data, giving a mean (± SEM) oral clearance of 14.3 l h−1 (± 1.0) and a volume of distribution of 347 l (± 62). Oral clearance was influenced by body weight, age, sex and disease diagnosis. A large proportion of the interindividual variability (36% of clearance and 63% of volume of distribution) remained unexplained by these demographic covariates. Plasma α1-acid glycoprotein concentrations had a marked influence on total imatinib concentrations. Moreover, we observed an intra/extracellular ratio of 8, suggesting substantial uptake of the drug into the target cells.
Conclusion
Because of the high pharmacokinetic variability of imatinib and the reported relationships between its plasma concentration and efficacy and toxicity, the usefulness of therapeutic drug monitoring as an aid to optimizing therapy should be further investigated. Ideally, such an approach should take account of either circulating α1-acid glycoprotein concentrations or free imatinib concentrations.
Keywords: chronic myeloid leukaemia, drug monitoring, gastrointestinal stromal tumours, imatinib, orosomucoid, protein binding
Introduction
Imatinib mesylate (Gleevec® or Glivec®; Novartis Pharma AG, Basel, Switzerland) has transformed the treatment and prognosis of chronic myeloid leukaemia (CML) [1, 2] and gastrointestinal stromal tumours (GIST) [3]. Imatinib was rationally designed to inhibit the Bcr-Abl tyrosine kinase. This fusion oncoprotein results from a t(9,22) translocation which gives rise to the Philadelphia chromosome, the hallmark of CML and of some acute lymphoblastic leukaemias (ALL) [4]. Imatinib was also found to be a potent inhibitor of the autophosphorylation of two additional tyrosine kinases, namely, c-Kit, involved in the oncogenesis of GIST [5], and platelet-derived growth factor receptor (PDGFR), involved, for example, in the pathogenesis of the hypereosinophilic syndrome [6].
However, imatinib must be taken indefinitely and is not devoid of toxicity. For CML and GISTs, the most frequent adverse events are fluid retention, nausea, skin rash, asthenia and muscle cramps, with an incidence of more than 50% (grades 1–4). A trend suggesting increased incidence of grade 3/4 adverse events with advancing disease was observed, but this rarely leads to discontinuation of therapy [7, 8]. Moreover, resistance or escape from disease control occurs in a significant number of patients. Resistance to imatinib is variable, especially in CML during the accelerated or blastic phase [4, 9, 10]. Cellular mechanisms of resistance include point mutations in the BCR-ABL and KIT genes or alternatively amplification of BCR-ABL. Activation of alternative survival signalling pathways can also arise [11, 12]. The probability of harbouring resistance mutations increases with disease progression as a consequence of increased tumour cell abundance [13]. Resistance could also be directly or indirectly caused by an increase in the cellular efflux of imatinib, mediated by the drug transporter P-glycoprotein (P-GP, the gene product of ABCB1, formerly MDR1) [14, 15], or by the breast cancer resistance protein (Bcrp1, ABCG2) [16–18]. In addition, imatinib has recently been shown to be a substrate of the organic cation influx transporter 1 (hOCT1) [19]. Host-dependent mechanisms of resistance have also been implicated [4, 20], including modulation of imatinib binding to α1-acid glycoprotein (AGP) [21–23] and/or enhanced drug metabolism [9, 24]. Imatinib is mainly metabolized by cytochrome P450 3A4 (CYP3A4) [25, 26] and AGP plasma concentrations have been reported to be higher in the resistant CML blastic phase (2.3 g l−1) than in the chronic phase (1.1 g l−1) [23].
The pharmacokinetics of imatinib has been studied during Phases I, II and III of its clinical development [26–31]. Two of these studies applied a population approach [28, 30], and one [30] has indicated that imatinib clearance tends to increase over a period of 12 months’ treatment with the drug. Accordingly, imatinib concentrations in patients whose dosage was progressively increased were substantially lower than those at the beginning of treatment. By contrast, a 25% decrease in imatinib clearance was reported in another study in CML patients [28], albeit followed up for only 1 month. Because of this discrepancy, further investigation of the pharmcokinetics of imatinib is merited, with particular respect to the identification of individual kinetic determinants that could modulate clinical response [32]. Weight, creatininaemia, albuminaemia, haemoglobinaemia and some other covariates have already been assessed [28, 30]. Data on the effect of MDR1 expression and CYP3A4 activity on imatinib AUC have been published only in abstract form [33]. Finally, only in vitro data are available about the intracellular concentrations of imatinib on the uptake of imatinib into cells [29].
Accordingly, the aims of the present study were (i) to characterize the population pharmacokinetics of imatinib in CML and GIST patients, (ii) to evaluate the influence of various demographic covariates on imatinib absorption and disposition, (iii) to assess the specific role of AGP on imatinib pharmacokinetics, and (iv) to explore the intracellular uptake of imatinib in vivo.
Methods
Study population
Data from 59 patients, providing a total of 321 plasma samples, were collected over 3 years for the population PK analysis. These patients included 38 with GIST, 20 with CML and one with ALL, all of whom received imatinib at daily doses ranging from 150 to 800 mg. All patients were pooled in our analysis, regardless of their medical history. Median age was 55 years (range 20–79), body weight was 71 kg (44–110) and height was 172 cm (152–189); 26 patients were female. Most peripheral blood samples were drawn at 1–6-month intervals on follow-up visits for routine laboratory tests. The median number of measurements for each patient was four (range 1–14). All samples were obtained under steady-state conditions (i.e. after unchanged dosage for at least 1 month). Additional measurements were taken in five patients over one dosing interval to obtain a detailed concentration—time profile, with nine peripheral blood samples drawn between 0 and 8 h after drug intake. In these five patients, intracellular concentrations of imatinib were also measured in peripheral blood mononuclear cells (PBMC) obtained from four blood samples drawn before and 2, 4 and 6 h after drug intake using Vacutainer® CPT (Cell Preparation Tubes; Becton Dickinson, Allschwil, Switzerland), according to the manufacturer’s recommended procedure and the method previously developed in our laboratory for the intracellular measurement of anti-HIV drugs [34]. All samples were processed at 4 °C and the collected cells were washed three times with cold phosphate-buffered saline, prior to cell counting and imatinib analysis (described below).
The following data were recorded for each patient: body weight, sex, age, height, creatinine concentration (CRT, in µmol l−1) and concomitant intake of medications that might influence the metabolism of imatinib (Table 1).
Table 1. Characteristics of the 59 patients evaluated in the population pharmacokinetics analysis of imatinib.
| Characteristic | Patients |
|---|---|
| Pathology diagnosis (n) | |
| GIST | 38 |
| CML | 20 |
| ALL | 1 |
| Sex (n) | |
| Men | 33 |
| Women | 26 |
| Age (years) | |
| Median | 55 |
| Range | 20–79 |
| Body weight (kg) | |
| Median | 71 |
| Range | 44–110 |
| Height (cm) | |
| Median | 172 |
| Range | 152–189 |
| AGP plasma levels (g l−1) | |
| Median | 0.9 |
| Range | 0.4–3.2 |
| MDR1 genotype (n) | |
| 3435CC | 5 |
| 3435CT | 22 |
| 3435TT | 9 |
| CYP3A4 ratio (UCR) | |
| Median | 3.6 |
| Range | 0.7–38.5 |
| CYP3A4 inducers coadmininstered | |
| Carbamazepine, rifampicine, dexamethasone | 3 |
| CYP3A4 inhibitors coadministered | |
| Verapamil, diltiazem, fluoxetine, fluvoxamine, amiodarone, ethinylestradiol, fluconazole, voriconazole | 9 |
UCR, Urinary cortisol ratio; GIST, gastrointestinal stromal tumour; CML, chronic myeloid leukaemia; ALL, acute lymphoblastic leukaemia.
MDR1
genotype was determined in 36 patients with respect to the 3435C→T single nucleotide polymorphism, known to be associated with P-GP expression [35]. CYP3A4 activity was assessed by measuring the 6β-hydroxycortisol/cortisol ratio in a 10-ml spot sample of urine collected at the same time as the 164 peripheral blood samples [36]. These urine samples were stored at − 20 °C until analysis (see below). Plasma AGP concentrations were measured in 278 plasma samples.
The study was approved by the Ethics Committee of the Lausanne Faculty of Medicine. Informed written consent was obtained from the participants.
Drug and metabolite analysis
Peripheral blood samples (5 ml) were collected into K-EDTA Monovette® syringes (Sarstedt, Nümbrecht, Germany). Plasma was isolated by centrifugation and stored at − 20 °C until analysis. Total plasma imatinib concentrations (Ctot) were determined by reversed-phase high-performance liquid chromatography (HPLC) according to a validated method [37]. The limit of quantification of the assay was 50 µg l−1 and its precision was < 2.5% between 100 µg l−1 and 10 000 µg l−1.
