Abstract
Aims
To determine the effect of CYP2C8 genotype and of fluvoxamine on the pharmacokinetics of rosiglitazone.
Methods
Twenty-three healthy subjects with the following genotypes were included in a two-phase, open-label, cross-over trial: CYP2C8*3/ *3 (n = 3), CYP2C8*1/ *3 (n = 10) and CYP2C8*1/ *1 (n = 10). In Phase A, the subjects were given 4 mg rosiglitazone as a single oral dose. In Phase B, the subjects were treated with multiple oral doses of 50 mg fluvoxamine maleate for 3 days prior to the single oral administration of 4 mg rosiglitazone. Plasma concentrations of rosiglitazone and relative amounts of N-desmethylrosiglitazone were measured in both phases for 24 h after drug administration.
Results
The pharmacokinetics of rosiglitazone and N-desmethylrosiglitazone were not significantly different between the CYP2C8 genotypic groups. Fluvoxamine caused a statistically significant (P= 0.0066) increase in the AUC0–∞ of rosiglitazone, with a geometric mean ratio of 1.21 [95% confidence interval (CI) 1.06–1.39]. The elimination half-life (t1/2) was also significantly higher (P= 0.0203) with a geometric mean ratio of 1.38 [95% CI 1.06–1.79]. The coadministration of fluvoxamine had no influence on the pharmacokinetics of N-desmethylrosiglitazone.
Conclusion
The importance of the CYP2C8*3 mutation in the in vivo metabolism of rosiglitazone could not be confirmed. Fluvoxamine increased the AUC0–∞ and t1/2 of rosiglitazone moderately and hence may be a weak inhibitor of CYP2C8.
Keywords: CYP2C8 polymorphisms, drug interaction, fluvoxamine, pharmacokinetics, rosiglitazone
Introduction
Rosiglitazone maleate is an oral agent in the thiazolidine dione class used in the treatment of Type 2 diabetes mellitus. The thiazolidine diones are highly selective and potent agonists for the peroxisome proliferator-activated receptor gamma (PPARγ), which decrease blood glucose levels by sensitizing peripheral tissues to insulin [1–3]. The bioavailability of rosiglitazone is approximately 99% after oral dosing in tablet form and the absorption is rapid, reaching maximal concentration (Cmax) within 1 h [4]. The major products of metabolism following incubation of rosiglitazone with human liver microsomes are para-hydroxy- and N-desmethylrosiglitazone. These pathways are catalysed mainly by CYP2C8, with a minor contribution from CYP2C9 [5]. The in vivo metabolism of rosiglitazone is complex, with many N-demethylated and hydroxylated metabolites being identified in both nonconjugated and conjugated form [4]. The quantitative importance of N-demethylation of rosiglitazone in vivo has been verified by several studies [6–9]. Thus, the CYP2C8- and CYP2C9-inhibitor gemfibrozil [6, 7] increases the AUC of rosiglitazone and prolongs its t1/2, decreases the N-desmethylrosiglitazone/rosiglitazone AUC ratio and prolongs the time to maximum concentration (tmax) of N-desmethylrosiglitazone [8]. Furthermore, trimethoprim, which is an inhibitor of CYP2C8, raises the AUC of rosiglitazone and decreases the formation of N-desmethylrosiglitazone [9]. It has also been shown that the antifungal agent ketoconazole causes a slight increase in the AUC and Cmax of rosiglitazone in humans [10] and that rifampicin decreases the AUC by inducing CYP2C8 [9, 11].
CYP2C8 catalyses the metabolism of a number of other clinically used drugs, such as the anticancer drug paclitaxel [12] and the antimalarial drug amodiaquine [13]. Several allelic variants of CYP2C8 have been described (http://www.imm.ki.se/cypalleles), namely CYP2C8*1A–C [14, 15], CYP2C8*2–3 [16], CYP2C8*4 [15] and CYP2C8*5–10 [17]. CYP2C8*3 has a reported allele frequency of 0.10–0.23 in the Caucasian population [18].
A study by Bidstrup et al. 2006 described no significant effect of the CYP2C8*3 genotypes on the in vivo metabolism of a therapeutic dose of repaglinide, a known substrate of CYP2C8 [19]. On the other hand, Niemi et al. [20, 21] reported that plasma concentrations of repaglinide were lower in subjects with the CYP2C8*1/*3 genotype. A study by Martinez et al. showed differences in the AUC and t1/2 of (R)-ibuprofen between subjects with the CYP2C8*3/ *3, CYP2C8*1/*1 and CYP2C8*1/ *3 genotypes [22] and the combined effect of CYP2C8 and CYP2C9 genotypes on racemic ibuprofen pharmacokinetics was described by Garcia-Martin et al. [23].
