Abstract
Aims
To investigate the association between torsemide renal clearance and genetic variation in the basolaterally expressed renal organic anion transporters OAT1 and OAT3 and in the luminally situated OAT4.
Methods
We analysed 22 polymorphisms in the OAT coding genes SLC22A6, SLC22A8 and SLC22A11 and their haplotypes and measured torsemide renal clearance in 95 healthy men. In addition, the effect of torsemide on the OAT-mediated transport was studied in vitro.
Results
In stably transfected HEK293 cells torsemide (100 µm) inhibited the uptake by human OAT1, OAT3 and OAT4 by 63.1, 58.1 and 68.0%, respectively. Torsemide renal clearance ranged from 6.5 to 43.1 ml min−1 with a log-normal distribution and a geometric mean of 15.6 ml min−1 (15.0–16.1 ± SEM). No clear outlier group was observed. AA carriers of the polymorphism rs11231809 in SLC22A11 had a torsemide renal clearance of 13.3 ml min−1 (12.7–13.9) compared with 15.1 ml min−1 (14.5–15.8) in AT and 18.0 ml min−1 (16.7–19.5) in TT carriers (P = 0.002). The two most frequent haplotypes at SLC22A11 showed an association with torsemide renal clearance. Homozygous carriage of these two haplotypes resulted in renal clearances of 21.2 ml min−1 (19.0–23.7) and 11.8 ml min−1 (10.5–13.5), respectively. No association between reanl clearance and genetic variation in SLC22A6 or SLC22A8 was observed.
Conclusions
Genetic variation in the gene encoding the luminally expressed OAT4 rather than in the basolaterally expressed OATs may affect the renal clearance of torsemide.
Keywords: haplotype, organic anion transporter, polymorphism, renal clearance, SLC22A11, torsemide
Introduction
Little is known about the clinical implications of genetic variation in drug transporters [1–4]. The solute carrier family 22 includes the genes SLC22A6, SLC22A8 and SLC22A11 coding for the organic anion transporters OAT1, OAT3 and OAT4. The basolaterally expressed OAT1 and OAT3 and the luminally expressed OAT4 contribute to the active renal organic anion secretion system, which is the major determinant of the renal secretion of organic anions including β-lactam antibiotics, methotrexate, nonsteroidal anti-inflammatory drugs and thiazide and loop diuretics [5–10]. The affinities of OAT1 and OAT3 for diuretics are generally higher than those of OAT4 [11].
Torsemide is a loop diuretic that is eliminated renally with a clearance of the unbound fraction (1–2%) of about 1200 ml min−1 [8]. Thus, torsemide is thought to be eliminated primarily via active anion secretion, and that its active excretion accounts for about 90% of torsemide renal clearance [8, 12]. However, the interaction of torsemide with the OATs has not been investigated. Extrarenal elimination accounts for about two-thirds of the total oral clearance of torsemide, mainly through metabolism by the genetically polymorphic expressed cytochrome P450 enzyme, CYP2C9 [13].
Few data exist on genetic variation in SLC22A6, SLC22A8 and SLC22A11 and its functional consequences [8]. Iida and colleagues identified eight polymorphisms in SLC22A6 and 14 polymorphisms in SLC22A8 in 48 healthy Japanese [14]. Most polymorphisms were intronic and none altered the amino acid sequence. Recently, 20 genetic variants were described in the human OAT1 gene in a genetically diverse sample from 92 individuals of African, Asian and caucasian origin [15]. Two variants were nonsynonymous amino acid substitutions, namely Arg50His and Lys525Ile. Both were observed in subjects of African ancestry, with allele frequencies of 0.17 and 0.02, respectively, but not in other ethnic groups.
In a sample of 96 individuals belonging to 11 different ethnic groups, Xu and colleagues identified 29 variants in the coding regions of the genes encoding OAT1, 2, 3, 4 and URAT1 [16]. Only two nonsynonymous amino acid substitutions were observed more than once, both in OAT4, namely Val155Met and Val339Met. These two variants were found exclusively in African populations and their allele frequencies were 0.11 and 0.14, respectively [16].
Databases such as the NCBI (http://www.ncbi.nlm.nih.gov) list numerous polymorphisms in SLC22A6, SLC22A8 and SLC22A11. Often the allele frequencies are unknown, as are the functional consequences. Listed nonsynonymous amino acid substitutions are His50Arg, Leu104Pro, Thr226Ile, Val256Ala, Trp293Arg and Gln454Arg in SLC22A6 (OAT1), Leu129Phe, Ser149Arg, Arg260Ile, Trp277Arg, Ala281Val, Phe305Ile, Val310Ala, Ser399Ala and Ile448Val in SLC22A8 (OAT3) and Pro29Leu, Val31Ile and Gly155Val in SLC22A11 (OAT4). The highest allele frequency was 2.6% (Ala281Val in SLC22A8).
Little is known about the functional consequences of genetic variation in SLC22A6, 8 or 11 [8]. A recent investigation concluded that the coding region of SLC22A6 may not contribute substantially to interindividual differences in the renal excretion of xenobiotics [17]. However, the design of this study limits the interpretation to amino acid exchanges with very low allele frequencies. Furthermore, another, earlier study did not find an association between the SLC22A8 polymorphism Ala389Val and the renal tubular secretion of pravastatin [18].
The primary aim of the present study was to characterize the associations between the genetic variation in the OAT1, 3 and 4 genes and the renal clearance of torsemide.
Methods
In vitro studies
The stably transfected human epithelial kidney cell lines T-REx™-HEK293-hOAT1, T-REx™-HEK293-hOAT3 and T-REx™-HEK293-hOAT4 were established by using the Flp-In™ expression system (Invitrogen, Karlsruhe, Germany) according to the manufacturer’s protocol as described elsewhere [19]. Briefly, the cDNAs including the open reading frames of hOAT1 (GenBank no. AF097490), hOAT3 (GenBank no. BI760120), and hOAT4 (GenBank AL514126) were subcloned into the Flp-In™ expression vector pcDNA5/FRT, containing a Flp recombination target (FRT) site linked to the hygromycin resistance gene. The constructs pcDNA5/FRT-hOAT1 or pcDNA5/FRT-hOAT3 or pcDNA5/FRT-hOAT4 were then cotransfected with the Flp recombinase expression vector pOG44 into Flp-In™ HEK293 cells. Cells stably expressing hOAT1, hOAT3, or hOAT4 were selected in hygromycin (200 µg ml−1) according to the manufacturer’s protocol. The cells were grown in flasks containing Dulbecco’s modified minimum essential medium (high glucose; Invitrogen) supplemented with 10% fetal bovine serum, 1% penicillin/streptomycin and blasticidine (5 mg l−1; Sigma-Aldrich, Munich, Germany). Cultures were maintained in a humidified atmosphere containing 5% CO2 at 37 °C. Cells were split in a 1 : 5 ratio every 3–4 days.