6β-Hydroxycortisol and cortisol urinary concentrations, and intracellular imatinib concentrations in PBMC cells were measured using validated LC-MS/MS methods developed in our laboratory, and which were adapted from previously published assays [38–40]. The equipment consisted of a liquid chromatograph (Agilent 1100 system; Agilent, Böblingen, Germany) coupled with a tandem-mass spectrometer (TSQ Quantum Discovery tandem triple-stage quadripole Thermo; Finnigan, San Jose, CA, USA). Prior to these analyses, urine samples were purified using a solid-liquid SPE technique (Chromabond® XTR; Macherey-Nagel, Düren, Germany) and PBMCs were extracted with 100 µl PBS buffer and 100 µl acetonitrile (in order to denature proteins). The limits of quantification of the assays were 1 µg l−1 for cortisol, 10 µg l−1 for 6β-hydroxycortisol and 10 µg l−1 for intra-PBMC imatinib. Mean interday precision and accuracy were 7.4 and 7.2%, and 9.1 and 3.7%, for cortisol and 6β-hydroxycortisol, respectively. Mean intraday and interday precision for intra-PBMC imatinib were 2.9 and 6.3%, respectively, and the mean intraday and interday accuracy were 1.9 and 5.9%, respectively.
Plasma AGP concentrations were determined by an immunoturbidimetric assay using a COBAS INTEGRA 400 system (Roche Diagnostics, Basel, Switzerland). This is based on the formation of a precipitate of AGP with a specific antiserum which is determined by turbidimetry at 340 nm [41]. The limit of quantification of the assay was 0.16 g l−1. Mean intraday and interday precision were 1.9 and 2.4%, respectively, for a 0.62 g l−1 concentration and 1.0 and 1.5%, respectively, for a 2.22 g l−1 concentration.
MDRI genotyping
DNA was isolated from blood using the QIAamp® DNA Blood Mini Kit (Qiagen, Basel, Switzerland) and was used for genotyping by polymerase chain reaction (PCR) and restriction fragment length polymorphism. The primer sequences were as follows: forward 5′-TGGCAAAGAAATAAAGCGAC-3′; and reverse 5′-GACTCGATGAAGGCATGTA-3′. The PCR products of 189 bp were restricted by MboI and separated on 3% MetaPhor® agarose gels (Cambrex, Verviers, Belgium). The expected restriction enzyme-digested fragments were 143 and 46 bp for the CC genotype at nucleotide 3435; 189, 143 and 46 bp for CT; and 189 bp for TT.
Pharmacokinetic modelling
Demographic covariate analysis
Imatinib PK was best characterized by a one-compartment model with first-order absorption. This model is described by the following differential equations:
| (1) |
| (2) |
where A1 and A2 are the amounts of imatinib in the absorption and central compartments, ka is the first-order absorption rate, Ctot the measured drug concentration (corresponding to the A2/Vd ratio, and where Vd is the volume of distribution) and CL the clearance.
Three demographic covariates (body weight, sex and age), as well as the disease diagnosis, were then sequentially incorporated into the model and tested for significance, leading to the following equations:
 |
(3) |
| (4) |
where body weight (BW) and age (AGE) are expressed as the relative deviation of the individual BW and AGE from the population mean (BWmean = 70 kg and AGEmean = 50 years, respectively). Dichotomous variable were used for sex (q = 0 for female and 1 for male) and for the diagnosis (p = 0 for CML and 1 for GIST).
Biological covariates analysis
Several models were tested to assess the effect of AGP concentration on the pharmacokinetic parameters. First, a linear relationship was tested:
| (5) |
where X is expressed as the relative deviation of the individual X from population mean Xmean. The population mean AGP concentration (AGPmean) was equal to 0.95 g l−1. was also used afterwards for testing other biological covariates (see below).
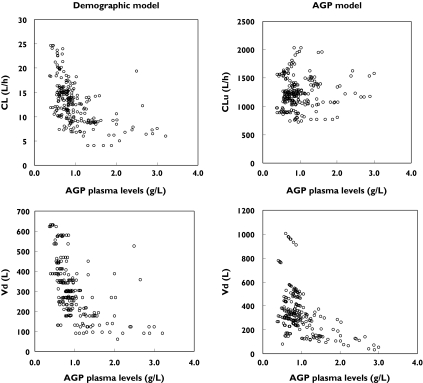
Based on an observed hyperbolic relationship between AGPtot and CL/Vd (see Results and Figure 2, left-hand panel), two simple hyperbolic models (Power and Emax-like, respectively) were also tested for CL and Vd:
Figure 2.
Pharmacokinetic parameters derived from the demographic covariates model (left panel) and from the final α1-acid glycoprotein (AGP) model (right panel), plotted according to AGP plasma concentrations
| (6) |
| (7) |
Additionally, a model derived from the expression of the free fraction ( fu), proposed by Rowland et al.[42]
| (8) |
was assessed:
| (9) |
where θ1 represents the unbound clearance (CLu) and θ2 accounts for both the association constant (Ka) and the fraction of the number of binding sites unoccupied ( fup). This model assumes a constant, nonsaturable free fraction.
Furthermore, a mechanistic approach was built up on the basis of physiological considerations. The model given by was thus rewritten, based on the assumption that only the unbound imatinib concentration Cu was able to undergo first-order elimination through an unbound clearance process:
| (10) |
The values of Cu were related to the predicted concentrations Ctot through the following equation, incorporating AGPtot, the AGP dissociation constant for imatinib Kd, and a scaling factor L:
 |
(11) |
The derivation of Equation 11 is described in Appendix II. A similar equation had been previously proposed for the analysis of the pharmacokinetics of sulphonamides by Bourne et al.[43, 44]. In our model we simply used the dissociation constant Kd instead of the association constant Ka. The L constant accounts for the difference in concentration unit between Ctot (µg l−1) and AGPtot (g l−1), assuming a one-to-one molar binding ratio [21]. It was fixed to 11.7 (assuming a molar mass of 493.6 g mol−1 for imatinib [45] and a mean molar mass of 42 000 g mol−1 for AGP [46]).
On the basis of similar considerations, another hyperbolic equation relating Vd to AGPtot was derived (see for details):
 |
(12) |
where VAGP represents the volume of distribution of the protein and Vu the volume of distribution of the unbound fraction of imatinib. Formally, makes Vd dependent on the predicted total concentration of imatinib (Ctot). However, to fit such an equation in NONMEM®, it was necessary to use the observed concentration instead.
The demographic covariates (BW, sex and age) and the disease diagnosis were tested again (see Equations 3 and 4), based on the new model incorporating AGP (Equations 10 and 11).
The effect of other biological covariates (MDR1 genotype, CYP3A4 activity and creatinine clearance) on imatinib PK was also assessed. For MDR1, the following expression was used:
| (13) |
where MDRT is equal to −1 for MDR1 genotype 3435CC, 0 for 3435CT and 1 for 3435TT. For CYP3A4 activity (3A4A, expressed as the urinary cortisol ratio) and creatinine clearance (CLCRT), the linear was used with 3A4Amean = 5.4 (6β-hydroxycortisol/cortisol ratio) and CLCRTmean = 75 ml min−1. CLCRT was calculated according to the Cockcroft and Gault formula [47].
Elimination half-life t1/2, absorption half-life t1/2a and imatinib free fraction fu were calculated as follows:
where Cu is the predicted unbound concentration and Ctot the predicted total concentration.
Statistical modelling
A hierarchical model was used to account for inter- and intraindividual variability. The individual pharmacokinetic parameters θj were modelled assuming a log normal distribution among the patients and were of the general form:
| (14) |
where θ is the population mean and ηj independent normally distributed random effects with a mean of zero and variance Ω.
A proportional model was used to describe intraindividual (residual) variability in imatinib pharmcokinetics. For the generic response Y and the corresponding prediction Ŷ,Y the ith measurement for the jth individual takes the form:
| (15) |
where ɛij is independent and normally distributed with a mean of zero and a variance σ2.
Parameter estimation and model selection
The analysis was performed using NONMEM® software (version V, with NM-TRAN version II) [48] running on a mainframe station (Sun Fire® 3800 server with UltraSPARC® III processors; Sun, Santa Clara, CA, USA). The program uses mixed (fixed and random) effects regression to estimate population means and variances of the pharmacokinetic parameters and to identify factors that may affect them. The data were fitted using a stepwise procedure and the first-order conditional method (FOCE INTERACTION and three significant digits) with the subroutine ADVAN 6. To determine the statistical significance between the two models, different statistical selection criteria can be used that require a minimal decrease of 2–10 points in the objective function (OF) [49, 50]. The decrease in OF corresponds to minus twice the logarithm of the linearized maximum likelihood of the model and is approximately χ2 distributed (based on the likelihood ratio test). Therefore, a decrease of > 3.8 points was considered significant for one additional parameter and >5.9 points for two additional parameters. Regression diagnostic plots generated with Excel® (version 11.2; Microsoft Co., Redmond, WA, USA) were used for comparison between models.