CYP2C9 is among the most important drug-metabolizing enzymes in humans. Substrates include phenytoin, tolbutamide, torsemide, S-warfarin and numerous nonsteroidal anti-inflammatory drugs [24, 25]. The two most significant mutant alleles are CYP2C9*2 (C430>T: Arg144→Cys) and CYP2C9*3 (A1075>C: Ile359→Leu) and have been shown to affect drug metabolism [26–28]. Garcia-Martin et al. found that ibuprofen clearance was determined not only by CYP2C8 genotype but also by the CYP2C9 genotypes *1/ *1, *1/ *2, *1/ *3, *2/ *2, *2/ *3 and *3/ *3 [23]. Despite the high degree of homology in amino acid sequence, the crystal structures of human CYP2C8 [29] and CYP2C9 [30] reveal structural differences in the active site, which determine substrate specificity. A linkage between the CYP2C8 and CYP2C9 genetic polymorphism in the Caucasian population has been observed. Yasar et al. have reported that approximately 96% of the subjects with the CYP2C8*3 allele also carried CYP2C9*2 and 85% of the subjects possessing the CYP2C9*2 variant also carried CYP2C8*3 [31].
Fluvoxamine is an antidepressant drug that belongs to the class of selective serotonin reuptake inhibitors (SSRI). It is a potent inhibitor of the in vitro CYP2C9-mediated 4-methylhydroxylation of tolbutamide and the 7-hydroxylation of (S)-warfarin in human liver microsomes [32]. In vivo, fluvoxamine is also a moderate inhibitor of CYP2C19 and CYP2C9 [33–35] and a potent inhibitor of CYP1A2 [34, 36], but its possible effect on the metabolism of CYP2C8 substrates has never been described. Walsky et al. investigated 209 drugs as potential inhibitors of CYP2C8, but fluvoxamine was not tested [37].
The aims of this study were to investigate the effect of (i) the CYP2C8*3 allele and (ii) multiple doses of fluvoxamine on the pharmacokinetics of rosiglitazone.
Methods
Subjects
Twenty-three healthy Caucasians gave written informed consent to participate in the trial. Subjects were divided into three groups based on their CYP2C8 genotype. Their demographic information and CYP2C8 and CYP2C9 genotypes are shown in Table 1. All subjects were free of cardiovascular, hepatic, renal or gastrointestinal disease, drug abuse or alcohol dependence assessed by physical examination and a review of medical history. In addition, blood pressure and the results of laboratory tests (blood chemistry, haematology and immunology) were required to be within the normal range.
Table 1.
Demographic information and CYP2C genotypes of the subjects
| Subject no. | Sex | Age, years | Weight, kg | Height, cm | CYP2C8 genotype | CYP2C9 genotype |
|---|---|---|---|---|---|---|
| 1 | M | 23 | 76 | 189 | *3/*3 | *2/*2 |
| 2 | F | 27 | 60 | 162 | *3/*3 | *2/*2 |
| 3 | M | 24 | 74 | 184 | *3/*3 | *2/*2 |
| Median (range) | – | 24 (23–27) | 74 (60–76) | 184 (162–189) | – | – |
| 4 | M | 25 | 80 | 180 | *1/*1 | *1/*1 |
| 5 | M | 26 | 80 | 184 | *1/*1 | *1/*1 |
| 6 | M | 29 | 78 | 176 | *1/*1 | *1/*1 |
| 7 | M | 26 | 78 | 174 | *1/*1 | *1/*1 |
| 8 | M | 26 | 98 | 199 | *1/*1 | *1/*1 |
| 9 | M | 26 | 66 | 175 | *1/*1 | *1/*1 |
| 10 | M | 27 | 84 | 183 | *1/*1 | *1/*1 |
| 11 | M | 25 | 117 | 185 | *1/*1 | *1/*1 |
| 12 | M | 26 | 97 | 190 | *1/*1 | *1/*1 |
| 13 | M | 25 | 88 | 190 | *1/*1 | *1/*1 |
| Median (range) | – | 26 (25–29) | 82 (66–117) | 183.5 (174–199) | – | – |
| 14 | M | 25 | 89 | 180 | *1/*3 | *1/*2 |
| 15 | M | 26 | 70 | 178 | *1/*3 | *1/*2 |
| 16 | M | 26 | 81 | 186 | *1/*3 | *1/*2 |
| 17 | M | 25 | 79 | 188 | *1/*3 | *1/*2 |
| 18 | M | 25 | 67 | 181 | *1/*3 | *1/*2 |
| 19 | M | 26 | 71 | 176 | *1/*3 | *1/*2 |
| 20 | M | 29 | 82 | 180 | *1/*3 | *1/*2 |
| 21 | M | 25 | 72 | 170 | *1/*3 | *1/*2 |
| 22 | M | 28 | 80 | 195 | *1/*3 | *1/*2 |
| 23 | F | 23 | 56 | 159 | *1/*3 | *1/*2 |
| Median (range) | – | 25.5 (23–29) | 75.5 (56–89) | 180 (159–195) | – | – |
Genotyping
The DNA was extracted from peripheral leucocytes using a PUREGENETM genomic DNA purification kit according to the guidelines of the manufacturers (Gentra Systems, Minneapolis, MN, USA). CYP2C8 genotype with regard to the *3 mutation was determined by Professor A. Rane (Department of Medical Laboratory Sciences and Technology, Division of Clinical Pharmacology, Karolinska Institute, Huddinge University Hospital, Stockholm, Sweden) by a previously published method [31]. The subjects were chosen from a Caucasian population [38], which had been screened with respect to their CYP2C9 genotype [39].
