Abstract
One of the metabolic fates of 3-deoxyglucosone, a product of protein deglycation and a potent glycating agent, is to be oxidized to 2-keto-3-deoxygluconate, but the enzyme that catalyses this reaction is presently unknown. Starting from human erythrocytes, which are known to convert 3-deoxyglucosone to 2-keto-3-deoxygluconate, we have purified to near homogeneity a NAD-dependent dehydrogenase that catalyses this last reaction at neutral pH. Sequencing of a 55 kDa band coeluting with the enzymatic activity in the last step indicated that it corresponded to aldehyde dehydrogenase 1A1 (ALDH1A1), an enzyme known to catalyse the oxidation of retinaldehyde to retinoic acid. Overexpression of human ALDH1A1 in HEK cells led to a more than 20-fold increase in 3-deoxyglucosone dehydrogenase activity. In mouse tissues 3-deoxyglucosone dehydrogenase activity was highest in liver, intermediate in lung and testis, and negligible or undetectable in other tissues, in agreement with the tissue distribution of ALDH1A1 mRNA. 3-Deoxyglucosone dehydrogenase activity was undetectable in tissues from ALDH1A1−/− mice. ALDH1A1 appears therefore to be the major if not the only enzyme responsible for the oxidation of 3-deoxyglucosone to 2-keto-3-deoxygluconate. The urinary excretion of 2-keto-3-deoxygluconate amounted to 16.7 μmol/g creatinine in humans, indicating that 3-deoxyglucosone may be quantitatively a more important substrate than retinaldehyde for ALDH1A1.
Keywords: Fructosamines, deglycation, oxoaldehyde, aldehyde dehydrogenase
1. Introduction
Recent work has led to the identification of a new protein repair mechanism that removes fructosamines from proteins [1–3]. Fructosamines arise through a spontaneous reaction of glucose with amines present at the amino terminus and at the lysine side-chains of proteins [4]. The enzyme that repairs fructosamines is an ATP-dependent kinase, fructosamine 3-kinase, which phosphorylates the third carbon of the sugar moiety of fructoselysine residues. Fructosamine 3-phosphates are unstable, decomposing to 3-deoxyglucosone, inorganic phosphate and a free amine, thus regenerating the amine in its original state [1–3]. 3-Deoxyglucosone, the carbohydrate product of fructosamine 3-phosphate decomposition, is itself a potent glycating agent that may react with lysine and arginine side-chains to form various AGEs, thought to be involved in the pathogenesis of diabetic complications [5]. Cells have, however, the ability of reducing 3-deoxyglucosone to 3-deoxyfructose and to oxidize it to 2-keto-3-deoxygluconate [6]. Several enzymes that catalyse the reduction to 3-deoxyfructose have been identified. These are dihydrodiol dehydrogenase, aldehyde reductase and aldose reductase [7–9]. The enzyme that catalyses the oxidation of 3-deoxyglucosone to 2-keto-3-deoxygluconate has, however, not yet been identified.
In the present paper, we report that human erythrocytes, which have been shown to convert 3-deoxyglucosone to 2-keto-3-deoxygluconate [6] contain an NAD-dependent 3-deoxyglucosone dehydrogenase acting at neutral pH. We have purified this enzyme from human erythrocytes, have partially characterized it and found that it corresponds to aldehyde dehydrogenase 1A1 (ALDH1A1).
2. Materials and methods
2.1. Purification of 3-DGDH from human erythrocytes
All steps were performed at 4°C. 300 ml of washed erythrocytes were thawed and diluted with 1.5 l of buffer A (10 mM Hepes, pH 7.1, 1 mM dithiothreitol, 1 μg/ml leupeptin, 1 μg/ml antipain). The resulting lysate was centrifuged for 1 h at 11 000 g. NaCl (100 mM final concentration) and poly(ethylene)glycol (14 %) were added to the supernatant (1.2 l), which was centrifuged for 45 min at 11 000 g. The resulting pellet was resuspended in 900 ml buffer B (25 mM Tris, pH 8.0, 1 mM dithiothreitol, 1 μg/ml leupeptin, 1 μg/ml antipain) and loaded onto a 125 ml DEAE Sepharose column. The column was washed with 250 ml buffer B and a linear NaCl gradient (0–500 mM in 500ml) was applied to elute 3-deoxyglucosone dehydrogenase (3-DGDH).