The general model was first evaluated with the data from the five patients who had undergone multiple blood sampling. An analysis of the whole population was then conducted using those initial estimates. The influence of each recorded patient characteristic on the Bayesian individual estimates of oral clearance (CL) and oral volume of distribution (Vd) was visually explored with Excel®. Patient characteristics showing potential influence on the pharmacokinetic parameters were then evaluated with NONMEM® by sequentially introducing them into the model. Finally, a simulation based on the final pharmacokinetic parameter estimates was performed with Excel® using first-order error propagation formulae to calculate the 90% prediction interval (Figure 1), encompassing the 5th and 95th expected concentration percentiles at each time point. The figures were generated with Prism® (version 4.03; Graphpad Software Inc., San Diego, CA, USA).
Figure 1.
Plasma imatinib concentrations observed in patients receiving imatinib, together with the mean population prediction (solid line) and 90% prediction interval (dashed lines). The graphs represent a once (upper part) or twice (lower part) daily regimen, based either on the demographic covariates model (left) or on the final α1-acid glycoprotein (AGP) model (right) (values adjusted to 400 mg q.d.; AGP model curves generated assuming mean values of AGP level and fu)
Results
The 321 imatinib plasma concentration values measured in the 59 patients ranged between 67 and 11 221 µg l−1. In the subpopulation of 36 patients (corresponding to 245 samples) five had the MDR1 3435CC and nine had the TT genotypes, theoretically associated with higher and lower P-GP expression, respectively, compared with the CT genotype found in 22 patients. CYP3A4 activity (evaluated as the urinary cortisol ratio and determined in 164 samples from 39 patients) ranged from 0.7 to 38.5. Finally, the plasma concentration of AGP in 51 patients (from 278 samples) ranged from 0.4 to 3.2 g l−1. The characteristics of the population are summarized in Table 1.
A one-compartment model with first-order absorption from the gastrointestinal tract appropriately described the data (Equations 1 and 2). A two-compartment model did not improve the fit, with a difference in the objective function (ΔOF) of zero. In the absence of intravenous data, the mean population bioavailability (F) was fixed to 1, in accordance with the almost complete absorption reported for imatinib [25, 51]. Significant interpatient variability could be assigned to both oral clearance (CV = 45%; ΔOF = −123.7) and oral volume of distribution (CV = 130%; ΔOF = −30.2), with the covariance between CL and Vd further decreasing the objective function (ΔOF = −18.4).
Individual estimates of CL and Vd were derived for each patient from this basic model and plotted against all the covariates to identify potential influences. Among the demographic factors, BW, height, age, sex, as well as the disease diagnosis, showed some influence on imatinib pharmacokinetics. Height, being correlated to BW, was not further investigated. CL increased significantly with BW (ΔOF = −63.4). The combination of BW and sex produced a slight improvement compared with when BW alone was used (ΔOF = −3.5). The addition of age and disease diagnosis provided some further improvement in fit (ΔOF = −4.5 and − 4.6, respectively). Finally, sex significantly affected Vd (ΔOF = −5.1). Although sex, age and disease diagnosis gave a fit of borderline statistical significance, these covariates were retained in the model, since they were always recorded and might be clinically relevant for selected patients (global ΔOF = −17.7).
Coadministration of CYP3A4 inhibitors (ΔOF =−0.8) or inducers (ΔOF = 0.0) did not affect imatinib pharmacokinetics to a statistically significant extent in this population of patients.
The final population estimates of CL and oral Vd from the demographic model were 14.3 l h−1 and 347 l, respectively. The derived elimination half-life was 17 h, and the absorption half-life 1.1 h. The interindividual variability in CL and Vd (CV = 36% and 63%) remained higher than the residual intraindividual variability (CV = 31%). CL increased by 99% on doubling of BW, decreased in female compared with male patients by 6%, decreased by 16% on doubling of age and decreased by 8% in GIST compared with CML patients. Details of the development of this demographic model can be found in and the pharmacokinetic data are shown in Table 2. The observed plasma concentrations of imatinib are presented in Figure 1 (left panel), along with the population mean and 90% prediction interval.
Table 2. Imatinib population pharmacokinetic parameters obtained from the two main models.
| Population mean | Inter-individual variability* | ||||
|---|---|---|---|---|---|
| Model | Parameter | Estimate | SE† | Estimate | SE† |
| Demographic | CL (l h−1) | 14.3 | 7.1% | 36% | 28.6%† |
| Vd (l) | 347 | 17.9% | 63% | 39.6%† | |
| ka (h−1) | 0.61 | 30.0% | – | ||
| σ (CV %)§ | 31% | 20.3%† | – | ||
| AGP | CLu (l h−1) | 1310 | 13.1% | 17% | 40.7%† |
| Vd (l) | 301 | 7.8% | 66% | 46.5%† | |
| ka (h−1) | 0.61 | 18.9% | – | ||
| Kd (mg l−1) | 0.090 | 14.3% | – | ||
| σ (CV %)§ | 23% | 20.3%† | – | ||
Estimates of variability expressed as coefficient of variation (CV %).
SE, Standard error of the estimates, expressed as CV %.
SE, Standard error of the variance components, taken as SEestimate/estimate, expressed as a percentage.
Residual intraindividual variability of the plasmaconcentration, expressed as CV %. AGP,α1-acid glycoprotein.
The plots in Figure 2 (left panel) clearly suggest a hyperbolic dependency of imatinib oral CL and Vd on plasma AGP concentrations. The use of a linear model to characterize the relationship between CL and AGP (Equation 5) improved significantly the fit (ΔOF =−170.5). Only a slightly better description of the data was detected when using a power function (; ΔOF = −5.6). The use of an Emax model (Equation 7) produced a substantial improvement in the fit (ΔOF =−267.2) and the constant free fraction approach described by was associated with an even better fit (ΔOF = −271.4). Finally, the mechanistic approach (Equation 12) provided the best improvement in the fit (ΔOF = −;284.1), and the plot of CLu vs. AGPtot showed an absence of a relationship between these two variables (Figure 2, right panel). As the apparent volume of distribution estimated from this model still showed some correlation with AGPtot, we tried to model this relationship (using Equations 5, 6, 7 and 12). The most significant improvement was obtained using a linear function (Equation 5; ΔOF =−11.7). The Emax-like relationship brought no significant improvement in the fit (Equation 7; ΔOF = −2.2). The mechanistic approach (Equation 12) was not superior to the model that did not take account of AGP (ΔOF =−0.0). This was probably due to the lack of very low plasma AGP concentrations that could have contributed to a residual hyperbolic relationship between AGPtot and Vd. As this mechanistic model could not be retained, Figure 2 (right panel) represents only the relationship between AGPtot and total volume of distribution (Vd).
Following the inclusion of AGP in the analysis (Equations 10 and 11), the four demographic covariates found to correlate significantly with CL in the previous analysis (see above) were again added to the AGP model and shown to affect CLu (rather than on CL) and Vd (linearly correlated to AGPtot by Equation 5). Whereas BW still improved the prediction of CLu (ΔOF = −27.0; change by 91% on doubling of BW), the addition of age and sex did not significantly improve the model compared with BW alone (ΔOF = −4.7). Disease diagnosis appeared to have no significant effect on Vd (ΔOF =−0.5), but significantly decreased CLu in GIST patients (ΔOF = −6.8; with a difference of 10.2% between GIST and CML patients), confirming the effect already observed with the demographic covariates model.
The influence of three other biological covariates were assessed. The MDR1 polymorphism was studied in the subpopulation of 36 patients for whom both MDR1 genotype and plasma AGP concentrations were available (corresponding to 245samples). The assignment of this genotype as a covariate on either CLu or Vd tended to improve the fit (ΔOF = −1.3 and − 1.0), albeit not significantly. Compared with the MDR1 3435CC genotype, CLu decreased by 10.6% in the TT genotype, whereas Vd decreased by 29.8%. No clear influence of CYP3A4 activity on CLu or Vd could be detected (ΔOF = −0.2 and −1.0) in this limited number of observations (154 samples in 39 patients). Doubling CYP3A4 activity only increased CLu by 1.1%. Creatinine clearance significantly influenced CLu (ΔOF =−5.7; with a change of 29.0% on a doubling of creatinine clearance) and Vd (ΔOF = −7.0; with a change of 53.2% on a doubling of creatinine clearance), in the corresponding subpopulation (271 samples in 47 patients). Such findings were in line with those obtained with the demographic model (data not shown).
The final parameters of the AGP model are given in Table 2. The value of the in vivo dissociation constant (Kd) of 8.97 × 10−2 mg l−1 (i.e. 1.82 × 10−4 mmol l−1), corresponds to an association constant of 5.5 × 106 l mol−1 and leads to a median free fraction ( fu) of 1.1% (range 0.3–2.3). Details of the development of the AGP model can be found in Appendix IV.
Figure 3 shows the intracellular and plasma imatinib concentration vs. time profiles observed in the five patients studied more intensively. As these patients were at steady state on a once-daily regimen, the concentration at 0 h (i.e. prior to drug administration) was plotted at a time corresponding to the period elapsed between the previous drug intake (i.e. the day before) and the time zero sample.
Figure 3.