Design
The study was registered in the European Clinical Trials Database (EudraCT no. 2004-003978-28). It was approved by The Danish Medicines Agency, The Danish Data Protecting Agency and the Regional Ethical Committee of Vejle and Funen Counties, and was conducted in accordance with Good Clinical Practice (GCP) and monitored by the GCP-Unit, Odense University Hospital. Drug use was monitored to ensure good compliance. Sample size calculations were based on the primary outcome represented by differences in AUC between subjects with CYP2C8*1/ *3 and CYP2C8*1/ *1 genotypes. It was estimated that a true difference of 35% could be detected, given a two-sided level of significance (α) of 0.05 and a power (β) of 80%, using 10 subjects in each group. The number of subjects with the CYP2C8*3/ *3 genotype is small and only three could be recruited.
The study was conducted as an open-label, two-phase, cross-over trial with a wash-out period of at least 2 weeks between the phases. The volunteers fasted from 8 h before to 1 h after rosiglitazone administration.
Phase A
At 08.00 h, the subjects were given a single 4-mg rosiglitazone tablet (Avandia®; GlaxoSmithKline Pharma A/S, Brondby, Denmark).
Phase B
The subjects were given 50-mg fluvoxamine maleate tablets (Fevarin®; Solvay Pharmaceuticals B.V., Weesp, Holland) at 08.00 h and 20.00 h for 3 days. On day 4, both a 4-mg Avandia® tablet and a 50-mg Fevarin® tablet were administered at 08.00 h.
Sample analysis
In both phases, blood samples were drawn from an i.v. cannula in a forearm vein at 0, 20, 40, 60, 80 min and at 2, 3, 4, 6, 8, 10 and 24 h after rosiglitazone administration. Blood samples containing ethylenediaminetetraacetic acid as anticoagulant were centrifuged for 10 min at 2400 g. Plasma was separated and stored at − 20 °C until analysis. The plasma concentrations of rosiglitazone were determined in duplicate by a validated high-performance liquid chromatography (HPLC) method which also detects N-desmethylrosiglitazone [40]. The limit of quantification was 1 ng ml−1 and the detection limit was 0.25 ng ml−1 for rosiglitazone in human plasma. The intra- and interday precision coefficients of variation did not exceed 8.7% [40]. Only relative concentrations of N-desmethylrosiglitazone could be determined, since the pure reference substance was not available. Structural identification of rosiglitazone and N-desmethylrosiglitazone was made by an IonSpec Ultima Fourier Transform Mass Spectrometer with a 4.7-T magnet using electrospray ionization. The molecular ions of rosiglitazone (m/z = 358) and N-desmethylrosiglitazone (m/z = 344) were found in fractions, collected from the HPLC eluent. This is similar to the LS/MS data described by Cox et al. [4].
Data analysis
The pharmacokinetic parameters were determined using a noncompartmental model (WinNonlin® Professional, version 4.1; Pharsight Corporation, Mountain View, CA, USA). The AUC0–∞ was calculated using the linear trapezoidal method. The values of Cmax and tmax were read directly from the concentration–time curve. The terminal elimination half-life (t1/2) was calculated from the equation t1/2 = ln2/λ, where λ is the terminal slope calculated by linear regression of the time vs. log concentration. Prior to statistical analysis, AUC0–∞, t1/2 and Cmax data were transformed to their natural logarithm to justify a Gaussian distribution. Statistical analysis was performed using GraphPad QuickCalcs (GraphPad Software, Inc., San Diego, CA, USA), StatXact-3 (Cytel Software Corporation, Cambridge, MA, USA) and Microsoft Office Excel 2003 (Microsoft Corporation, Redmond, WA, USA).
AUC0–∞, Cmax and t1/2 values were compared between genotypes using unpaired t-test. The geometric ratio of means with their 95% confidence intervals (CIs) and P-values were then calculated. Effects on tmax were determined using unpaired Hodges–Lehman estimates of median differences with exact 95% CIs and P-values. The effects of fluvoxamine on the AUC0–∞, Cmax and t1/2 of rosiglitazone were analysed using the paired test.
Results
One subject with the CYP2C8*3/ *3 genotype (no. 1) developed a superficial venous thrombosis 1 day after the first trial day following i.v. catheterization. He was successfully treated with locally applied heparinoid, but was excluded from the study. One of the CYP2C8*1/ *3 genotypes (no. 15) experienced general malaise, nausea, tiredness, insomnia, constipation, accommodation and miction difficulties 1 day after the initiation of fluvoxamine treatment. The latter was stopped immediately, the subject withdrew from the study and the symptoms gradually disappeared.