The fractions containing 3DGDH (50 ml) were pooled, diluted to 500 ml in buffer B and loaded on a 25 ml Q Sepharose column. The column was washed with 50 ml buffer B and 3-DGDH was eluted with a linear NaCl gradient (0–300 mM in 400 ml). The active fractions (42 ml) were pooled and concentrated to 4 ml in an Amicon cell equipped with a YM-10 membrane. Two ml of this preparation were loaded onto a 1.6 × 60 cm Sephacryl S-200 column equilibrated with 25 mM Hepes, pH 7.1, 100 mM NaCl, 1 mM dithiothreitol, 1 μg/ml leupeptin and 1 μg/ml antipain (buffer C). The active fractions from two such columns (9 ml) were pooled and diluted to a final wolume of 50 ml in buffer D (25 mM Hepes, pH 7.1, 1 mM dithiothreitol, 1 mM MgCl2, 1 μg/ml leupeptin and 1 μg/ml antipain) and applied on a 10 ml Blue Sepharose column. The column was washed with 3 volumes of buffer D; 3DGDH was recovered in the flow-through and washing fractions (40 ml) and concentrated to 10 ml; 5 ml of this preparation was applied onto a UnoQ (1 ml) column equilibrated in buffer B. The column was washed with buffer B and the retained protein was eluted with a linear NaCl gradient (0–400 mM in 20 ml). Protein identification was performed by mass spectrometry as previously described [10].
2.2. Over-expression of 3-DGDH in HEK293 cells
The sequence encoding human ALDH1A1 was PCR-amplified from human liver cDNA using Taq polymerase, a 5′ primer (CTGACCGAATTCCACCATGTCATCCTCAGGCACG) containing the putative ATG codon flanked by an EcoRI site (underlined), and a 3′ primer (GCTAGCCAAGCTTGTTATGAGTTCTTCTGAGAGAT) containing the putative stop codon flanked by an HindIII site. The amplification product (≈1550 bp) was cloned in pBluescript and checked by sequencing. An EcoRI-HindIII fragment was removed from this plasmid and ligated in pCMV5, which was used for transfections. HEK-293 cells were cultured under 5% CO2 at 37°C in Dulbecco’s minimal essential medium containing 10% (vol/vol) FCS in 10-cm diameter Sarstedt dishes. The cells were transfected using JetPEI™ (Poly-Plus Transfection) with 8 μg pCMV5 DNA, incubated for 54 hours and harvested in 0.75 ml of buffer D. After three cycles of freezing and thawing, the cell lysates were centrifuged for 30 min at 10 000 g and 4°C. The supernatant obtained from 20 dishes was diluted with two volumes of buffer B and loaded onto a 10-ml DEAE-Sepharose column at 4°C. The column was washed with 30 ml of buffer B and then a linear NaCl gradient (0–500 mM in 100ml buffer B) was applied to elute 3-deoxyglucosone dehydrogenase. The latter represented 40% of total proteins in the purest fractions used for enzymatic assays.
2.3. Preparation of mouse tissues extracts
Tissues were collected from 30 g male NMRI or from ALDH1A1−/− mice [11], and immediately frozen in liquid nitrogen and maintained at −70°C until use. The frozen samples were homogenized at 4°C in a Potter-Elvehjem device with 3 ml/g tissue of buffer E (25 mM Hepes, pH 7.1, 100 mM KCl, 1 mM dithiotreitol, 5 μg/ml leupeptin and 5 μg/ml antipain). The homogenates were centrifuged for 30 min at 10 000 g. The supernatant was collected and proteins were precipitated by adding poly(ethylene)glycol to a final concentration of 25% (which quantitatively precipitates 3-DGDH). Samples were centrifuged for 30 min at 10 000 g. The resulting pellet was resuspended in the same volume of buffer B and used for 3-DGDH assays.