For uptake measurements the stably transfected HEK293 cells (and untransfected control cells) were initially cultivated in Petri dishes. After 2–3 days (confluence growing) the cells were harvested and plated into 24-well plastic dishes (Sarstedt, Nümbrecht, Germany) at a density of 2 × 105 cells per well. Transport assays were performed after 3 days in mammalian Ringer solution (130 mm NaCl, 4 mm KCl, 1 mm CaCl2, 1 mm MgSO4, 1 mm NaH2PO4, 20 mm HEPES and 18 mm glucose at pH 7.4). The cells were washed twice with 500 µl Ringer solution and for the uptake measurement the cells were incubated for 5 min at room temperature in Ringer solution containing 10 µm 3H-PAH in the case of hOAT1, or 10 nm 3H-estrone sulphate in the case of hOAT3- and hOAT4-transfected HEK293 cells. For the inhibition studies the test solutions included additionally 100 µm torsemide. Based on published data on other loop diuretics, we anticipated 100 µm torsemide as a near-maximal inhibitory concentration [11]. The incubation of the cells with radioactively labelled compounds was stopped after 5 min and the extracellular tracer was removed by washing the monolayer three times with 750 µl of ice-cold Ringer solution. The cells were dissolved in 0.5 ml 1 N NaOH and the 3H content was determined after neutralization of the probes with 0.5 ml 1 N HCl by liquid scintillation counting (Canberra-Packard, Dreieich, Germany). Successful transfection was confirmed by comparison with identically treated untransfected HEK293 cells. Uptake of the respective tritiated transporter substrate into the untransfected cells allowed the determination of the limit of maximal inhibition of transport via the transporters in the transfected cells.
Clinical study
An open-label, single-dose pharmacokinetic study was performed in 95 unselected healthy subjects. The study was approved by the Ethics Committee of the Georg August University, Göttingen, Germany. All subjects provided written informed consent after a full explanation of the protocol before the study. Nonsmoking healthy caucasian males aged between 18 and 50 years were enrolled. To avoid effects caused by variation in hormone status, we restricted our study to male subjects. All were healthy as confirmed by medical examination, blood pressure measurement, red and white blood cell counts and measurements of serum transaminases, γ-glutamyltransferase, serum total bilirubin, serum albumin, serum creatinine and electrolytes. Subjects taking any drugs were excluded from the study.
The subjects were instructed how to manage a salt-restricted diet with <200 mmol of sodium per day. Salt restriction started 48 h before drug administration and continued for 24 h thereafter. The subjects fasted overnight and until 4 h after drug administration. In the morning the subjects emptied their bladder and then ingested a single oral dose of 10 mg torsemide (Unat®; Roche, Mannheim, Germany). Blood was drawn immediately before dosage and at 0.5, 1, 2, 3, 4, 6, 8, 10 and 24 h thereafter. For genotyping, a separate sample was drawn into tubes containing ethylenediaminetetraacetic acid as the anticoagulant. Urine fractions were collected in intervals between defined time points.
Drug analysis
Measurement of torsemide was performed as described previously [13, 20]. Analysis was by high-performance liquid chromatography with ultraviolet detection after liquid-solid extraction. Plasma (1 ml) was mixed with 50 µl of a methanolic solution containing trifluorotorsemide as internal standard, acidified with 500 µl of 0.5 mol l−1 phosphoric acid and then applied to BondElut cartridges (Varian, Inc., Palo Alto, CA, USA) containing 500 mg C2 modified silica. The cartridges were washed with 0.5 ml of 0.5 mol l−1 phosphoric acid, 0.5 ml distilled water and finally with 1.5 ml methanol. The sample was eluted with 2 ml methanol/water [75 : 25 (v/v)]. Chromatographic separation was performed on a LiChroCART CN column (VWR, Darmstadt, Germany). The mobile phase was 0.02 mol l−1 perchloric acid, pH 2.5, and acetonitrile [90 : 10 (v/v)] delivered at a flow rate of 1.5 ml min−1. The eluent was monitored at 290 nm. The limit of detection was 5 ng ml−1 and the limit of quantification was 20 ng ml−1. At 20 and 1000 ng ml−1 the biases (difference between measured and true concentration) were 4.1% and 4.0% and the coefficients of variation were 8.0% and 10.1%, respectively.
We developed a more sensitive assay using LC-MS/MS for the detection of torsemide in urine. Seventy-five microlitres of urine were mixed with 225 µl water and then acidified with 200 µl of 0.5 mol l−1 phosphoric acid. The samples were transferred to preconditioned BondElut cartridges filled with 100 mg C2 modified silica. After washing with phosphoric acid, elution was performed with methanol/water [75 : 25 (v/v)]. Samples were chromatographed on a LiChroCART CN column with an eluent consisting of acetic acid and methanol [45 : 55 (v/v)]. The analytes were detected using a API 4000® (Applied Biosystems, Darmstadt, Germany) triple quadrupole mass spectrometer equipped with a turbo ion spray source. The positive ion mode was used. The limit of detection was 0.5 ng ml−1 and the limit of quantification was 1 ng ml−1. At 5, 10 and 500 ng ml−1 the biases (difference between measured and true concentration) were 18.7%, 11.4% and −1.7% and the coefficients of variation were 11.9%, 9.9% and 4.4%, respectively.
Genotyping
Genotyping was performed in 110 subjects including the study population plus additional subjects. To describe the major haplotypes, polymorphisms were selected on the basis of frequency (preferentially above 5%), even distribution at the gene loci and the availability of assays. None of the selected polymorphisms coded for an amino acid substitution. SNP genotyping was performed by Pyroseqencing® on the PSQ® HS96A System (Biotage AB, Uppsala, Sweden). Polymerase chain reaction (PCR) primers were designed using Primer SNP Design Version 1.01 software (http://www.Pyrosequencing.com). All PCRs were carried out in a reaction volume of 15 µl with 6 ng genomic DNA, 10 mm dNTP, 10 µm of each forward and reverse oligonucleotide PCR primers (one of which is biotinylated), Taq polymerase (5 U µl−1), 1.5 mm MgCl2 (25 mm). Primer details and PCR conditions are described in Table 1. Pyrosequencing® protocols were standard as defined by the supplier (Biotage AB), using an internal primer diluted in 1 × annealing buffer (20 mmol l−1 Tris-acetate, 2 mmol l−1 MgAc2), 2 × binding wash buffer II pH 7.6 (10 mmol l−1 Tris–HCl, 2 m NaCl, 1 mmol l−1 EDTA, 0.1% Tween 20), Streptavidin Sepharose Beads (Amersham Biosciences, Uppsala, Sweden), 0.2 m NaOH, 70% ethanol and a PSQ HS96A SNP reagent kit (Biotage AB). Samples were analysed on a PSQ HS96A instrument with proprietary pyrosequencing software (Biotage AB).
Table 1.