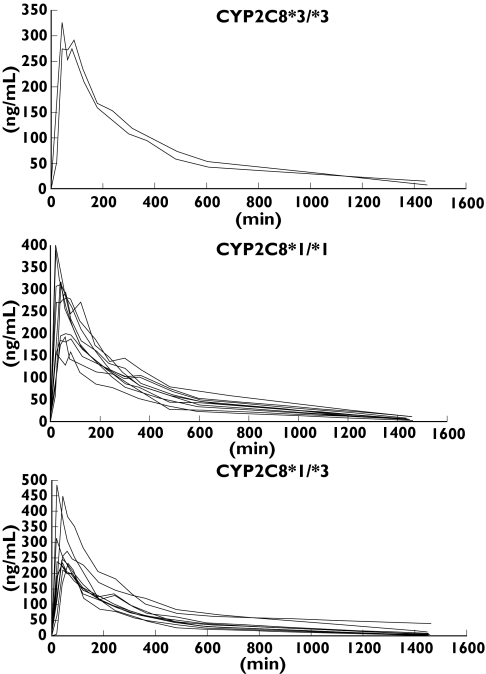
The plasma concentration–time curves of the three genotypes in each phase are shown in Figures 1 and 2. Large variations are seen within each genotype and, in particular, one of the CYP2C8*1/ *3 subjects (no. 23) displayed a higher t1/2 (19.4 h) and AUC0–∞ (3455 h ng ml−1) than all the other subjects in phase B. Table 2 shows the median and range of the pharmacokinetic parameters of rosiglitazone and the metabolite N-desmethylrosiglitazone. In Phase A, the median AUC0–∞ of 1479 h ng ml−1 in CYP2C8*3/ *3 subjects was 22% higher than in CYP2C8*1/ *1 (1210 h ng ml−1) and CYP2C8*1/ *3 (1208 h ng ml−1) subjects. The median Cmax in Phase A was 28% and 26% higher in CYP2C8*3/ *3 (335 ng ml−1) compared with CYP2C8*1/ *1 (265 ng ml−1) and CYP2C8*1/ *3 (260 ng ml−1) subjects, respectively. The same trends were seen during Phase B. Thus, the median AUC0–∞ in CYP2C8*3/ *3 subjects was 1851 h ng ml−1, 1478 h ng ml−1 in CYP2C8*1/ *1 and 1353 h ng ml−1 in CYP2C8*1/ *3 subjects. The corresponding median values for Cmax were 309 ng ml−1, 303 ng ml−1 and 245 ng ml−1. However, these differences between genotypes were not statistically significant. Furthermore, no differences in AUC0–∞ and tmax were found for the metabolite N-desmethylrosiglitazone. The longer tmax and t1/2 of N-desmethylrosiglitazone made it impossible to calculate precise values of the latter within the 24-h sampling period.
Figure 1.
Phase A, plasma drug concentration–time curves in subjects with the CYP2C8*3/ *3 (n = 3), CYP2C8*1/ *1 (n = 10) and CYP2C8*1/ *3 (n = 10) genotypes after a single oral dose of 4-mg rosiglitazone
Figure 2.
Phase B, plasma drug concentration–time curves in subjects with the CYP2C8*3/ *3 (n = 2), CYP2C8*1/ *1 (n = 10) and CYP2C8*1/ *3 (n = 9) genotypes after a single oral dose of 4-mg rosiglitazone and coadministration of fluvoxamine
Table 2.
Median and range of the pharmacokinetic parameters of rosigltazone and N-desmethylrosiglitazone in each genotype in study Phase A and B
| Phase A | Phase B | |||||
|---|---|---|---|---|---|---|
| CYP2C8*3/*3 n = 3 | CYP2C8*1/*1 n = 10 | CYP2C8*1/*3 n = 10 | CYP2C8*3/*3 n = 2 | CYP2C8*1/*1 n = 10 | CYP2C8*1/*3 n = 9 | |
| Rosiglitazone | ||||||
| t1/2, h | 3.2 (2.9–5.0) | 3.9 (2.9–7.8) | 3.1 (1.7–6.3) | 6.7 (4.9–8.6) | 5.0 (4.2–6.4) | 4.2 (3.1–19.4) |
| tmax, h | 1.1 (0.7–1.1) | 1.0 (0.6–1.4) | 0.7 (0.3–1.0) | 1.1 (0.7–1.5) | 0.9 (0.3–1.4) | 0.7 (0.3–1.0) |
| Cmax, ng ml−1 | 335 (285–374) | 265 (124–361) | 260 (179–435) | 309 (292–326) | 303 (158–400) | 245 (210–485) |
| AUC0–∞, h ng ml−1 | 1479(1221–1537) | 1210(980–1903) | 1208(852–1288) | 1851(1838–1863) | 1478(918–2160) | 1353(876–3455) |
| N-desmethyl-rosiglitazone | ||||||
| t1/2, h | 16.7(12.4–20.4) | 16.4(12.6–22.4) | 15.4(7.0–35.5) | 24.7(15.6–33.7) | 34.6(20.1–79.0) | 19.3(12.2–23.6) |
| tmax, h | 6.0 (5.1–8.0) | 6.1 (4.0–10.1) | 6.0 (3.1–8.0) | 6.1 (6.0–6.1) | 8.0 (4.0–10.0) | 8.0 (4.1–10.0) |
| AUC0−24, relative area units | 400 (333–478) | 399 (325–476) | 456 (136–544) | 435 (379–490) | 395 (248–446) | 450 (376–627) |
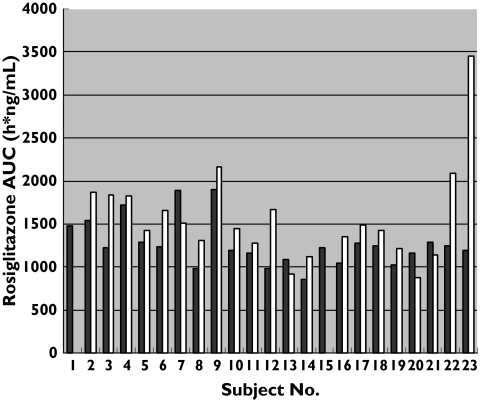
The interaction with fluvoxamine in Phase B resulted in higher rosiglitazone median AUC0–∞ in all three genotypes (Figure 3). Values were increased by 25% in CYP2C8*3/ *3, 22% in CYP2C8*1/ *1 and 12% in CYP2C8*1/ *3 subjects. The overall (n = 21) geometric mean ratio was 1.21 with a 95% CI of 1.06, 1.39, a result that is statistically significant (P= 0.0066). The difference in AUC0–∞ was also significantly different without the outlier (subject no. 23) with an overall (n = 20) geometric mean ratio of 1.16, a 95% CI of 1.05–1.29 and a P-value of 0.0067. The t1/2 was also significantly higher in Phase B with a geometric mean ratio of 1.38, a 95% CI of 1.06–1.79 and a P-value of 0.0203. Without subject no. 23 (n = 20) the difference in t1/2 was also significant (P= 0.0078), giving a geometric mean ratio of 1.24 (1.07–1.44).