Intracellular and plasma pharmacokinetic profiles of imatinib in five individual patients (solid lines, plasma concentrations; broken lines, peripheral blood mononuclear cell concentrations)
Except for one subject, the intra/extracellular ratio appeared to be reasonably constant and indicated a mean eightfold accumulation of imatinib in the cell. The atypical appearance of the intracellular curve in the subject represented by open triangles is most probably due to inaccurate cell counting in the 24-h sample.
Discussion
This study enabled the development of two types of population pharmacokinetic model for imatinib. The use of a demographic covariates model confirmed the large interindividual variability in imatinib disposition, which can be explained only partly by BW, sex, age and disease diagnosis. Together with the residual intrapatient variability (31%), this translates into a mean fivefold width of the prediction interval around the mean data (or sixfold at Cmax; Figure 1, left panel). Our estimates of CL and Vd are in good agreement with those recently published in GIST [30] and CML [28] patients, as well as in healthy subjects after a single oral dose [31]. Our data also confirm that disease diagnosis has only a small influence on total and free clearance, in agreement with the initial assumption of the manufacturer that pharmacokinetics of imatinib was unaffected by CML or GIST [32]. The elimination half-life of 17 h is also in good agreement with previously reported values of 17–19 h in CML patients [25, 27, 29], 12–16 h in GIST patients [30] and 14 h in healthy subjects [31]. The relationship between CL and BW suggests that the latter could be used to guide dosing. Moreover, the substantial variability in disposition observed implies that a given dose of imatinib may give rise to, in some patients, plasma concentrations that depart markedly from those expected based on the mean pharmacokinetic profile (established in Phase I and II clinical studies). Figure 1 shows typical concentration—time curves usable for Bayesian-type dosage adjustment in patients receiving imatinib.
Unlike other previously studied covariates, AGP plasma concentrations proved to have a marked influence on imatinib pharmacokinetics and explained about one-half of the interpatient variability in total CL. Using the model incorporating this covariate, the estimates of Vd and ka remained similar, but the 90% prediction interval around the mean plasma concentration—time data was reduced to a threefold width (Figure 1, right panel). This model is derived from mechanistic considerations, assuming saturable protein binding because of the limited amount of circulating AGP. The disappearance of the effects of sex and age in this AGP model can probably be explained by the known correlation between age and sex and AGP concentrations [52]. The role of AGP in imatinib disposition has been reported [22, 26] and the marked relationship between protein binding and the pharmacokinetics of imatinib again suggests the possibility that high concentrations of AGP might predispose to resistance to the drug [53–55]. However, because imatinib has a low hepatic extraction, a change in protein binding should not translate into significant variations in free drug concentration, since CLu remains constant in our model. This implies that the free AUC, and thus the cellular disposition to the drug, will remain unaffected. Furthermore, AGP concentrations might be expected to fall in parallel with response to imatinib treatment, as the disease burden is reduced.
Because protein binding affects the total concentration of imatinib, this factor should be taken into account when monitoring and interpreting total concentrations. Our findings indeed indicate that total plasma imatinib concentrations do not simply reflect the free (and thus the target) concentrations of this molecule [22]. Either the measurement of free concentration or the correction of the total concentration for binding to AGP should be considered. Although free drug concentration monitoring might be more appropriate, the technology to do so is not widely available. Thus, using plasma AGP concentrations instead to deduce the free fraction of the drug could represent a more convenient approach. In day-to-day practice, the determination of total plasma AGP and total imatinib concentrations would enable the calculation of a free concentration of imatinib from Equation 10. This value could then be compared with the typical free concentration profile shown in Figure 1 (right panel) and provide guidance for dose adjustment.
Our model incorporating AGP also provides estimates of the in vivo association constant and fu. Our value of the association constant of 5.5 × 106 l mol−1 determined in vivo is similar to that of 4.9 × 106 l mol−1 previously reported in vitro[21]. This denotes a very high affinity of imatinib for AGP and could explain the marked effect of AGP on the pharmacokinetics of the drug, which has also been reported for other anticancer agents [56]. The median in vivo free fraction ( fu) of 1.1% is of the same order of magnitude as the value determined in vitro (3.1% at an AGP concentration of 1 g l−1 and imatinib concentrations of 0.3–0.5 µg ml−1[57]). Such findings clarify in part the in vivo effect of protein binding on imatinib disposition [26]. The effect of albumin was not considered because its concentration varies much less than that of AGP, and imatinib is known to bind preferentially to AGP [21, 57]. A recent population PK study reported no significant relationship between albumin concentrations and imatinib pharmacokinetics [30].
The MDR1 polymorphism and CYP3A4 activity seemed to have a limited influence on imatinib disposition, although the 3435C→T MDRI variant is known to be correlated to the AUC of imatinib [33]. The analysis of other single nucleotide polymorphisms (in ABCB1 and ABCG2) may enable additional information to be gained on imatinib transport. Furthermore, the use of other markers of CYP3A4, such as the erythromycin breath test or midazolam clearance, may help to define better any relationship between imatinib pharmacokinetics and the activity of this enzyme [33]. Despite the concerns over the validity of the urinary cortisol ratio, it has the advantage of representing a non-invasive test [58]. The lack of apparent effect of CYP3A4 inhibitors or inducers on imatinib disposition is probably due to the limited number of our patients exposed to such drugs. Most were outpatients not receiving any comedication. However, the observed effect of creatinine clearance on imatinib clearance and volume of distribution suggests some influence, either direct or indirect, of renal function on imatinib disposition, despite its predominantly hepatic clearance [25].
The present study reports the first in vivo assessment of the intracellular accumulation of imatinib. A mean eightfold cell/plasma ratio was observed, which is in agreement with in vitro literature data, giving approximately a corresponding fivefold factor in leukaemia cell cultures incubated with imatinib and human plasma [29]. This is in line with the recent demonstration that imatinib undergoes active transport inside leukaemia cells via the solute liquid transporter hOCT1 [19]. It has been recently reported that hOCT1 expression varies between CML patients who are responders and nonresponders [59]. The relative stability of the cell/plasma ratio suggests that measuring plasma imatinib concentration (either total, AGP-corrected or free) probably provides a good surrogate for intracellular exposure to the drug. The intracellular concentration of imatinib would be better termed the ‘cell-associated amount’, as the drug may be embedded in membrane lipid bilayers, be complexed to cytoplasmic proteins or sequestrated in specific subcellular fractions. Such uptake has been reported for anti-HIV drugs [60, 61] and only a fraction of the so-called ‘intracellular amount’ of the drug is available to exert its pharmacological activity.
A pharmacokinetic—pharmacodynamic (PK—PD) analysis is currently being carried out in our centre to determine whether the side-effects or effectiveness of imatinib correlate with plama drug concentration among subgroups of patients. Several cases have been reported where imatinib treatment had to be discontinued because of the occurrence of serious adverse events [62–64]. However, in only one case were plasma drug concentrations measured and found to be elevated [64]. Furthermore, a number of studies have suggested that the administration of doses higher than the typical 400-mg daily regimen could improve response in some patient groups. A better response was observed in the accelerated and blastic phases of CML when the drug was given at a dose of 600 mg daily [65] and a 800-mg daily regimen was associated with longer progression-free survival in GIST patients [66]. About 10% of imatinib is metabolized by CYP3A4 to a N-demethylated piperazine derivative (CGP74588), which has similar in vitro potency to the parent drug [26]. At steady-state, this represents less than 20% of the dose [29].
However, such a small amount of active metabolite should not impact significantly on any PK—PD analysis of the drug.
In conclusion, the high interpatient and limited intrapatient variability in imatinib pharmacokinetics together with the potential relationship between exposure and efficacy and toxicity suggest that therapeutic monitoring of the drug may aid dosage adjustment. Such a measure may help to limit the incidence of side-effects and delay the emergence of tumour resistance, which seems to be favoured by prolonged subtherapeutic drug concentration exposure both in vitro[23, 67, 68]. In addition, therapeutic drug monitoring may improve compliance to therapy. However, before individualization of imatinib therapy based on routine drug concentration monitoring can be recommended, further PK—PD analysis in well-controlled trials is required.
Conflict of interest
The manufacturer of imatinib, Novartis, played no role in the initiation of this study. None of the authors has received commercial research funding or has relationships that could be perceived as having influenced the present research. However, S.L. and M.A.D. have been reimbursed by Novartis Pharma AG for attending a symposium. S.L. and T.B. have been paid by Novartis Pharma AG for speaking at a conference. L.A.D. and M.A.D. have received a grant-in-aid from Novartis Pharma AG. T.B. has received financial support from Novartis Pharma AG for two unrelated clinical studies.
The authors thank the program of the Master of Advanced Studies in Hospital Pharmacy (Professor A. Pannatier, Pharmacy Service, University Hospital of Lausanne, Switzerland), Fondation pour la Recherche et l’Enseignement en Pharmacologie clinique (Lausanne), Professor F. Widmer (Department of Ecology, University of Lausanne) for his valued editing of the manuscript and O. Woringer (Preparatory Mathematics Courses, Federal Polytechnic School, Lausanne) for his appreciated mathematical advice.