Primer sequences and PCR conditions of the Pyrosequencing™ assays
| Identification* | Forward primer (5′-3′) | Reverse primer (5′-3′) | Internal primer (5′-3′) | PCR thermoprofile |
|---|---|---|---|---|
| rs736342 | ATCCCATGATCATCTCCCTTC | †CTGGCACCAGGATATGTGTG | GCTAAAGTTTCAATGCCT | 95 5′, 45(95 30 s, 62 30 s, 72 30 s) 72 5′, 4 for ever |
| rs2276300 | †CTCCCAGCCTAATCCCTTAC | AACTGAAGCCATGCTGACAC | TGAGACTTCCCATGATAAC | 95 5′, 45(95 15 s, 58 30 s, 72 15 s) 72 5′, 4 for ever |
| rs10897312 | †GCTTCAGAGAATCCCCTTCC | GAAGATGGGGGCCTTTGTTA | AAAAGAAAAATGCTCTCAG | 95 5′, 45(95 15 s, 58 30 s, 72 15 s) 72 5′, 4 for ever |
| rs955434 | GGAGTCCCTGCTTAGAGACAGA | †CCTTCTCTCCATTGCTGCTTAT | CGGAGCTGGGGTT | 95 5′, 45(95 15 s, 60 30 s, 72 15 s) 72 5′, 4 for ever |
| rs953894 | †GATGATAGGGGCTGCTTGAG | TACTGCCTCCTTTGGTCTCC | CACGTCAGACATGACCA | 95 5′, 45(95 15 s, 60 30 s, 72 15 s) 72 5′, 4 for ever |
| OAT3_goe2 | ACTGGCTGTTGATGTTCTTCC | †CTGAGCCTTTCTCCCTCTTC | TCTTGTCTGGAAAGTCCT | 95 5′, 45(95 15 s, 60 30 s, 72 15 s) 72 5′, 4 for ever |
| rs2276299 | †GAGGAGAGGGCCACATACC | GCTACACCTTTGGCCAGTTC | CGTTGGCTGCAGTT | 95 5′, 45(95 15 s, 60 30 s, 72 15 s) 72 5′, 4 for ever |
| rs2187383 | †TGCATTCCTGGATCCCTAGA | ATTGGCCCCAGAACACATAG | CATCCTCTCTGGTCCTT | 95 5′, 45(95 15 s, 60 30 s, 72 15 s) 72 5′, 4 for ever |
| OAT3_goe1 | †ATACCTCTCCCTGGCCTATG | AAGATCTGCAGCAGGTTGTG | GGGAGGCCCAGTATG | 95 5′, 45(95 15 s, 60 30 s, 72 15 s) 72 5′, 4 for ever |
| rs948979 | TAGTGTTAGGGCACACCCAGA | †ATTGAGGGTGGGATGGAAG | CCAGACAGAAAATGTCACT | 95 5′, 45(95 15 s, 60 30 s, 72 15 s) 72 5′, 4 for ever |
| rs3759053 | †CTCCAGCACCAAAAGTGAAAG | AGCCCAGTTGATATTTCTTCCA | TTCAGTAACATCAAAGCAC | 95 5′, 45(95 15 s, 58 30 s, 72 15 s) 72 5′, 4 for ever |
| rs3782099 | †CTGCCTGGTGAGAGGAGA | ACACTCACCTGGCTCTTCC | CACAGCATCCCCTTG | 95 5′, 45(95 15 s, 58 30 s, 72 15 s) 72 5′, 4 for ever |
| rs1783811 | CCGAGGAGTACCACTGTGT | †GGTAAGCCAAGTGTTAGCTC | AAGGAACAAGCCCAA | 95 5′, 45(95 15 s, 58 30 s, 72 15 s) 72 5′, 4 for ever |
| rs4963326 | †TCCCCATCTGAGAGAGAGAATT | GCAGAGGCAGGATTTGAATTTA | CCTTCCTGAACACACTG | 95 5′, 45(95 15 s, 58 30 s, 72 15 s) 72 5′, 4 for ever |
| rs10792369 | CTTTCTGTGGGCCATCAAC | †ACGGGGCCTCAGATATACAA | CCCTCCATCCAATGT | 95 5′, 45(95 15 s, 58 30 s, 72 15 s) 72 5′, 4 for ever |
| rs11231809 | †TAACTCACCTGTTCCGTGATT | ATCAGACGTATCGTTTGTAAGG | ATCGTTTGTAAGGACTCA | 95 5′, 45(95 15 s, 58 30 s, 72 15 s) 72 5′, 4 for ever |
| rs4930423 | †TCCAGGAAACTCCTTTGTCAT | AGATTGAATAATGAGGGGAATACA | ATGAGGGGAATACAGTG | 95 5′, 45(95 15 s, 58 30 s, 72 15 s) 72 5′, 4 for ever |
| rs2078267 | †AGCCCAACAACATCAGAGTA | ACCCAGTTGTTATCTGTCCA | AGGCTCCAGTGTCGG | 95 5′, 45(95 15 s, 58 30 s, 72 15 s) 72 5′, 4 for ever |
| rs528211 | TTAGGGAGGTGTGGCAAGTT | †GGCAAAATCTCATCACAGACC | TTTGAAGACAGAGAGGC | 95 5′, 45(95 15 s, 58 30 s, 72 15 s) 72 5′, 4 for ever |
| rs476037 | GTTAAGTGGAGTCGGTCAGG | †TCAGCTCCACAGCTTTACC | GCAGGCAGAGGATGT | 95 5′, 45(95 15 s, 58 30 s, 72 15 s) 72 5′, 4 for ever |
Rs number as in the NCBI database SNP. Two putative polymorphisms, identified when comparing genomic databases, had no reference number (OAT3_goe 1 and 2).
Biotinylated primer.
Controls were included to avoid errors during genotyping. Thus, all of the 96-well plates with DNA samples contained one well with a DNA-free reaction mix to detect possible contamination of the reagents with DNA. Another well contained a DNA sample that was expected to yield identical genotypes for all plates tested for a given genetic variant. Using these controls, no genotyping errors were observed.
Pharmacokinetic and data analysis
Pharmacokinetic analysis was performed using the nonparametric analysis module of the WinNonlin® software, version 2.1. Renal clearance was calculated from the expression Ae/AUC and AUC using the linear trapezoidal rule and extrapolating to infinity. Ae was the amount of torsemide excreted in urine extrapolated to infinity.
Linkage disequilibrium blocks were identified using Haploview®, version 3.2 software [21]. The linkage plot relied on the D’ value of Lewontin [22]. More than one group of haplotypes contributed to variation at the loci investigated, causing automatic linkage block definition to fail. Taking the linkage between the haplotypes of otherwise separated smaller linkage blocks into account, definition of the linkage blocks was performed by hand, based on the ‘n + 1’ method of Zeggini [23]. The individual haplotypes were characterized using Phase®, version 2.1.1 software [24, 25]. The output assigning pairs of haplotypes to the individuals was used. Ten runs were each performed and the haplotype pair most often assigned was used for the genotype–phenotype association.
Statistical analyses were performed using the software package SPSS®, version 12.0.0 (SPSS Inc., Chicago, IL, USA). Normal distribution was tested using the Shapiro–Wilk test, which is more sensitive to outliers than the Kolmogorov–Smirnov test. However, the results of the two tests were qualitatively identical. Geometric (logarithmic) means were calculated as arithmetic means after logarithmic transformation. The respective statistical spread was calculated as SEM of the log-transformed data. Relationships between genotypes/haplotypes and the renal torsemide clearance were tested using two-sided linear regression analysis. In the absence of any other hypothesis, we assumed a linear gene–dose effect as the simplest alternative to the null hypothesis of no genotype effect. The independent variables genotype and haplotype were coded as the number of variant alleles or the number of haplotypes, respectively. A P-value <0.05 was considered significant.