Figure 3.
The area under the rosiglitazone concentration–time curves (AUC0–∞) for each subject in Phase A (▪, baseline) and Phase B (with fluvoxamine, □)
Discussion
The pharmacokinetic characteristics of rosiglitazone and N-desmethylrosiglitazone were found to be similar to previously published data [4, 8]. One explanation for the large interindividual variation seen in the pharmacokinetic parameters could be the prsence of other known and unknown mutations of CYP2C8. However, since the frequencies of the other known allelic variants are relatively low in the Caucasian population, other factors probably contribute to the variability. Another explanation may be that other enzymes or excretory mechanisms are involved in the disposition of rosiglitazone. In the present study, the subjects in each group of CYP2C8 genotypes where chosen to have the same CYP2C9 genotype to ensure that any possible contribution by CYP2C9 to rosiglitazone metabolism was approximately the same in all three groups. This was made easy because of the allelic linkage between CYP2C8*3 and CYP2C9*2 [31]. Thus, since the CYP2C9 genotypes were identical in each group, differences in CYP2C9 activity probably did not contribute to the variation in rosiglitazone pharmacokinetics within each group.
Although the median values of AUC0–∞ and Cmax were ≥ 20% higher in the subjects homozygous for the CYP2C8*3 allele, these findings were not statistically significant. A greater number of homozygous CYP2C8*3 subjects are probably needed to detect a possible real difference between subjects with CYP2C8*3/ *3 compared with CYP2C8*1/ *1 and/or CYP2C8*1/ *3 genotypes. Only three subjects homozygous for the CYP2C8*3 allele were included in the study due to its low frequency in the population. According to the Hardy–Weinberg equation, only 1–5% of the Caucasian population have the CYP2C8*3/ *3 genotype and thus the recruitment of sufficient numbers of subjects is problematic. The study revealed no significant pharmacokinetic differences between the groups with the CYP2C8*1/ *1 and CYP2C8*1/ *3 genotypes. Data have been published on the in vivo effect of the CYP2C8*3 genotypes on the metabolism of CYP2C8 substrates. Thus, ibuprofen clearance has been linked to the CYP2C8 and the CYP2C9 polymorphisms [22, 23]. Furthermore, it has been reported that the CYP2C8*3 allele is associated with a lower AUC of repaglinide (compared with CYP2C8*1), whereas polymorphism in the SLCO1B1 gene results in substantially higher plasma concentrations of the drug, suggesting that the activity of this transporter is an important determinant of repaglinide pharmacokinetics [20, 21]. In contrast, Bidstrup et al. found no significant differences in pharmacokinetics between subjects carrying the CYP2C8*3 allele and subjects who were homozygous wild-type given therapeutic (2 mg) and subtherapeutic (0.25 mg) doses of repaglinide [19].
The present study does not demonstrate conclusively whether rosiglitazone is metabolized exclusively by CYP2C8 alone and that the CYP2C8*3 allele expresses a totally inactive form of CYP2C8. If this were the case, larger differences in AUC and t1/2 between the CYP2C8*3 genotypes would be expected. Overall, the data for rosiglitazone, repaglinide and ibuprofen suggest that the CYP2C8*3 allele does not affect the metabolism of these CYP2C8 substrates to an equal extent. The crystal structure of CYP2C8 reveals a large active site capable of binding substrates in multiple orientations [29], and site-directed mutagenesis studies show that active site residues differentially affect the binding and turnover of different substrates [41].
The higher rosiglitazone AUC and t1/2 found in the present study during coadministration of fluvoxamine is much less than the effect of the CYP2C8 inhibitor gemfibrozil, which increased the AUC by 2.3-fold and prolonged the t1/2 from 3.6 to 7.6 h. In addition, the N-desmethylrosiglitazone/rosiglitazone AUC ratio was decreased by 38% and the tmax of N-desmethylrosiglitazone prolonged [8]. Compared with the present study, the effects of the CYP2C8 inhibitors trimethoprim and ketoconazole were also larger, increasing the rosiglitazone AUC by 38% and 47%, respectively [9, 10]. These studies with the inhibitors gemfibrozil, trimethoprim and ketoconazole confirmed that rosiglitazone is a CYP2C8 substrate, but they also demonstrated a difference in the degree of inhibition of rosiglitazone metabolism [6–10].