Appendix I
The following table summarizes the development of the demographic population pharmacokinetic model for imatinib.
| Hypothesis | Model | θa | θb | θc | θ1 | θ2 | θ3 | θ4 | ΔOF |
|---|---|---|---|---|---|---|---|---|---|
| Demographic covariates model (n = 59) | |||||||||
| Basic model | 12.1 | 299 | 0.467 | — | |||||
| Does BW influence CL? | θa + θ1 · BW | 12.1 | 255 | 0.433 | 13.5 | −63.4 | |||
| Vd? | θb + θ1 · BW | 12.1 | 299 | 0.466 | 0.00 | +0.0 | |||
| Does sex influence CL? | θa + θ2 · q − θ2 · (1 − q) | 12.2 | 267 | 0.433 | 2.3 | −36.1 | |||
| Vd? | θb + θ2 · q − θ2 · (1 − q) | 12.1 | 281 | 0.389 | −77.9 | −8.6 | |||
| Does age influence CL? | θa + θ3 · AGE | 12.3 | 307 | 0.470 | −1.6 | −1.6 | |||
| Vd? | θb + θ3 · AGE | 12.1 | 299 | 0.466 | 0.0 | +0.0 | |||
| Does pathology diagnosis influence CL? | θa + θ4 · p − θ4 · (1 − p) | 12.8 | 295 | 0.442 | −1.6 | −9.9 | |||
| Vd? | θb + θ4 · p − θ4 · (1 − p) | 12.1 | 255 | 0.372 | 53.1 | −2.5 | |||
| Does CYP3A4 inh. influence CL? | θa + θ6 · INH | 12.0 | 295 | 0.454 | +0.9 | ||||
| Does CYP3A4 ind. influence CL? | θb + θ7 · IND | 12.1 | 299 | 0.467 | +0.0 | ||||
| Does BW + sex influence CL? | θa + θ1 · BW + θ2 · q − θ2 · (1 − q) | 12.1 | 259 | 0.451 | 11.2 | 0.8 | −66.9 | ||
| Does BW + sex + age influence CL? | θa + θ1 · BW + θ2 · q − θ2 · (1 − q) + θ3 · AGE | 12.4 | 268 | 0.466 | 11.7 | 0.7 | −2.2 | −71.4 | |
| Does BW + sex + age + pathology diagnosis influence CL? | θa + θ1 · BW + θ2 · q − θ2 · (1 − q) + θ3 · AGE + θ4 · p − θ4 · (1 − p) | 12.8 | 271 | 0.471 | 11.4 | 0.7 | −1.7 | −0.9 | −76.0 |
| Does BW + sex + age + pathology diagnosis influence CL, and sex Vd? | θa + θ1 · BW + θ2 · q − θ2 · (1 − q) + θ3 · AGE + θ4 · p − θ4 · (1 − p)θb + θ5 · q − θ5 · (1 − q) | 12.8 | 258 | 0.437 | 12.7 | 0.8 | −2.1 | −1.0 | −81.1 |
θa,b,c, Tested PK parameters (clearance, CL, volume of distribution, Vd and absorption constant, ka, respectively); θ1,2,3,4,5, covariate coefficient estimates. BW (body weight) and age are expressed as the relative deviation of the individual BW and age from the population mean (70 kg and 50 years, respectively). INH or IND, 1 if concomitant inhibitor/inducer drug is present; q, 0 if female and 1 if male; p, 0 if chronic myeloid leukaemia, 1 if gastrointestinal stromal tumour; n, number of patient in the dataset; ΔOF, difference in the NONMEM objective function (OF) compared with baseline model.
Appendix II
Derivation of Cu (imatinib free concentration; Equation 11), assuming saturable binding of imatinib to AGP (considered as the main binding protein [21]):
| (16) |
Applying the law of mass action gives (u = unbound; b = bound):
| (17) |
and
| (18) |
Combining Equations 17 and 18 provides an expression for Cu:
| (19) |
| (20) |
Finally, solving this equation provides the expression for Cu given in Equation 11.
Appendix III
To express Vd as a function of AGPtot and Ctot, it is useful to determine the distribution of imatinib according to Figure 4 (assuming that AGP is the main binding protein [21]).
Figure 4.
Representation of the distribution of imatinib, based on concentrations and volumes (Vtot = Vd = total imatinib volume of distribution, VAGP = total α1-acid glycoprotein (AGP) plasma volume; Vu = free imatinib volume, Ctot = total imatinib concentration, Cu = free imatinib concentration, Cb = concentration of imatinib bound to AGP, AGPtot = total AGP concentration, AGPu = free AGP concentration, AGPb = concentration of AGP bound to imatinib, L = scaling factor)
It follows that the total unbound and AGP-bound volumes are given by:
| (21) |
| (22) |
| (23) |
Assuming that the total quantity of imatinib (Atot) is equal to the sum of the unbound (Au) and bound (Abound) forms:
| (24) |
Therefore, considering that Ctot is the sum of Cu and Cb (i.e. Cu = Ctot − Cb) it follows that:
| (25) |
Furthermore, assuming the law of mass action and rewriting Equation 19 according to Cb gives:
| (26) |
Because Cu is Ctot − Cb, Equation 26 can be expressed as:
| (27) |
Substituting Equation 25 into Equation 27 and solving according to Vtot gives Equation 12, which expresses Vd (the ratio between Atot and CLtot) as a function of AGPtot and Ctot, with parameters VAGP, Vu, Kd and LEquation 12 depicts an oblique hyperbole with one horizontal asymptote.
Appendix IV
The following table summarizes the models used to examine the influence of biological covariates on imatinib CL and Vd.
| Hypothesis | Model | θa | θb | θc | θ1 | θ2 | θ3 | ΔOF |
|---|---|---|---|---|---|---|---|---|
| AGP model (n = 51) | ||||||||
| 1-compartment, 1st order | 11.8 | 322 | 0.660 | – | ||||
| Does AGP influence CL (eq.5)? | CL = f(eq.5) | 12.5 | 308 | 0.820 | −170.5 | |||
| Does AGP influence CL (eq. 6)? | CL = f(eq. 6) | 12.2 | 320 | 0.672 | −5.6 | |||
| Does AGP influence CL (eq. 7)? | CL = f(eq. 7) | 12.2 | 356 | 0.797 | −267.2 | |||
| Does AGP influence CL (eq. 8)? | CL = f(eq. 8) | 12.1 | 365 | 0.767 | −271.4 | |||
| Does AGP influence CL (eq. 10–11)? | CLu = f(eq. 10–11) | 1200 | 355 | 0.741 | −284.1 | |||
| Does AGP influence Vd (eq. 5)? | CLu = f(eq. 10–11), Vd = f(eq. 5) | 1110 | 327 | 0.707 | −295.8 | |||
| Does AGP influence Vd (eq. 6)? | CLu = f(eq. 10–11), Vd = f(eq. 6) | 1230 | 354 | 0.742 | −267.2 | |||
| Does AGP influence Vd (eq. 7)? | CLu = f(eq. 10–11), Vd = f(eq. 7) | 1040 | 338 | 0.732 | −298.0 | |||
| Does AGP influence Vd (eq. 11)? | CLu = f(eq. 10–11), Vd = f(eq. 11) | 1290 | 355 | 0.740 | −267.2 | |||
| Does BW influence CLu? | CLu = f(eq. 10–11), θa + θ1 · BW, | 1100 | 320 | 0.672 | 1000 | 61 | −325.0 | |
| Vd = f(eq. 5) | ||||||||
| Does BW + sex influence CLu? | CLu = f(AGP, eq. 10–11) | 1100 | 306 | 0.656 | 844 | −327.2 | ||
| θa + θ1 · BW + θ2 · q − θ2 · | ||||||||
| (1−q), Vd = f(eq. 5) | ||||||||
| Does BW influence CLu and sex | CLu = f(AGP, eq. 10–11), | 1040 | 317 | 0.661 | 964 | 28 | −325.6 | |
| influence Vd? | θa + θ1 · BW, Vd = f(eq. 5), | |||||||
| θb + θ2 · q − θ2 · (1−q) | −325.6 | |||||||
| Does BW + sex + age influence CLu? | CLu = f(AGP, eq. 10–11), | 1110 | 305 | 0.640 | 858 | 51 | −151 | −328.6 |
| θa+ θ1·BW + θ2· q − θ2 | ||||||||
| (1 − q) + θ3 · AGE, Vd = f(eq.5) | ||||||||
| Does BW + pathology diagnosis influence CLu? | CLu = f(AGP,eq. 10–11) | 1180 | 324 | 0.698 | −120 | −332.0 | ||
| θa + θ1 · BW + θ2 · p − θ2 · | ||||||||
| (1 − p), Vd = f(eq. 5) | ||||||||
| Vd? | CLu = f(AGP,eq. 10–11;), | 905 | 313 | 0.654 | −8 | −325.7 | ||
| θa + θ1 · BW, Vd = f(eq. 5), | ||||||||
| θb + θ2 · p − θ2 · (1 − p) | ||||||||
| MDR1 genotype (n = 36) | ||||||||
| AGP model | 1170 | 289 | 0.699 | – | ||||
| Does MDR1 influence CLu? | θa + θ1 · MDRT | 1240 | 297 | 0.709 | −66 | −1.3 | ||
| Vd? | θb + θ1 · MDRT | 11240 | 344 | 0.705 | −51 | −1.0 | ||
| CYP3A4 activity (n = 39) | ||||||||
| AGP model | 1180 | 257 | 0.460 | – | ||||
| Does CYP3A4 influence CLu? | θa + θ2 · 3A4A | 1220 | 256 | 0.455 | 13 | −0.2 | ||
| Vd? | θb + θ3 · 3A4A | 1090 | 278 | 0.489 | 11 | −1.0 | ||
| CRT clearance (n = 47) | ||||||||
| AGP model | 1120 | 311 | 0.695 | – | ||||
| Does CLCRT influence CLu? | θa + θ3 · CLCRT | 1160 | 306 | 0.692 | 336 | −5.7 | ||
| Vd? | θb + θ4 · CLCRT | 1300 | 316 | 0.702 | 168 | −7.0 | ||
θa,b,c, Tested PK parameters (clearance CL, free clearance CLu, volume of distribution Vd and absorption constant, ka, respectively); θ1,2,3, covariate estimates (BW, sex, age or pathology diagnosis, or, respectively, MDRT, 3A4A and CLCRT); BW (body weight), age, AGP (AGP plasma levels), 3A4A (CYP3A4 cortisol ratio), CLCRT (creatinine clearance): expressed as the relative deviation of the individual BW, AGE, AGPtot, 3A4A and CRT from the population mean (70 kg, 50 years, 0.95 g l−1, 5.4 and 75 ml min−1, respectively); q = 0 if female, 1 if male; MDRT = −1 if 3435CC, 0 if 3435CT, 1 if 3435TT; p = 0 if CML, 1 if GIST; n, number of patients in the dataset; ΔOF, difference in the NONMEM objective function (OF) compared with baseline model. In Equation 11, the Ctot variable was defined as equal to the DV variable and was thus simply duplicated from this one.