2.4. Enzymatic assays
For the assay of 3-DGDH activity, the enzyme preparation (50 μl) was incubated at 30°C in a reaction mixture (150 μl) containing 25 mM Hepes, pH 7.1, 1 mM dithiotreitol, 5 mM NAD and 1 mM 3-deoxyglucosone. After appropriate times (15–120 min), 50 μl perchloric acid was added and the mixture centrifuged for 10 min at 16 000 g at 4°C. The supernatant was neutralised with 3 M K2CO3 and 2-keto-3-deoxygluconate was assayed enzymatically with E.coli 2-keto-3-deoxygluconate kinase [2]. The activity of purified 3-DGDH was also determined by following A340 with a Beckman spectrophotometer. Typically, 10 μl of enzyme preparation was incubated at room-temperature in a reaction mixture (50 μl in a 100 μl micro-cuvette) containing 25 mM Hepes, pH 7.1, 1 mM dithiotreitol, 1 mM NAD and the indicated concentrations of cofactor and substrate.
Urine samples were mixed with 0.5 volumes of 10 % perchloric acid, washed with two volumes 5% charcoal, heated 10 min at 96°C and neutralised with 3 M K2CO3 before determination of 2-keto-3-deoxygluconate. Creatinine was determined using the Jaffé method.
3. Results
3.1. Purification and identification of 3-deoxyglucosone dehydrogenase
We purified the NAD-linked 3-deoxyglucosone dehydrogenase reported to be present in human erythrocytes [6]. This enzyme was assayed at neutral pH (pH 7.1) by measuring the NAD-dependent conversion of 3-deoxyglucosone to 2-keto-3-deoxygluconate with a specific assay based on E. coli 2-keto-3-deoxygluconate kinase. The purification procedure involved fractionation with poly(ethylene)glycol and chromatographic steps on DEAE-Sepharose, Q-Sepharose, Sephacryl S-200, Blue-Sepharose and UnoQ. 3-Deoxyglucosone dehydrogenase was eluted as a single peak from each of the columns (not shown), suggesting that a single enzyme was responsible for this activity in erythrocytes. The enzyme was purified more than 400-fold with a yield of about 1 % (Table 1). This low yield was largely due to the fact that only the most active fractions were selected after each step.
Table 1.
Purification of 3-deoxyglucosone dehydrogenase from human erythrocytes
| Proteins (g) | Activity (mU/mg) | Total Activity (mU) | Purification | Yield (%) | |
|---|---|---|---|---|---|
| Hemolysate | 78 | 0.029 | 2250 | - | 100 |
| PEG (pellet) | 66 | 0.029 | 1881 | 1 | 83.6 |
| DEAE-Sepharose | 1.905 | 0.21 | 400.1 | 7 | 17.8 |
| Q-Sepharose | 0.312 | 0.641 | 200.1 | 22 | 8.9 |
| Sephacryl S-200 | 0.053 | 2.195 | 115.8 | 76 | 5.1 |
| Blue-Sepharose | 0.016 | 2.68 | 42.9 | 93 | 1.9 |
| UnoQ | 0.0017 | 12.57 | 22.0 | 436 | 1 |
SDS-PAGE analysis of the fractions of the last chromatographic step (Figure 1b) revealed that three bands were still present in the preparation. However, only the 55 kDa co-eluted with 3-deoxyglucosone dehydrogenase. This band was cut from the gel, digested with trypsin, and the resulting peptides were analyzed by tandem mass spectrometry. Eleven of them (Fig. 1c) matched with aldehyde dehydrogenase 1A1 (ALDH1A1; gi-21361176), an enzyme known to act on retinaldehyde, benzaldehyde and other aldehydes [12].
Figure 1. Identification of 3-deoxyglucosone dehydrogenase as aldehyde dehydrogenase 1A1 (ALDH1A1).