The coadministration of fluvoxamine increased the AUC0–∞ of rosiglitazone in 17 of the 21 subjects studied. This moderate effect was overall significant and is comparable to a previous study, where a daily dose of 75 mg fluvoxamine decreased the median clearance of the CYP2C9 substrate tolbutamide from 845 ml h−1 to 688 ml h−1 [36]. If steady state had been achieved for fluvoxamine (requiring 4 days of dosing), its effect on rosiglitazone pharmacokinetics might have been more pronounced.
We conclude that the presence of the CYP2C8*3 and CYP2C9*2 alleles did not influence the pharmacokinetics of rosiglitazone significantly at a dose used clinically. Fluvoxamine moderately increased the AUC0–∞ of rosiglitazone in subjects with the CYP2C8*1/ *1, CYP2C8*1/ *3 and CYP2C8*3/ *3 genotypes.
Acknowledgments
Competing interests: None declared.
We gratefully thank The Alfred Benzon Foundation, The Lundbeck Foundation and The Danish Medical Research Council for financial support.
References
- 1.Patel J, Miller E, Patwardhan R. Rosiglitazone (BRL49653) monotherapy has significant glucose lowering effect in type 2 diabetic patients. Diabetes. 1998;47:A17. [Google Scholar]
- 2.Patel J, Anderson RJ, Rappaport EB. Rosiglitazone monotherapy improves glycaemic control in patients with type 2 diabetes: a twelve-week, randomized, placebo-controlled study. Diabetes Obes Metab. 1999;1:165–72. doi: 10.1046/j.1463-1326.1999.00020.x. [DOI] [PubMed] [Google Scholar]
- 3.Young PW, Buckle DR, Cantello BCC, Chapman H, Clapham JC, Coyle PJ, Haigh D, Hindley RM, Holder JC, Kallender H, Latter AJ, Lawrie KWM, Mossakowska D, Murphy GJ, Cox LR, Smith S. Identification of high-affinity binding sites for the insulin sensitizer rosiglitazone (BRL-49653) in rodent and human adipocytes using a radioiodinated ligand for peroxisomal proliferator-activated receptor gamma. J Pharmacol Exp Ther. 1998;284:751–9. [PubMed] [Google Scholar]
- 4.Cox PJ, Ryan DA, Hollis FJ, Harris AM, Miller AK, Vousden M, Cowley H. Absorption, disposition, and metabolism of rosiglitazone, a potent thiazolidinedione insulin sensitizer, in humans. Drug Metab Dispos. 2000;28:772–80. [PubMed] [Google Scholar]
- 5.Baldwin SJ, Clarke SE, Chenery RJ. Characterization of the cytochrome P450 enzymes involved in the in vitro metabolism of rosiglitazone. Br J Clin Pharmacol. 1999;48:424–32. doi: 10.1046/j.1365-2125.1999.00030.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang JS, Neuvonen M, Wen X, Backman JT, Neuvonen PJ. Gemfibrozil inhibits CYP2C8-mediated cerivastatin metabolism in human liver microsomes. Drug Metab Dispos. 2002;30:1352–6. doi: 10.1124/dmd.30.12.1352. [DOI] [PubMed] [Google Scholar]
- 7.Wen X, Wang JS, Backman JT, Kivisto KT, Neuvonen PJ. Gemfibrozil is a potent inhibitor of human cytochrome P4502C9. Drug Metab Dispos. 2001;29:1359–61. [PubMed] [Google Scholar]
- 8.Niemi M, Backman JT, Granfors M, Laitila J, Neuvonen M, Neuvonen PJ. Gemfibrozil considerably increases the plasma concentrations of rosiglitazone. Diabetologia. 2003;46:1319–23. doi: 10.1007/s00125-003-1181-x. [DOI] [PubMed] [Google Scholar]
- 9.Niemi M, Backman JT, Neuvonen PJ. Effects of trimethoprim and rifampin on the pharmacokinetics of the cytochrome P4502C8 substrate rosiglitazone. Clin Pharmacol Ther. 2004;76:239–49. doi: 10.1016/j.clpt.2004.05.001. [DOI] [PubMed] [Google Scholar]
- 10.Park JY, Kim KA, Shin JG, Lee KY. Effect of ketoconazole on the pharmacokinetics of rosiglitazone in healthy subjects. Br J Clin Pharmacol. 2004;58:397–402. doi: 10.1111/j.1365-2125.2004.02161.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Park JY, Kim KA, Kang MH, Kim SL, Shin JG. Effect of rifampin on the pharmacokinetics of rosiglitazone in healthy subjects. Clin Pharmacol Ther. 2004;75:157–62. doi: 10.1016/j.clpt.2003.10.003. [DOI] [PubMed] [Google Scholar]
- 12.Rahman A, Korzekwa KR, Grogan J, Gonzalez FJ, Harris JW. Selective biotransformation of taxol to 6-alpha-hydroxytaxol by human cytochrome-P450 2C8. Cancer Res. 1994;54:5543–6. [PubMed] [Google Scholar]