Results
In HEK293 cells, stable transfection with hOAT1, hOAT3 and hOAT4 was confirmed by 7.5-fold, 6.0-fold and 4.2-fold increases in the uptake of 3H-PAH and 3H-estrone sulphate, respectively. Accordingly, 100% inhibition of the transfected transporters would result in 87%, 83% and 76% decreases of total 3H uptake in the respective cells. In transfected HEK293 cells, torsemide (100 µm) inhibited the uptake of the respective 3H-substrate by 63.1 ± 3.9% (hOAT1), 58.1 ± 6.2% (hOAT3) and 68.0 ± 5.1% (hOAT4, all mean ± SEM of n = 3 independent experiments, all P < 0.001 compared with transfected cells without torsemide, t-test).
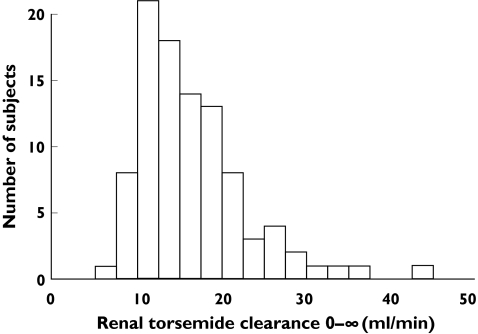
Ninety-five healthy male caucasians with a mean (range) age of 28 years (19–50 years) took part in the clinical study. The mean (range) body weight and height were 78 kg (57–93 kg) and 182 cm (170–198 cm) and the mean body surface was 2.0 m2 (range 1.7–2.3 m2). In this population, renal torsemide clearance ranged from 6.5 ml min−1 to 43.1 ml min−1. Although the maximum renal clearance was 6.6-fold higher than the minimum value, no clear outlier group, or a bi- or trimodal distribution was observed (Figure 1). The pattern of the renal clearance was significantly different from a normal distribution (Shapiro–Wilk test P < 0.001) but the values were log-normally distributed (P = 0.368). The geometric mean of the renal clearance of torsemide was 15.6 ml min−1 (15.0–16.1, ± SEM). Torsemide is 98–99% bound to plasma protein, giving a mean renal clearance of unbound torsemide of 830–1560 ml min−1. Clearance remained constant over the 24-h period, and there appeared to be subgroups of high and low ‘organic anion tubular secretors’. No effect on clearance of time after dosage or urinary flow was observed. Neither body weight, body surface, serum albumin concentration nor creatinine clearance explained this interindividual variation in renal clearance. Normalizing for these parameters did not affect the difference between the maximum and minimum values.
Figure 1.
Distribution pattern of torsemide renal clearance
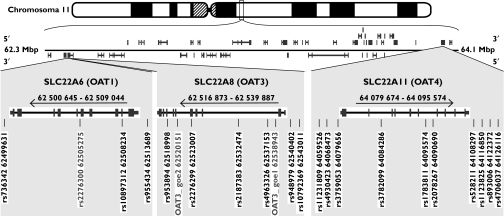
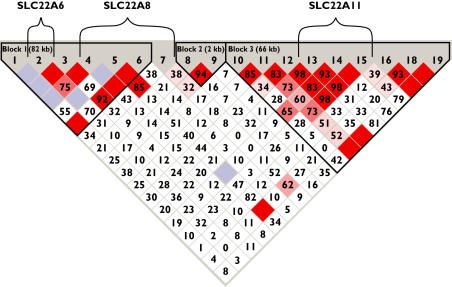
Twenty-two putative polymorphisms were investigated (Figure 2). Three of them were monomorphic and will not be discussed further. The remaining 19 polymorphisms are described in Table 2; all were in Hardy–Weinberg equilibrium. Three blocks of linkage disequilibrium were identified (Figure 3). One block starts 3′ of SLC22A6 with a border not determined in the present work. This block includes SLC22A6 and most of SLC22A8 but excludes the most 5′ polymorphism of SLC22A8 (rs4963326). Its analysis resulted in 11 of 64 possible haplotypes separated into two distinct groups (Table 3). Six frequent haplotypes (defined as an allele frequency of at least 5%) accounted for up to 96% of all alleles.
Figure 2.
Genomic location of SLC22A6, SLC22A8 and SLC22A11 on chromosome 11 as given by the ‘Map Viewer’ of the NCBI showing that there are several other genes between the SLC22A6-SLC22A8 locus and the SLC22A11 locus. The arrows denote the transcription direction. The intron–exon structure of the genes is given according to the ‘Gene’ database of the NCBI; the grey boxes denote exons and the narrow boxes untranslated regions. The polymorphisms studied are identified by their rs-number and their position on chromosome 11 according to the ‘SNP’ database of the NCBI March 2005. Two putative polymorphisms, identified when comparing genomic databases had no reference number (OAT3_goe 1 and 2). The three putative polymorphisms that turned out to be monomorphic are given in grey letters
Table 2.