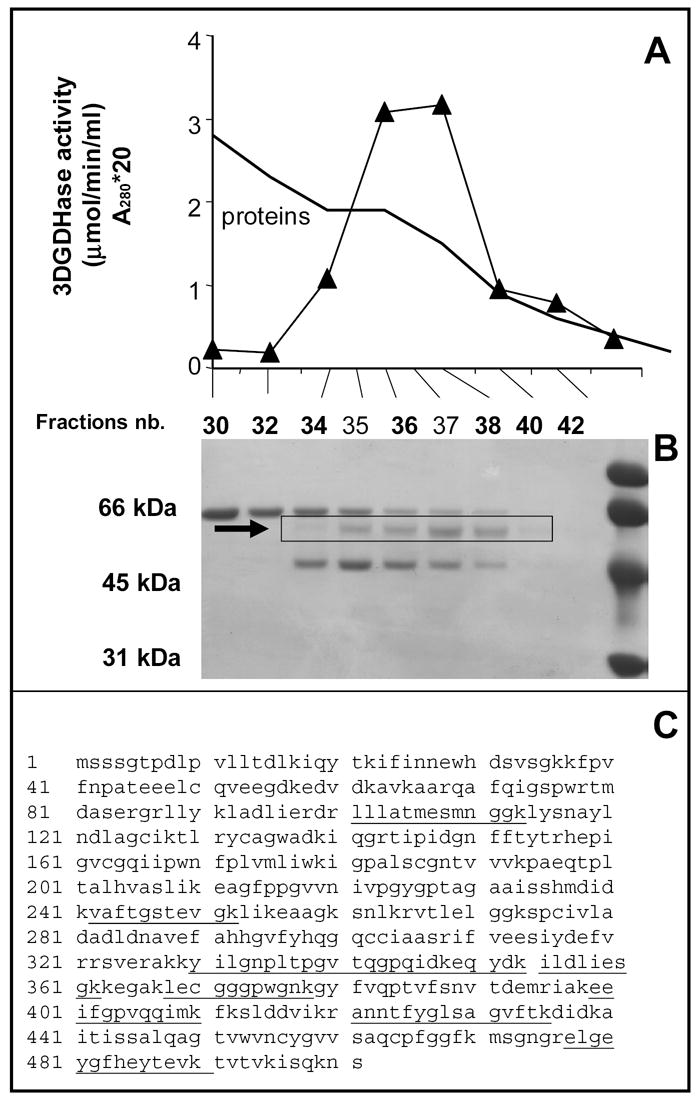
Panel A shows the elution profile of 3-DGDH from the UnoQ column, the last purification step. Twenty μl of the indicated fractions were loaded on a SDS-PAGE gel, which was stained with Coomassie blue (panel B). The indicated band was cut from the gel, submitted to trypsin digestion and analyzed by mass spectrometry. Panel C shows the identified peptides (underlined) in the sequence of ALDH1A1 (gi-21361176).
To confirm that ALDH1A1 was endowed with 3-deoxyglucosone dehydrogenase activity, HEK-293 cells were transfected with an eukaryotic pCMV5 expression vector containing or not the open reading frame of human ALDH1A1. Extracts of cells transfected with pCMV5-ALDH1A1 construct displayed a 3-deoxyglucosone dehydrogenase activity of 1.6 ± 0.1 nmol/min/mg of protein (n = 3) whereas no activity could be detected (< 0.05 mU/mg protein) in cells transfected with a control plasmid.
At pH 7.1 and in the presence of 100 μM 3-deoxyglucosone, partially purified recombinant ALDH1A1 displayed a Km for NAD of 85 μM and a Vmax of 18 nmol/min/mg protein) whereas the activity with 1 mM NADP was < 5 % of that observed with 1 mM NAD. In the presence of 1 mM NAD, recombinant ALDH1A1 displayed a Km of 95 μM for 3-deoxyglucosone and a Vmax of 45 nmol/min/mg protein. The enzyme acted also on benzaldehyde with a Km value ≤ 2 μM and a Vmax of 72 nmol/min/mg protein. The pH curve of the 3-deoxyglucosone dehydrogenase activity showed a broad pH optimum between 7.1 and 9, with nearly identical activities at these values (not shown).