- 13.Li XQ, Bjorkman A, Andersson TB, Ridderstrom M, Masimirembwa CM. Amodiaquine clearance and its metabolism to N-desethylamodiaquine is mediated by CYP2C8. A new high affinity and turnover enzyme-specific probe substrate. J Pharmacol Exp Ther. 2002;300:399–407. doi: 10.1124/jpet.300.2.399. [DOI] [PubMed] [Google Scholar]
- 14.Klose TS, Blaisdell JA, Goldstein JA. Gene structure of CYP2C8 and extrahepatic distribution of the human CYP2Cs. J Biochem Mol Toxicol. 1999;13:289–95. doi: 10.1002/(sici)1099-0461(1999)13:6<289::aid-jbt1>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- 15.Bahadur N, Leathart JBS, Mutch E, Steimel-Crespi D, Dunn SA, Gilissen R, Van Houdt J, Hendrickx J, Mannens G, Bohets H, Williams FM, Armstrong M, Crespi CL, Daly AK. CYP2C8 polymorphisms in Caucasians and their relationship with paclitaxel 6 alpha-hydroxylase activity in human liver microsomes. Biochem Pharmacol. 2002;64:1579–89. doi: 10.1016/s0006-2952(02)01354-0. [DOI] [PubMed] [Google Scholar]
- 16.Dai D, Zeldin DC, Blaisdell JA, Chanas B, Coulter SJ, Ghanayem BI, Goldstein JA. Polymorphisms in human CYP2C8 decrease metabolism of the anticancer drug paclitaxel and arachidonic acid. Pharmacogenetics. 2001;11:597–607. doi: 10.1097/00008571-200110000-00006. [DOI] [PubMed] [Google Scholar]
- 17.Hichiya H, Tanaka-Kagawa T, Soyama A, Jinno H, Koyano S, Katori N, Matsushima E, Uchiyama S, Tokunaga H, Kimura H, Minami N, Katoh M, Sugai K, Goto Y, Tamura T, Yamamoto N, Ohe Y, Kunitoh H, Nokihara H, Yoshida T, Minami H, Saijo N, Ando M, Ozawa S, Saito Y, Sawada J. Functional characterization of five novel CYP2C8 variants, G171S, R186X, R186G, K247R, and K383N, found in a Japanese population. Drug Metab Dispos. 2005;33:630–6. doi: 10.1124/dmd.105.003830. [DOI] [PubMed] [Google Scholar]
- 18.Totah RA, Rettie AE. Cytochrome P4502C8: substrates, inhibitors, pharmacogenetics, and clinical relevance. Clin Pharmacol Ther. 2005;77:341–52. doi: 10.1016/j.clpt.2004.12.267. [DOI] [PubMed] [Google Scholar]
- 19.Bidstrup TB, Damkier P, Olsen AK, Ekblom M, Karlsson A, Brosen K. The impact of CYP2C8 polymorphism and grapefruit juice on the pharmacokinetics of repaglinide. Br J Clin Pharmacol. 2006;61:49–57. doi: 10.1111/j.1365-2125.2005.02516.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Niemi M, Leathart JB, Neuvorten A, Backman JT, Daly AK, Neuvonen PJ. Polymorphism in CYP2C8 is associated with the reduced plasma concentrations of repaglinide. Clin Pharmacol Ther. 2003;74:380–7. doi: 10.1016/S0009-9236(03)00228-5. [DOI] [PubMed] [Google Scholar]
- 21.Niemi M, Backman JT, Kajosaari LI, Leathart JB, Neuvonen M, Daly AK, Eichelbaum M, Kivisto KT, Neuvonen PJ. Polymorphic organic anion transporting polypeptide 1B1 is a major determinant of repaglinide pharmacokinetics. Clin Pharmacol Ther. 2005;77:468–78. doi: 10.1016/j.clpt.2005.01.018. [DOI] [PubMed] [Google Scholar]
- 22.Martinez C, Garcia-Martin E, Blanco G, Gamito FJG, Ladero JM, Agundez JAG. The effect of the cytochrome P450CYP2C8 polymorphism on the disposition of (R)-ibuprofen enantiomer in healthy subjects. Br J Clin Pharmacol. 2005;59:62–9. doi: 10.1111/j.1365-2125.2004.02183.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Garcia-Martin E, Martinez C, Tabares B, Frias J, Agundez JAG. Interindividual variability in ibuprofen pharmacokinetics is related to interaction of cytochrome P4502C8 and 2C9 amino acid polymorphisms. Clin Pharmacol Ther. 2004;76:119–27. doi: 10.1016/j.clpt.2004.04.006. [DOI] [PubMed] [Google Scholar]
- 24.Goldstein JA. Clinical relevance of genetic polymorphisms in the human CYP2C subfamily. Br J Clin Pharmacol. 2001;52:349–55. doi: 10.1046/j.0306-5251.2001.01499.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miners JO, Birkett DJ. Cytochrome P4502C9: an enzyme of major importance in human drug metabolism. Br J Clin Pharmacol. 1998;45:525–38. doi: 10.1046/j.1365-2125.1998.00721.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rettie AE, Wienkers LC, Gonzalez FJ, Trager WF, Korzekwa KR. Impaired (S)-warfarin metabolism catalyzed by the R144C allelic variant of Cyp2C9. Pharmacogenetics. 1994;4:39–42. doi: 10.1097/00008571-199402000-00005. [DOI] [PubMed] [Google Scholar]