References
- 1.Druker BJ. Imatinib mesylate in the treatment of chronic myeloid leukaemia. Expert Opin Pharmacother. 2003;4:963–71. doi: 10.1517/14656566.4.6.963. [DOI] [PubMed] [Google Scholar]
- 2.Tothova E, Kafkova A, Fricova M, Benova B, Kirschnerova G, Tothova A. Imatinib mesylate in Philadelphia chromosome-positive, chronic-phase myeloid leukemia after failure of interferon alpha. Neoplasma. 2005;52:63–7. [PubMed] [Google Scholar]
- 3.Steinert DM, McAuliffe JC, Trent JC. Imatinib mesylate in the treatment of gastrointestinal stromal tumour. Expert Opin Pharmacother. 2005;6:105–13. doi: 10.1517/14656566.6.1.105. [DOI] [PubMed] [Google Scholar]
- 4.Capdeville R, Buchdunger E, Zimmermann J, Matter A. Glivec (STI571, imatinib), a rationally developed, targeted anticancer drug. Nat Rev Drug Discov. 2002;1:493–502. doi: 10.1038/nrd839. [DOI] [PubMed] [Google Scholar]
- 5.Demetri GD, von Mehren M, Blanke CD, Van den Abbeele AD, Eisenberg B, Roberts PJ, Heinrich MC, Tuveson DA, Singer S, Janicek M, Fletcher JA, Silverman SG, Silberman SL, Capdeville R, Kiese B, Peng B, Dimitrijevic S, Druker BJ, Corless C, Fletcher CD, Joensuu H. Efficacy and safety of imatinib mesylate in advanced gastrointestinal stromal tumors. N Engl J Med. 2002;347:472–80. doi: 10.1056/NEJMoa020461. [DOI] [PubMed] [Google Scholar]
- 6.Cools J, DeAngelo DJ, Gotlib J, Stover EH, Legare RD, Cortes J, Kutok J, Clark J, Galinsky I, Griffin JD, Cross NC, Tefferi A, Malone J, Alam R, Schrier SL, Schmid J, Rose M, Vandenberghe P, Verhoef G, Boogaerts M, Wlodarska I, Kantarjian H, Marynen P, Coutre SE, Stone R, Gilliland DG. A tyrosine kinase created by fusion of the PDGFRA and FIP1L1 genes as a therapeutic target of imatinib in idiopathic hypereosinophilic syndrome. N Engl J Med. 2003;348:1201–14. doi: 10.1056/NEJMoa025217. [DOI] [PubMed] [Google Scholar]
- 7.Cohen MH, Johnson JR, Pazdur R. U.S. Food and Drug Administration drug approval summary: conversion of imatinib mesylate (STI571; Gleevec) tablets from accelerated approval to full approval. Clin Cancer Res. 2005;11:12–19. [PubMed] [Google Scholar]
- 8.Zalcberg JR, Verweij J, Casali PG, Le Cesne A, Reichardt P, Blay JY, Schlemmer M, Van Glabbeke M, Brown M, Judson IR Eortc Soft Tissue and Bone Sarcoma Group, the Italian Sarcoma Group, Australasian Gastrointestinal Trials Group. Outcome of patients with advanced gastro-intestinal stromal tumours crossing over to a daily imatinib dose of 800 mg after progression on 400 mg. Eur J Cancer. 2005;41:1751–7. doi: 10.1016/j.ejca.2005.04.034. [DOI] [PubMed] [Google Scholar]
- 9.Nimmanapalli R, Bhalla K. Mechanisms of resistance to imatinib mesylate in Bcr-Abl-positive leukemias. Curr Opin Oncol. 2002;14:616–20. doi: 10.1097/00001622-200211000-00005. [DOI] [PubMed] [Google Scholar]
- 10.Hochhaus A, La Rosee P. Imatinib therapy in chronic myelogenous leukemia: strategies to avoid and overcome resistance. Leukemia. 2004;18:1321–31. doi: 10.1038/sj.leu.2403426. [DOI] [PubMed] [Google Scholar]
- 11.Sawyers CL, Hochhaus A, Feldman E, Goldman JM, Miller CB, Ottmann OG, Schiffer CA, Talpaz M, Guilhot F, Deininger MW, Fischer T, O’Brien SG, Stone RM, Gambacorti-Passerini CB, Russell NH, Reiffers JJ, Shea TC, Chapuis B, Coutre S, Tura S, Morra E, Larson RA, Saven A, Peschel C, Gratwohl A, Mandelli F, Ben-Am M, Gathmann I, Capdeville R, Paquette RL, Druker BJ. Imatinib induces hematologic and cytogenetic responses in patients with chronic myelogenous leukemia in myeloid blast crisis: results of a phase II study. Blood. 2002;99:3530–9. doi: 10.1182/blood.v99.10.3530. [DOI] [PubMed] [Google Scholar]
- 12.Weisberg E, Griffin JD. Resistance to imatinib (Glivec): update on clinical mechanisms. Drug Resist Updat. 2003;6:231–8. doi: 10.1016/s1368-7646(03)00062-1. [DOI] [PubMed] [Google Scholar]
- 13.Michor F, Hughes TP, Iwasa Y, Branford S, Shah NP, Sawyers CL, Nowak MA. Dynamics of chronic myeloid leukaemia. Nature. 2005;435(7046):1267–70. doi: 10.1038/nature03669. [DOI] [PubMed] [Google Scholar]
- 14.Mahon FX, Belloc F, Lagarde V, Chollet C, Moreau-Gaudry F, Reiffers J, Goldman JM, Melo JV. MDR1 gene overexpression confers resistance to imatinib mesylate in leukemia cell line models. Blood. 2003;101:2368–73. doi: 10.1182/blood.V101.6.2368. [DOI] [PubMed] [Google Scholar]
- 15.Widmer N, Colombo S, Buclin T, Decosterd LA. Functional consequence of MDR1 expression on imatinib intracellular concentrations. Blood. 2003;102:1142. doi: 10.1182/blood-2003-03-0993. [DOI] [PubMed] [Google Scholar]
- 16.Breedveld P, Pluim D, Cipriani G, Wielinga P, van Tellingen O, Schinkel AH, Schellens JH. The effect of Bcrp1 (Abcg2) on the in vivo pharmacokinetics and brain penetration of imatinib mesylate (Gleevec): implications for the use of breast cancer resistance protein and P-glycoprotein inhibitors to enable the brain penetration of imatinib in patients. Cancer Res. 2005;65:2577–82. doi: 10.1158/0008-5472.CAN-04-2416. [DOI] [PubMed] [Google Scholar]
- 17.Burger H, van Tol H, Boersma AW, Brok M, Wiemer EA, Stoter G, Nooter K. Imatinib mesylate (STI571) is a substrate for the breast cancer resistance protein (BCRP)/ABCG2 drug pump. Blood. 2004;104:2940–2. doi: 10.1182/blood-2004-04-1398. [DOI] [PubMed] [Google Scholar]
- 18.Burger H, Nooter K. Pharmacokinetic resistance to imatinib mesylate: role of the ABC drug pumps ABCG2 (BCRP) and ABCB1 (MDR1) in the oral bioavailability of imatinib. Cell Cycle. 2004;3:1502–5. doi: 10.4161/cc.3.12.1331. [DOI] [PubMed] [Google Scholar]
- 19.Thomas J, Wang L, Clark RE, Pirmohamed M. Active transport of imatinib into and out of cells: implications for drug resistance. Blood. 2004;104:3739–45. doi: 10.1182/blood-2003-12-4276. [DOI] [PubMed] [Google Scholar]
- 20.Hegedus T, Orfi L, Seprodi A, Varadi A, Sarkadi B, Keri G. Interaction of tyrosine kinase inhibitors with the human multidrug transporter proteins, MDR1 and MRP1. Biochim Biophys Acta. 2002;1587:318–25. doi: 10.1016/s0925-4439(02)00095-9. [DOI] [PubMed] [Google Scholar]
- 21.Gambacorti-Passerini C, Barni R, le Coutre P, Zucchetti M, Cabrita G, Cleris L, Rossi F, Gianazza E, Brueggen J, Cozens R, Pioltelli P, Pogliani E, Corneo G, Formelli F, D’Incalci M. Role of alpha1 acid glycoprotein in the in vivo resistance of human BCR-ABL(+) leukemic cells to the abl inhibitor STI571. J Natl Cancer Inst. 2000;92:1641–50. doi: 10.1093/jnci/92.20.1641. [DOI] [PubMed] [Google Scholar]