The polymorphisms studied and their association with torsemide renal clearance
| No. | Identification* | MAF† | Nucleotides | N | ClTor (ml min−1)‡ | P-value§ |
|---|---|---|---|---|---|---|
| Polymorphisms at SLC22A6 and SLC22A8 | ||||||
| 1 | rs736342 | 0.12 | C/C | 75 | 15.3 (14.7–15.9) | |
| C/T | 17 | 17.3 (15.7–19.1) | ||||
| T/T | 3 | 14.7 (13.5–15.9) | ||||
| 2 | rs10897312 | 0.12 | C/C | 76 | 15.3 (14.7–15.9) | |
| C/T | 16 | 16.5 (15.1–17.9) | ||||
| T/T | 3 | 21.9 (18.6–25.9) | ||||
| 3 | rs955434 | 0.29 | G/G | 48 | 15.3 (14.6–16.2) | |
| G/A | 37 | 15.6 (14.8–16.5) | ||||
| A/A | 9 | 17.5 (15.5–19.7) | ||||
| 4 | rs953894 | 0.25 | G/G | 53 | 15.3 (14.5–16.0) | |
| G/A | 36 | 16.0 (15.1–17.0) | ||||
| A/A | 6 | 16.8 (14.9–18.9) | ||||
| 5 | rs2276299 | 0.13 | T/T | 70 | 15.6 (15.0–16.3) | |
| T/A | 23 | 16.0 (14.9–17.1) | ||||
| A/A | 1 | 8.8 | ||||
| 6 | rs2187383 | 0.36 | G/G | 36 | 16.3 (15.5–17.2) | |
| G/T | 49 | 15.4 (14.6–16.2) | ||||
| T/T | 10 | 14.5 (13.1–16.0) | ||||
| 7 | rs4963326 | 0.31 | C/C | 46 | 14.8 (14.1–15.5) | |
| C/T | 40 | 16.4 (15.6–17.3) | ||||
| T/T | 9 | 16.9 (14.4–19.9) | ||||
| 8 | rs948979 | 0.23 | G/G | 57 | 14.9 (14.3–15.6) | |
| G/T | 32 | 17.8 (16.8–19.0) | ||||
| T/T | 6 | 12.0 (10.7–13.5) | ||||
| 9 | rs10792369 | 0.29 | T/T | 49 | 14.7 (14.0–15.5) | |
| T/C | 35 | 17.2 (16.3–18.1) | ||||
| C/C | 10 | 15.0 (12.9–17.4) | ||||
| Polymorphisms at SLC22A11 | ||||||
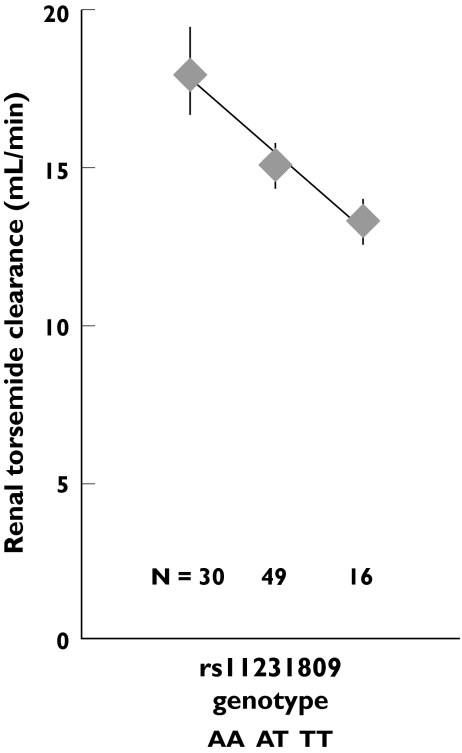
| 10 | rs11231809 | 0.43 | A/A | 30 | 18.0 (16.7–19.5) | 0.002 |
| A/T | 49 | 15.1 (14.5–15.8) | ||||
| T/T | 16 | 13.3 (12.7–13.9) | ||||
| 11 | rs4930423 | 0.44 | A/A | 28 | 13.6 (13.0–14.2) | 0.034 |
| A/C | 48 | 16.8 (16.1–17.6) | ||||
| C/C | 17 | 16.4 (14.7–18.4) | ||||
| 12 | rs3759053 | 0.45 | T/T | 27 | 16.9 (15.7–18.3) | 0.009 |
| T/C | 49 | 16.2 (15.5–16.9) | ||||
| C/C | 18 | 12.7 (11.9–13.4) | ||||
| 13 | rs3782099 | 0.47 | C/C | 25 | 17.0 (15.7–18.4) | 0.009 |
| C/T | 51 | 16.2 (15.5–17.0) | ||||
| T/T | 19 | 12.8 (12.1–13.5) | ||||
| 14 | rs1783811 | 0.31 | G/G | 46 | 17.4 (16.5–18.4) | 0.007 |
| G/A | 40 | 14.1 (13.4–14.8) | ||||
| A/A | 9 | 14.2 (13.3–15.1) | ||||
| 15 | rs2078267 | 0.48 | T/T | 23 | 16.6 (15.2–18.1) | 0.020 |
| T/C | 53 | 16.4 (15.7–17.1) | ||||
| C/C | 19 | 12.8 (12.1–13.5) | ||||
| 16 | rs528211 | 0.35 | G/G | 39 | 15.8 (14.9–16.7) | |
| G/A | 46 | 16.0 (15.2–16.8) | ||||
| A/A | 10 | 13.8 (12.7–14.9) | ||||
| 17 | rs11231825 | 0.01 | A/A | 93 | 15.7 (15.1–16.3) | |
| A/C | 2 | 13.4 | ||||
| C/C | 0 | – | ||||
| 18 | rs893006 | 0.35 | T/T | 39 | 15.8 (14.9–16.7) | |
| T/G | 46 | 16.1 (15.3–16.9) | ||||
| G/G | 10 | 13.4 (12.5–14.5) | ||||
| 19 | rs476037 | 0.09 | C/C | 77 | 15.5 (14.9–16.2) | |
| C/T | 18 | 16.1 (15.1–17.2) | ||||
| T/T | 0 | – | ||||
rs number as in the NCBI database SNP.
MAF, Minor allele frequency.
ClTor, renal torsemide clearance in ml min−1, the geometric mean ± SEM is given.
P-values of respective linear regression analyses are given when P < 0.05.
Figure 3.
The strength of the linkage between the polymorphisms as calculated by the software package Haploview®. Polymorphisms are numbered as given in Table 2. Dark red boxes without a value represent a D′-value of 1.00 and light blue boxes without a value represent a value that is not determinable. Numbers in the remaining diamond boxes are the 100-fold of the respective D′-values. The higher the value, the darker the fill colour of the boxes and the stronger the linkage
Table 3.
Frequent haplotypes formed by the polymorphisms studied; letters in bold denote the variant alleles
| Linkage blocks including SLC22A6 and SLC22A8 | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Polymorphism no. according to Table 2 | Allele | |||||||||
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | frequency | |
| Block 1 | ||||||||||
| H1 | C | C | G | G | T | T | 0.355 | |||
| H2 | C | C | G | G | T | G | 0.150 | |||
| H3 | T | C | G | G | T | G | 0.105 | |||
| H4 | C | C | G | G | A | G | 0.095 | |||
| Block 2 | ||||||||||
| H5 | C | C | A | A | T | G | 0.155 | |||
| H6 | C | T | A | A | T | G | 0.100 | |||
| Block 5′ end of SLC22A8 | ||||||||||
| H7 | G | T | 0.714 | |||||||
| H8 | T | C | 0.223 | |||||||
| H9 | G | C | 0.055 | |||||||
| Linkage block including SLC22A11 | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Polymorphism no. according to Table 2 | ||||||||||
| 10 | 11 | 12 | 13 | 14 | 15 | 16 | 18 | 19 | ||
| Group 1 | ||||||||||
| H10 | A | C | T | C | G | T | G | T | C | 0.286 |
| H11 | T | A | T | C | G | T | G | T | C | 0.068 |
| H12 | T | A | T | C | G | T | A | G | C | 0.064 |
| Group 2 | ||||||||||
| H13 | T | A | C | T | A | C | G | T | C | 0.177 |
| H14 | A | A | C | T | G | C | A | G | T | 0.077 |
| H15 | A | A | C | T | G | C | A | G | C | 0.064 |
The polymorphism rs4963326 was not assigned to a linkage block, the second of which thereby consisted of only rs9498979 and rs10792369, 3′ of SLC22A8. All four possible haplotypes were observed in this block but with a different distribution than expected by random coupling of the polymorphism alleles (Table 3).
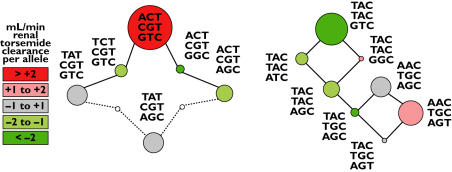
Haplotype analysis of the SLC22A11 locus was performed excluding only rs11231825 due to its low minor allele frequency. Analysis resulted in 26 of 512 possible haplotypes (Table 3, Figure 4). The six frequent haplotypes accounted for up to 74% of all alleles and were split into two haplotype groups (Table 3, Figure 4). The most prevalent haplotype H10 consisted of wild-type alleles except that it was the only frequent haplotype including the C-allele of rs4930423. The second block differs from the first by the four middle polymorphisms. Only the second most prevalent haplotype H13 included the T-allele of rs11231809 in this block.