- 27.SullivanKlose TH, Ghanayem BI, Bell DA, Zhang ZY, Kaminsky LS, Shenfield GM, Miners JO, Birkett DJ, Goldstein JA. The role of the CYP2C9-Leu (359) allelic variant in the tolbutamide polymorphism. Pharmacogenetics. 1996;6:341–9. doi: 10.1097/00008571-199608000-00007. [DOI] [PubMed] [Google Scholar]
- 28.Lee CR, Goldstein JA, Pieper JA. Cytochrome P4502C9 polymorphisms: a comprehensive review of the in-vitro and human data. Pharmacogenetics. 2002;12:251–63. doi: 10.1097/00008571-200204000-00010. [DOI] [PubMed] [Google Scholar]
- 29.Schoch GA, Yano JK, Wester MR, Griffin KJ, Stout CD, Johnson EF. Structure of human microsomal cytochrome P4502C8—evidence for a peripheral fatty acid binding site. J Biol Chem. 2004;279:9497–503. doi: 10.1074/jbc.M312516200. [DOI] [PubMed] [Google Scholar]
- 30.Williams PA, Cosme J, Ward A, Angova HC, Vinkovic DM, Jhoti H. Crystal structure of human cytochrome P4502C9 with bound warfarin. Nature. 2003;424:464–8. doi: 10.1038/nature01862. [DOI] [PubMed] [Google Scholar]
- 31.Yasar U, Lundgren S, Eliasson E, Bennet A, Wiman B, de Faire U, Rane A. Linkage between the CYP2C8 and CYP2C9 genetic polymorphisms. Biochem Biophys Res Commun. 2002;299:25–8. doi: 10.1016/s0006-291x(02)02592-5. [DOI] [PubMed] [Google Scholar]
- 32.Hemeryck A, De Vriendt C, Belpaire FM. Inhibition of CYP2C9 by selective serotonin reuptake inhibitors: in vitro studies with tolbutamide and (S)-warfarin using human liver microsomes. Eur J Clin Pharmacol. 1999;54:947–51. doi: 10.1007/s002280050580. [DOI] [PubMed] [Google Scholar]
- 33.Jeppesen U, Rasmussen BB, Brosen K. Fluvoxamine inhibits the CYP2C19-catalyzed bioactivation of chloroguanide. Clin Pharmacol Ther. 1997;62:279–86. doi: 10.1016/S0009-9236(97)90030-8. [DOI] [PubMed] [Google Scholar]
- 34.Jeppesen U, Gram LF, Vistisen K, Loft S, Poulsen HE, Brosen K. Dose dependent inhibition of CYP1A2, CYP2C19 and CYP2D6 by citalopram, fluoxetine, fluvoxamine and paroxetine. Eur J Clin Pharmacol. 1996;51:73–8. doi: 10.1007/s002280050163. [DOI] [PubMed] [Google Scholar]
- 35.Madsen H, Enggaard TP, Hansen LL, Klitgaard NA, Brosen K. Fluvoxamine inhibits the CYP2C9 catalyzed biotransformation of tolbutamide. Clin Pharmacol Ther. 2001;69:41–7. doi: 10.1067/mcp.2001.112689. [DOI] [PubMed] [Google Scholar]
- 36.Brosen K, Skjelbo E, Rasmussen BB, Poulsen HE, Loft S. Fluvoxamine is a potent inhibitor of cytochrome-P4501A2. Biochem Pharmacol. 1993;45:1211–4. doi: 10.1016/0006-2952(93)90272-x. [DOI] [PubMed] [Google Scholar]
- 37.Walsky RL, Gaman EA, Obach RS. Examination of 209 drugs for inhibition of cytochrome P450 2C8. J Clin Pharmacol. 2005;45:68–78. doi: 10.1177/0091270004270642. [DOI] [PubMed] [Google Scholar]
- 38.Pedersen RS, Verstuyft C, Becquemont L, Jaillon P, Brosen K. Cytochrome P4502C9 (CYP2C9) genotypes in a Nordic population in Demmark. Basic Clin Pharmacol Toxicol. 2004;94:151–2. doi: 10.1111/j.1742-7843.2004.pto940309.x. [DOI] [PubMed] [Google Scholar]
- 39.Verstuyft C, Morin S, Yang J, Loriot MA, Barbu V, Kerb R, Brinkmann U, Beaune P, Jaillon P, Becquemont L. Rapid and robust genotyping strategy for cytochrome P4502C9 and MDR1 single nucleotide polymorphism identification. Ann Biol Clin. 2003;61:305–9. [PubMed] [Google Scholar]
- 40.Pedersen RS, Brosen K, Nielsen F. HPLC method for determination of rosiglitazone in plasma. Chromatographia. 2005;62:197–201. [Google Scholar]
- 41.Kerdpin O, Elliot DJ, Boye SL, Birkett DJ, Yoovathaworm K, Miners JO. Differential contribution of active site residues in substrate recognition sites 1 and 5 to cytochrome p450 2C8 substrate selectivity and regioselectivity. Biochem. 2004;43:7834–42. doi: 10.1021/bi0496844. [DOI] [PubMed] [Google Scholar]