- 22.Gambacorti-Passerini C, Zucchetti M, Russo D, Frapolli R, Verga M, Bungaro S, Tornaghi L, Rossi F, Pioltelli P, Pogliani E, Alberti D, Corneo G, D’Incalci M. Alpha1 acid glycoprotein binds to imatinib (STI571) and substantially alters its pharmacokinetics in chronic myeloid leukemia patients. Clin Cancer Res. 2003;9:625–32. [PubMed] [Google Scholar]
- 23.Larghero J, Leguay T, Mourah S, Madelaine-Chambrin I, Taksin AL, Raffoux E, Bastie JN, Degos L, Berthaud P, Marolleau JP, Calvo F, Chomienne C, Mahon FX, Rousselot P. Relationship between elevated levels of the alpha 1 acid glycoprotein in chronic myelogenous leukemia in blast crisis and pharmacological resistance to imatinib (Gleevec) in vitro and in vivo. Biochem Pharmacol. 2003;66:1907–13. doi: 10.1016/s0006-2952(03)00469-6. [DOI] [PubMed] [Google Scholar]
- 24.Rochat B. Role of cytochrome P450 activity in the fate of anticancer agents and in drug resistance: focus on tamoxifen, paclitaxel and imatinib metabolism. Clin Pharmacokinet. 2005;44:349–66. doi: 10.2165/00003088-200544040-00002. [DOI] [PubMed] [Google Scholar]
- 25.Cohen MH, Williams G, Johnson JR, Duan J, Gobburu J, Rahman A, Benson K, Leighton J, Kim SK, Wood R, Rothmann M, Chen GUKM, Staten AM, Pazdur R. Approval summary for imatinib mesylate capsules in the treatment of chronic myelogenous leukemia. Clin Cancer Res. 2002;8:935–42. [PubMed] [Google Scholar]
- 26.Peng B, Lloyd P, Schran H. Clinical pharmacokinetics of imatinib. Clin Pharmacokinet. 2005;44:879–94. doi: 10.2165/00003088-200544090-00001. [DOI] [PubMed] [Google Scholar]
- 27.Peng B, Hayes M, Resta D, Racine-Poon A, Druker BJ, Talpaz M, Sawyers CL, Rosamilia M, Ford J, Lloyd P, Capdeville R. Pharmacokinetics and pharmacodynamics of imatinib in a phase I trial with chronic myeloid leukemia patients. J Clin Oncol. 2004;22:935–42. doi: 10.1200/JCO.2004.03.050. [DOI] [PubMed] [Google Scholar]
- 28.Schmidli H, Peng B, Riviere GJ, Capdeville R, Hensley M, Gathmann I, Bolton AE, Racine-Poon A. Population pharmacokinetics of imatinib mesylate in patients with chronic-phase chronic myeloid leukaemia: results of a phase III study. Br J Clin Pharmacol. 2005;60:35–44. doi: 10.1111/j.1365-2125.2005.02372.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Le Coutre P, Kreuzer KA, Pursche S, Bonin M, Leopold T, Baskaynak G, Dorken B, Ehninger G, Ottmann O, Jenke A, Bornhauser M, Schleyer E. Pharmacokinetics and cellular uptake of imatinib and its main metabolite CGP74588. Cancer Chemother Pharmacol. 2004;53:313–23. doi: 10.1007/s00280-003-0741-6. [DOI] [PubMed] [Google Scholar]
- 30.Judson I, Peiming M, Peng B, Verweij J, Racine A, di Paola ED, van Glabbeke M, Dimitrijevic S, Scurr M, Dumez H, van Oosterom A. Imatinib pharmacokinetics in patients with gastrointestinal stromal tumour: a retrospective population pharmacokinetic study over time. EORTC Soft Tissue and Bone Sarcoma Group. Cancer Chemother Pharmacol. 2005;55:379–86. doi: 10.1007/s00280-004-0876-0. [DOI] [PubMed] [Google Scholar]
- 31.Gschwind HP, Pfaar U, Waldmeier F, Zollinger M, Sayer C, Zbinden P, Hayes M, Pokorny R, Seiberling M, Ben-Am M, Peng B, Gross G. Metabolism and disposition of imatinib mesylate in healthy volunteers. Drug Metab Dispos. 2005;33:1503–12. doi: 10.1124/dmd.105.004283. [DOI] [PubMed] [Google Scholar]
- 32.Leveque D, Maloisel F. Clinical pharmacokinetics of imatinib mesylate. In Vivo. 2005;19:77–84. [PubMed] [Google Scholar]
- 33.Gurney H, Wong M, Rivory L, Wilcken N, Hoskins J, Collins M, Dellaforce SE, Lynch K, Schran H. Imatinib elimination: characterisation by in vivo testing of phenotype and genotype. Proc Am Soc Clin Oncol. 2003;22:193. [Google Scholar]
- 34.Colombo S, Beguin A, Telenti A, Biollaz J, Buclin T, Rochat B, Decosterd LA. Intracellular measurements of anti-HIV drugs indinavir, amprenavir, saquinavir, ritonavir, nelfinavir, lopinavir, atazanavir, efavirenz and nevirapine in peripheral blood mononuclear cells by liquid chromatography coupled to tandem mass spectrometry. J Chromatogr B Anal Technol Biomed Life Sci. 2005;819:259–76. doi: 10.1016/j.jchromb.2005.02.010. [DOI] [PubMed] [Google Scholar]
- 35.Marzolini C, Paus E, Buclin T, Kim RB. Polymorphisms in human MDR1 (P-glycoprotein): recent advances and clinical relevance. Clin Pharmacol Ther. 2004;75:13–33. doi: 10.1016/j.clpt.2003.09.012. [DOI] [PubMed] [Google Scholar]
- 36.Galteau MM, Shamsa F. Urinary 6beta-hydroxycortisol: a validated test for evaluating drug induction or drug inhibition mediated through CYP3A in humans and in animals. Eur J Clin Pharmacol. 2003;59:713–33. doi: 10.1007/s00228-003-0690-3. [DOI] [PubMed] [Google Scholar]
- 37.Widmer N, Béguin A, Rochat B, Buclin T, Kovacsovics T, Duchosal MA, Leyvraz S, Rosselet A, Biollaz J, Decosterd LA. Determination of imatinib (Gleevec) in human plasma by solid-phase extraction-liquid chromatography-ultraviolet absorbance detection. J Chromatogr B Anal Technol Biomed Life Sci. 2004;803:285–92. doi: 10.1016/j.jchromb.2004.01.006. [DOI] [PubMed] [Google Scholar]
- 38.Ohno M, Yamaguchi I, Saiki K, Yamamoto I, Azuma J. Specific determination of urinary 6beta-hydroxycortisol and cortisol by liquid chromatography-atmospheric pressure chemical ionization mass spectrometry. J Chromatogr B Biomed Sci App. 2000;746:95–101. doi: 10.1016/s0378-4347(00)00122-5. [DOI] [PubMed] [Google Scholar]
- 39.Taylor RL, Machacek D, Singh RJ. Validation of a high-throughput liquid chromatography-tandem mass spectrometry method for urinary cortisol and cortisone. Clin Chem. 2002;48:1511–9. [PubMed] [Google Scholar]
- 40.Bakhtiar R, Lohne J, Ramos L, Khemani L, Hayes M, Tse F. High-throughput quantification of the anti-leukemia drug STI571 (Gleevec) and its main metabolite (CGP 74588) in human plasma using liquid chromatography-tandem mass spectrometry. J Chromatogr B Anal Technol Biomed Life Sci. 2002;768:325–40. doi: 10.1016/s1570-0232(01)00611-0. [DOI] [PubMed] [Google Scholar]
- 41.COBAS INTEGRA 400/700/800, Alpha 1-Acid Glycoprotein. Mannheim: Roche Diagnostics; 2003. [specifications] [Google Scholar]
- 42.Rowland M, Tozer TN. 3. Baltimore: Wiliams & Wilkins; 1995. Distribution Clinical PharmacokineticsConcept and Applications; p. 146. [Google Scholar]
- 43.Bourne DW, Bialer M, Dittert LW, Hayashi M, Rudawsky G, Koritz GD, Bevill RF. Disposition of sulfadimethoxine in cattle: inclusion of protein binding factors in a pharmacokinetic model. J Pharm Sci. 1981;70:1068–72. doi: 10.1002/jps.2600700926. [DOI] [PubMed] [Google Scholar]