Figure 4.
The structure of the two haplotype groups at the SLC22A11 locus (Table 3). The six frequent haplotypes and less frequent haplotypes that link them are shown as circles with areas proportional to the haplotype frequency. Continuous lines indicate convertibility by the exchange of one nucleotide, and dotted lines convertibility by the exchange of two nucleotides. Phenotypes are represented by the circle colour as given by the legend to the left. The numbers in the legend denote differences compared to the mean torsemide renal clearance in ml min−1 per haplotypic allele from respective linear regression analyses
Among the polymorphisms, the renal clearance of torsemide was most significantly associated with rs11231809 (linear regression P = 0.002, Jonkheere–Terpstra P = 0.006, Table 2, Figure 5). It was the only association remaining significant after Bonferroni correction for the multiple testing of 19 polymorphisms. Rs11231809 was also significantly associated with the fraction of the drug excreted renally, calculated from the expression clearance/creatinine clearance, but not with creatinine clearance itself. Rs11231809 denotes an A→T exchange at the position 64 059 526 in chromosome 11, about 20 kb 5′ from the start of SLC22A11.
Figure 5.
Association of torsemide renal clearance with the rs11231809 genotype. Symbols and bars denote the geometric mean ± SEM
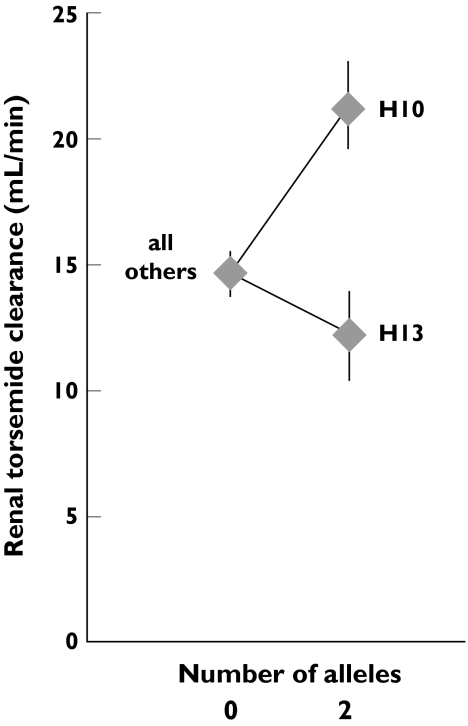
The A-allele of the polymorphism rs11231809 determined the haplotypes H10, H14 and H15 (Table 2) but only H10 was haplotype-dose dependently associated with renal torsemide clearance (linear regression P = 0.0001, Jonkheere–Terpstra P = 0.002, Figure 4, Table 4). The relationship was still significant after Bonferroni correction for multiple testing of 15 haplotypes. H10 was also associated with the fraction of torsemide excreted renally but not with creatinine clearance itself. Carriers of two H10 alleles had a 53% higher mean renal clearance than subjects not possessing the H10 allele. The second most prevalent haplotype, H13, was associated with a lower renal clearance (linear regression P = 0.015, Jonkheere–Terpstra P = 0.014, Table 4), but the relationship was not significant after Bonferroni correction for multiple testing of 15 haplotypes. Carriers of two H13 alleles had a 16% lower mean renal clearance than subjects not possessing the H13 allele. Multiple linear regression analysis including H10 and H13 as predictors of the renal clearance revealed a geometric mean of 21.2 ml min−1 (19.0–23.7 ± SEM) for carriers of two H10 alleles and of 11.9 ml min−1 (10.5–13.5) for carriers of two H13 alleles compared with a geometric mean of 14.5 ml min−1 (13.7–15.3) in carriers of alleles other than H10 and H13 (Figure 6). Thus the renal clearance of torsemide in homozygous H10 carriers was nearly 80% higher than that of homozygous H13 carriers.
Table 4.
Association of the haplotypes with torsemide renal clearance
| Renal torsemide clearance (ml min−1) geometric mean (± SEM) N | |||||
|---|---|---|---|---|---|
| 0* | 1 | 2 | (0/1/2*) | P† | |
| Haplotypes at SLC22A6 and SLC22A8 | |||||
| H1 | 16.4 (15.6–17.3) | 15.2 (14.4–16.0) | 15.0 (13.5–16.7) | 38/48/9 | |
| H2 | 15.9 (15.3–16.6) | 14.8 (13.6–16.0) | 16.5 | 71/22/2 | |
| H3 | 15.2 (14.7–15.8) | 17.2 (15.7–18.8) | 20.1 | 77/16/2 | |
| H4 | 15.8 (15.2–16.5) | 15.3 (14.4–16.3) | 9.2 | 75/19/1 | |
| H5 | 15.6 (14.9–16.2) | 16.0 (15.0–17.1) | 14.0 | 68/25/2 | |
| H6 | 15.3 (14.8–16.0) | 17.0 (15.5–18.6) | 16.8 | 77/17/1 | |
| H7 | 15.0 (12.9–17.4) | 17.0 (16.2–17.9) | 14.8 (14.1–15.6) | 10/36/49 | |
| H8 | 14.9 (14.3–15.5) | 18.1 (17.0–19.2) | 12.0 (10.7–13.5) | 58/31/6 | |
| H9 | 15.4 (14.8–16.0) | 18.2 (16.1–20.5) | 13.1 | 84/10/1 | |
| Haplotypes at SLC22A11 | |||||
| H10 | 13.8 (13.3–14.3) | 17.1 (16.2–18.1) | 21.1 (17.1–26.1) | 45/44/6 | 0.0001 |
| H11 | 15.7 (15.1–16.3) | 15.5 (14.5–16.5) | – | 83/12/0 | |
| H12 | 15.6 (15.1–16.3) | 15.6 (14.2–17.2) | – | 83/12/0 | |
| H13 | 16.6 (15.9–17.4) | 13.7 (12.9–14.5) | 13.9 (11.7–16.5) | 65/27/3 | 0.015 |
| H14 | 15.5 (14.9–16.1) | 16.5 (15.3–17.8) | – | 80/15/0 | |
| H15 | 15.7 (15.1–16.2) | 15.4 (13.5–17.7) | – | 83/12/0 | |
Number of respective haplotype alleles.
P-values of respective single linear regression analyses are given, when P < 0.05.
Figure 6.
Association of torsemide renal clearance with the SLC22A11 haplotypes H10 and H13. Symbols and bars denote the geometric mean ± SEM. ‘All others’ includes all subjects with neither an H10 nor an H13 allele. Values were derived from a multiple linear regression analysis
The number of H10 alleles was significantly (P = 0.042) associated with the AUC of torsemide extrapolated to infinity. Carriers of zero, 1 and 2 H10 alleles had AUC mean values of 3.6, 3.3 and 2.4 mg h−1 l−1 (± SEM of 0.2, 0.2 and 0.3, respectively). The association remained significant in a multiple linear regression analysis including CYP2C9 genotype as independent predictor (P = 0.035 and P < 0.0001 for the association with CYP2C9 genotype). Only a trend was observed between the AUC and rs11231809 (P = 0.054 and 0.072 controlled for CYP2C9 genotype). The amount of torsemide renally excreted was weakly associated with CYP2C9 genotype but not with genetic variation in SLC22A11. Including H10 or rs11231809 and CYP2C9 genotype as independent predictors in multiple linear regression analyses confirmed the independent association of the renal clearance of torsemide with both the H10 haplotype and CYP2C9 genotype (P = 0.00013 and P = 0.015), and with rs11231809 and CYP2C9 genotype (P = 0.003 and P = 0.023).
Discussion
We confirmed that torsemide interacts with the human OATs 1, 3 and 4 in vitro as was expected from an earlier clinical study and in vitro studies with other loop diuretics [11, 12]. We observed a 6.6-fold interindividual variation in the renal clearance of torsemide, compared with about a twofold difference in creatinine clearance. The torsemide renal clearance was log-normally distributed and neither a clear outlier group, nor a multimodal distribution was identified. These data provide no evidence that the renal clearance of torsemide displays polymodality in the population.
We identified one polymorphism (rs11231809) about 20 kb upstream of SLC22A11 that was significantly linked in a linear manner with torsemide renal clearance. Carriers of two A-alleles had a 35% higher clearance compared with TT subjects. Consecutive haplotype–phenotype analysis revealed that some haplotypes, including the A-allele, were associated with higher and some with lower torsemide renal clearance. This inconsistency and the long distance to the gene border imply that rs11231809 is a marker of rather than a truly functional polymorphism of the gene. A detailed functional characterization of the polymorphism was hampered by the fact that OAT4 is almost exclusively expressed in the kidney according to cDNA clone and SAGE analysis (http://www.weizmann.il/Gencards/..). Polymorphisms localized in 5′ upstream regions are most likely to affect the expression of a gene, which apparently does not carry nonsynonymous polymorphisms in the caucasian population. It is not yet clear where the truly functional polymorphism identified in this study is localized in or near SLC22A11.
We identified three blocks of linkage disequilibrium among the polymorphisms investigated. At two of the loci two groups of haplotypes were observed. Analysing the phenotype–genotype relationship, the most prominent finding was a linear association of the haplotype H10 at the SLC22A11 locus with torsemide renal clearance. In carriers of two H10 alleles, renal clearance was about 50% higher compared with those not possessing the H10 allele and nearly 80% higher compared with carriers of two H13 alleles. Thereby, the extent of the relationship between H10 and the phenotype was stronger than that for rs11231809. The renal clearance of torsemide is within the range of other loop diuretics and about 10 times that of creatinine clearance [8]. Active secretion accounts for about 90% of the renal excretion of torsemide. Thus, the extent of the association between the genetic variation and the active secretion of the drug may be about 10% higher than that for the measured total renal clearance of torsemide.
Earlier we reported an influence of the CYP2C9 genotype on torsemide pharmacokinetics [13]. The renal clearance of the drug accounts for about one-third of its total oral clearance. We now show that genetic variation in SLC22A11 also affects the pharmacokinetics of torsemide with a 33% decreased AUC in carriers of two H10 alleles compared with noncarriers and a correspondingly inverse trend with rs11231809. However, we found no relationship between genetic variation in SLC22A11 and the amount of torsemide renally excreted, which is known to be the determinant of the diuretic effect. In the earlier report we observed an unexpected association of the CYP2C9 genotype with the renal clearance of torsemide. However, inclusion of rs11231809 or H10 together with the CYP2C9 genotype confirmed the independent association of the phenotype with the variation in both genes.
At the SLC22A6–SLC22A8 locus the association with torsemide renal clearance was not statistically significant. This is in accordance with the results of recent studies [17, 18].
The transporter encoded by SLC22A11, OAT4, is localized at the luminal membrane in the renal proximal tubulus cells [8]. OAT4 is thought to be a major mediator of the renal excretion of anions from inside the renal tubulus cell into the urine, whereas the basolaterally located OAT1 and OAT3 are thought to mediate the uptake of organic anions from the blood into the renal cell. This hypothesis is supported by the observation that a number of (nephro-)toxic substrates, including diuretics, have lower affinities for OAT4 than for OAT1 or OAT3 [11]. When OAT4 is the rate-limiting step for the renal excretion, subjects with a genetically impaired clearance mediated by OAT4 may be prone to nephrotoxicity. Thus, studies are warranted on the influence of genetically determined variation in OAT4 on drug-induced nephrotoxicity.
A recent study suggests that OAT4 (but not OAT1, 2 and 3 and URAT1) is under accelerated selection pressure [16], which might reflect environmental exposure during evolution specific to humans compared with chimpanzees. Our finding of an association between torsemide renal clearance and genetic variation in SLC22A11 but not in SLC22A6 and SLC22A8 is in accordance with the purported selection pressure on OAT4. The properties of the selecting agent may fit into the picture of OAT4 as the bottleneck for the elimination of nephrotoxic anions [7]. The fact that the amino acid sequence has been purified during the selection process [16] may imply a need for an altered substrate specificity. A parallel development to higher OAT4 expression may not have gone through resulting in the observed genetically determined differences in the renal torsemide clearance.
In conclusion, we found large variation in the renal clearance of torsemide, which was partially explained by genetic variation in the luminally expressed OAT4 rather than by polymorphisms in the genes coding for basolaterally located OAT1 and OAT3.
Acknowledgments
The study was supported in part by the ‘Nationales Genomforschungsnetzwerk’ NGFN grants 01 GS 0107 and 01 GR 0416. The skilful contributions of Franziska Tuchen, Michaela Torn and Jan Westermann to the clinical study are gratefully acknowledged.