- 44.Bevill RF, Koritz GD, Rudawsky G, Dittert LW, Huang CH, Hayashi M, Bourne DW. Disposition of sulfadimethoxine in swine: inclusion of protein binding factors in a pharmacokinetic model. J Pharmacokinet Biopharm. 1982;10:539–50. doi: 10.1007/BF01059036. [DOI] [PubMed] [Google Scholar]
- 45.O'Neil MJ, Smith A, Heckelman PE. 13. Whitehouse Station: Merck Research Laboratories; 2001. The Merck Index. [Google Scholar]
- 46.Fournier T, Medjoubi NN, Porquet D. Alpha-1-acid glycoprotein. Biochim Biophys Acta. 2000;1482:157–71. doi: 10.1016/s0167-4838(00)00153-9. [DOI] [PubMed] [Google Scholar]
- 47.Cockcroft DW, Gault MH. Prediction of creatinine clearance from serum creatinine. Nephron. 1976;16:31–41. doi: 10.1159/000180580. [DOI] [PubMed] [Google Scholar]
- 48.Boeckmann AJ, Beal SL, Sheiner LB. San Francisco: NONMEM Project Group. San Francisco: University of California; 1992. NONMEM Users’ Guide. [Google Scholar]
- 49.Akaike H. New look at statistical-model identification. IEEE T Automatic Con. 1974;19:716–23. [Google Scholar]
- 50.Davidian M, Gallant AR. Durham: Duke University; 1992. Nlmix: A Program for Maximum Likelihood Estimation of the Nonlinear Mixed Effects Model with a Smooth Random Effects Density. [computer program] [Google Scholar]
- 51.Peng B, Dutreix C, Mehring G, Hayes MJ, Ben-Am M, Seiberling M, Pokorny R, Capdeville R, Lloyd P. Absolute bioavailability of imatinib (Glivec) orally versus intravenous infusion. J Clin Pharmacol. 2004;44:158–62. doi: 10.1177/0091270003262101. [DOI] [PubMed] [Google Scholar]
- 52.Denko CW, Gabriel P. Age and sex related levels of albumin, ceruloplasmin, alpha 1 antitrypsin, alpha 1 acid glycoprotein, and transferrin. Ann Clin Lab Sci. 1981;11:63–8. [PubMed] [Google Scholar]
- 53.Jorgensen HG, Elliott MA, Allan EK, Carr CE, Holyoake TL, Smith KD. Alpha1-acid glycoprotein expressed in the plasma of chronic myeloid leukemia patients does not mediate significant in vitro resistance to STI571. Blood. 2002;99:713–5. doi: 10.1182/blood.v99.2.713. [DOI] [PubMed] [Google Scholar]
- 54.Gambacorti-Passerini C, le Coutre P, Zucchetti M, D’Incalci M. Binding of imatinib by alpha(1)-acid glycoprotein. Blood. 2002;100:367–8. doi: 10.1182/blood-2002-02-0518. [DOI] [PubMed] [Google Scholar]
- 55.Jorgensen H, Elliott M, Paterson S, Holyoake T, Smith K. Further observations on the debated ability of AGP to bind imatinib—Response. Blood. 2002;100:368–9. [Google Scholar]
- 56.Fuse E, Hashimoto A, Sato N, Tanii H, Kuwabara T, Kobayashi S, Sugiyama Y. Physiological modeling of altered pharmacokinetics of a novel anticancer drug, UCN-01 (7-hydroxystaurosporine), caused by slow dissociation of UCN-01 from human alpha1-acid glycoprotein. Pharm Res. 2000;17:553–64. doi: 10.1023/a:1007512832006. [DOI] [PubMed] [Google Scholar]
- 57.Kretz O, Weiss HM, Schumacher MM, Gross G. In vitro blood distribution and plasma protein binding of the tyrosine kinase inhibitor imatinib and its active metabolite, CGP74588, in rat, mouse, dog, monkey, healthy humans and patients with acute lymphatic leukaemia. Br J Clin Pharmacol. 2004;58:212–6. doi: 10.1111/j.1365-2125.2004.02117.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Watkins PB. Noninvasive tests of CYP3A enzymes. Pharmacogenetics. 1994;4:171–84. doi: 10.1097/00008571-199408000-00001. [DOI] [PubMed] [Google Scholar]
- 59.Crossman LC, Druker BJ, Deininger MW, Pirmohamed M, Wang L, Clark RE. hOCT1 and resistance to imatinib. Blood. 2005;106:1133–4. doi: 10.1182/blood-2005-02-0694. author reply 4. [DOI] [PubMed] [Google Scholar]
- 60.Becher F, Pruvost A, Goujard C, Guerreiro C, Delfraissy JF, Grassi J, Benech H. Improved method for the simultaneous determination of d4T, 3TC and ddl intracellular phosphorylated anabolites in human peripheral-blood mononuclear cells using high-performance liquid chromatography/tandem mass spectrometry. Rapid Commun Mass Spectrom. 2002;16:555–65. doi: 10.1002/rcm.605. [DOI] [PubMed] [Google Scholar]
- 61.Hoggard PG, Owen A. The mechanisms that control intracellular penetration of the HIV protease inhibitors. J Antimicrob Chemother. 2003;51:493–6. doi: 10.1093/jac/dkg137. [DOI] [PubMed] [Google Scholar]
- 62.Brouard M, Saurat JH. Cutaneous reactions to STI571. N Engl J Med. 2001;345:618–9. doi: 10.1056/NEJM200108233450814. [DOI] [PubMed] [Google Scholar]
- 63.Elliott MA, Mesa RA, Tefferi A. Adverse events after imatinib mesylate therapy. N Engl J Med. 2002;346:712–3. doi: 10.1056/NEJM200202283460919. [DOI] [PubMed] [Google Scholar]
- 64.Gambillara E, Laffitte E, Widmer N, Decosterd LA, Duchosal MA, Kovacsovics T, Panizzon RG. Severe pustular eruption associated with imatinib and voriconazole in a patient with chronic myeloid leukemia. Dermatology. 2005;211:363–5. doi: 10.1159/000088510. [DOI] [PubMed] [Google Scholar]
- 65.Talpaz M, Silver RT, Druker BJ, Goldman JM, Gambacorti-Passerini C, Guilhot F, Schiffer CA, Fischer T, Deininger MW, Lennard AL, Hochhaus A, Ottmann OG, Gratwohl A, Baccarani M, Stone R, Tura S, Mahon FX, Fernandes-Reese S, Gathmann I, Capdeville R, Kantarjian HM, Sawyers CL. Imatinib induces durable hematologic and cytogenetic responses in patients with accelerated phase chronic myeloid leukemia: results of a phase 2 study. Blood. 2002;99:1928–37. doi: 10.1182/blood.v99.6.1928. [DOI] [PubMed] [Google Scholar]
- 66.Verweij J, Casali PG, Zalcberg J, LeCesne A, Reichardt P, Blay JY, Issels R, van Oosterom A, Hogendoorn PC, Van Glabbeke M, Bertulli R, Judson I. Progression-free survival in gastrointestinal stromal tumours with high-dose imatinib: randomised trial. Lancet. 2004;364(9440):1127–34. doi: 10.1016/S0140-6736(04)17098-0. [DOI] [PubMed] [Google Scholar]
- 67.Mahon FX, Deininger MW, Schultheis B, Chabrol J, Reiffers J, Goldman JM, Melo JV. Selection and characterization of BCR-ABL positive cell lines with differential sensitivity to the tyrosine kinase inhibitor STI571: diverse mechanisms of resistance. Blood. 2000;96:1070–9. [PubMed] [Google Scholar]
- 68.Le Coutre P, Tassi E, Varella-Garcia M, Barni R, Mologni L, Cabrita G, Marchesi E, Supino R, Gambacorti-Passerini C. Induction of resistance to the Abelson inhibitor STI571 in human leukemic cells through gene amplification. Blood. 2000;95:1758–66. [PubMed] [Google Scholar]