References
- 1.Mizuno N, Niwa T, Yotsumoto Y, Sugiyama Y. Impact of drug transporter studies on drug discovery and development. Pharmacol Rev. 2003;55:425–61. doi: 10.1124/pr.55.3.1. [DOI] [PubMed] [Google Scholar]
- 2.van Montfoort JE, Hagenbuch B, Groothuis GM, Koepsell H, Meier PJ, Meijer DK. Drug uptake systems in liver and kidney. Curr Drug Metab. 2003;4:185–211. doi: 10.2174/1389200033489460. [DOI] [PubMed] [Google Scholar]
- 3.Hediger MA, Romero MF, Peng JB, Rolfs A, Takanaga H, Bruford EA. The ABCs of solute carriers: physiological, pathological and therapeutic implications of human membrane transport proteins. Introduction. Pflugers Arch. 2004;447:465–8. doi: 10.1007/s00424-003-1192-y. [DOI] [PubMed] [Google Scholar]
- 4.Leabman MK, Huang CC, DeYoung J, Carlson EJ, Taylor TR, de la Cruz M, Johns SJ, Stryke D, Kawamoto M, Urban TJ, Kroetz DL, Ferrin TE, Clark AG, Risch N, Herskowitz I, Giacomini KM. Natural variation in human membrane transporter genes reveals evolutionary and functional constraints. Proc Natl Acad Sci USA. 2003;100:5896–901. doi: 10.1073/pnas.0730857100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Koepsell H, Endou H. The SLC22 drug transporter family. Pflugers Arch. 2004;447:666–76. doi: 10.1007/s00424-003-1089-9. [DOI] [PubMed] [Google Scholar]
- 6.Burckhardt G, Bahn A, Wolff NA. Molecular physiology of renal p-aminohippurate secretion. News Physiol Sci. 2001;16:114–8. doi: 10.1152/physiologyonline.2001.16.3.114. [DOI] [PubMed] [Google Scholar]
- 7.Miyazaki H, Sekine T, Endou H. The multispecific organic anion transporter family: properties and pharmacological significance. Trends Pharmacol Sci. 2004;25:654–62. doi: 10.1016/j.tips.2004.10.006. [DOI] [PubMed] [Google Scholar]
- 8.Vormfelde SV, Burckhardt G, Zirk A, Wojnowski L, Brockmoller J. Pharmacogenomics of diuretic drugs: data on rare monogenic disorders and on polymorphisms and requirements for further research. Pharmacogenomics. 2003;4:701–34. doi: 10.1517/phgs.4.6.701.22817. [DOI] [PubMed] [Google Scholar]
- 9.Burckhardt BC, Burckhardt G. Transport of organic anions across the basolateral membrane of proximal tubule cells. Rev Physiol Biochem Pharmacol. 2003;146:95–158. doi: 10.1007/s10254-002-0003-8. [DOI] [PubMed] [Google Scholar]
- 10.Wright SH, Dantzler WH. Molecular and cellular physiology of renal organic cation and anion transport. Physiol Rev. 2004;84:987–1049. doi: 10.1152/physrev.00040.2003. [DOI] [PubMed] [Google Scholar]
- 11.Hasannejad H, Takeda M, Taki K, Shin HJ, Babu E, Jutabha P, Khamdang S, Aleboyeh M, Onozato ML, Tojo A, Enomoto A, Anzai N, Narikawa S, Huang XL, Niwa T, Endou H. Interactions of human organic anion transporters with diuretics. J Pharmacol Exp Ther. 2004;308:1021–9. doi: 10.1124/jpet.103.059139. [DOI] [PubMed] [Google Scholar]
- 12.Lohrmann E, Burhoff I, Greger R. Tubular effects of the diuretic torasemide. Cardiology. 1994;84(Suppl. 2):135–42. doi: 10.1159/000176466. [DOI] [PubMed] [Google Scholar]
- 13.Vormfelde SV, Engelhardt S, Zirk A, Meineke I, Tuchen F, Kirchheiner J, Brockmoller J. CYP2C9 polymorphisms and the interindividual variability in pharmacokinetics and pharmacodynamics of the loop diuretic drug torsemide. Clin Pharmacol Ther. 2004;76:557–66. doi: 10.1016/j.clpt.2004.08.024. [DOI] [PubMed] [Google Scholar]
- 14.Iida A, Saito S, Sekine A, Mishima C, Kondo K, Kitamura Y, Harigae S, Osawa S, Nakamura Y. Catalog of 258 single-nucleotide polymorphisms (SNPs) in genes encoding three organic anion transporters, three organic anion-transporting polypeptides, and three NADH:ubiquinone oxidoreductase flavoproteins. J Hum Genet. 2001;46:668–83. doi: 10.1007/s100380170019. [DOI] [PubMed] [Google Scholar]
- 15.Bleasby K, Hall LA, Perry JL, Mohrenweiser HW, Pritchard JB. Functional consequences of single nucleotide polymorphisms in the human organic anion transporter hOAT1 (SLC22A6) J Pharmacol Exp Ther. 2005;314:923–31. doi: 10.1124/jpet.105.084301. [DOI] [PubMed] [Google Scholar]
- 16.Ahmadi KR, Weale ME, Xue ZY, Soranzo N, Yarnall DP, Briley JD, Maruyama Y, Kobayashi M, Wood NW, Spurr NK, Burns DK, Roses AD, Saunders AM, Goldstein DB. A single-nucleotide polymorphism tagging set for human drug metabolism and transport. Nat Genet. 2005;37:84–9. doi: 10.1038/ng1488. [DOI] [PubMed] [Google Scholar]
- 17.Fujita T, Brown C, Carlson EJ, Taylor T, de la Cruz M, Johns SJ, Stryke D, Kawamoto M, Fujita K, Castro R, Chen CW, Lin ET, Brett CM, Burchard EG, Ferrin TE, Huang CC, Leabman MK, Giacomini KM. Functional analysis of polymorphisms in the organic anion transporter, SLC22A6 (OAT1) Pharmacogenet Genomics. 2005;15:201–9. doi: 10.1097/01213011-200504000-00003. [DOI] [PubMed] [Google Scholar]
- 18.Nishizato Y, Ieiri I, Suzuki H, Kimura M, Kawabata K, Hirota T, Takane H, Irie S, Kusuhara H, Urasaki Y, Urae A, Higuchi S, Otsubo K, Sugiyama Y. Polymorphisms of OATP-C (SLC21A6) and OAT3 (SLC22A8) genes: consequences for pravastatin pharmacokinetics. Clin Pharmacol Ther. 2003;73:554–65. doi: 10.1016/S0009-9236(03)00060-2. [DOI] [PubMed] [Google Scholar]
- 19.Bakhiya N, Stephani M, Bahn A, Ugele B, Seidel A, Burckhardt G, Glatt H. Uptake of chemically reactive, DNA-damaging sulfuric esters into renal cells by human organic anion transporters. J Am Soc Nephrol. doi: 10.1681/ASN.2005080801. in press. [DOI] [PubMed] [Google Scholar]
- 20.Engelhardt S, Meineke I, Brockmoller J. Improved solid-phase extraction and HPLC measurement of torasemide and its important metabolites. J Chromatogr B Analyt Technol Biomed Life Sci. 2006;831:31–5. doi: 10.1016/j.jchromb.2005.11.013. [DOI] [PubMed] [Google Scholar]
- 21.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–5. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 22.Lewontin RC. The Interaction of selection and linkage. Ii Optimum models. Genetics. 1964;50:757–82. doi: 10.1093/genetics/50.4.757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zeggini E, Barton A, Eyre S, Ward D, Ollier W, Worthington J, John S. Characterisation of the genomic architecture of human chromosome 17q and evaluation of different methods for haplotype block definition. BMC Genet. 2005;6:21. doi: 10.1186/1471-2156-6-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stephens M, Donnelly P. A comparison of bayesian methods for haplotype reconstruction from population genotype data. Am J Hum Genet. 2003;73:1162–9. doi: 10.1086/379378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stephens M, Smith NJ, Donnelly P. A new statistical method for haplotype reconstruction from population data. Am J Hum Genet. 2001;68:978–89. doi: 10.1086/319501. [DOI] [PMC free article] [PubMed] [Google Scholar]