3.2. Tissue distribution of 3-deoxyglucosone dehydrogenase
Tissue distribution studies of the mouse ALDH1A1 mRNA indicates that it is mostly expressed in liver and that intermediate level are found in lung and testis. No mRNA is detectable in other tissues (brain, kidneys, muscle, heart) [13]. Using our specific enzymatic assay, we determined the distribution of 3-deoxyglucosone dehydrogenase in mouse tissues. As shown in Figure 2, the highest activity was observed in liver, followed by testis, lung and erythrocytes, whereas no activity was detectable in other tissues. No 3-deoxyglucosone dehydrogenase activity could be detected (< 0.05 mU/mg protein) in brain, lung, liver, kidneys, testis, and erythrocytes (assays in triplicates, results not shown) from ALDH1A1−/− mouse [11].
Figure 2. Distribution of 3-deoxyglucosone dehydrogenase in mouse tissues.
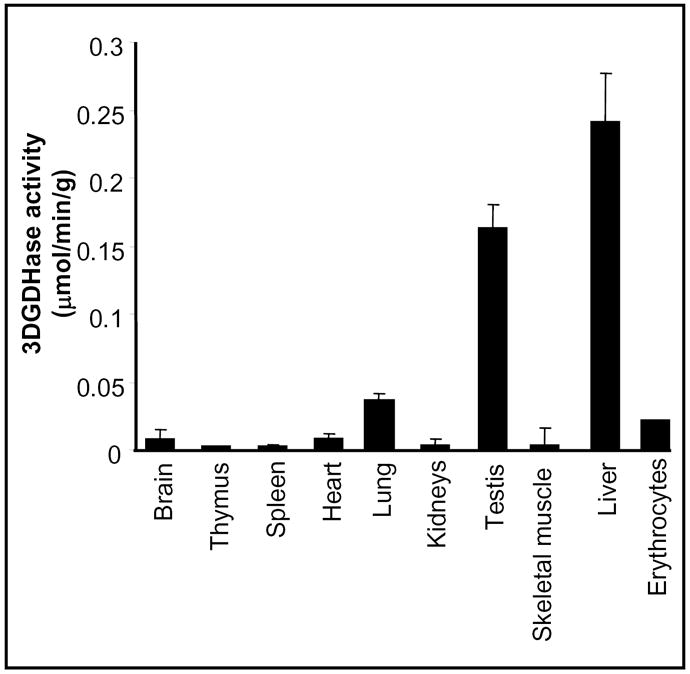
Results shown are the means of three experiments ± SEM.
3.4. Urinary excretion of 2-keto-3-deoxygluconate
No information is presently available on the presence of 2-keto-3-deoxygluconate in urine. To address this point, we determined the concentration of this compound in urine from 4 healthy subjects (male, 30–50 year old) using the assay based on 2-keto-3-deoxygluconate kinase [2]. A mean value of 16.7 ± 0.9 μmol/g creatinine was found, indicating that the daily excretion of 2-keto-3-deoxygluconate is about 21.7 μmol/day (assuming a daily creatinine excretion of 1.3 g).
4. Discussion
Erythrocytes have been shown to contain an NAD-dependent dehydrogenase catalyzing the oxidation of 3-deoxyglucosone to 2-keto-3-deoxygluconate. Purification of this enzyme to near homogeneity led to its identification as ALDH1A1. This identification was confirmed by showing that recombinant ALDH1A1 had similar properties as erythrocyte 3-deoxyglucosone dehydrogenase. The findings that the tissue distribution of 3-deoxyglucosone dehydrogenase parallels that ALDH1A1 mRNA and that this activity in undetectable in ALDH1A1 knockout mice indicate that ALDH1A1 is the major if not the only enzyme responsible for 3-deoxyglucosone oxidation. This conclusion is also supported by the finding that only one peak of 3-deoxyglucosone dehydrogenase activity was detected in each of the chromatographic steps that we carried out to purify this enzyme.
The enzyme described in this paper markedly differs from ‘ketoaldehyde dehydrogenase’ [14–16], an enzyme that has been mostly studied with methylglyoxal as a substrate, but the name of which suggests that it acts also on other ketoaldehydes such as 3-deoxyglucosone. Unlike 3DGDH/ALDH1A1, ketoaldehyde dehydrogenase displays a higher affinity for NADP than for NAD, is not active at neutral pH but well at alkaline pH, and requires high concentrations of amines to be active. Furthermore, this enzyme purified from rat liver does not appear to catalyse the oxidation of 3-deoxyglucosone to 2-keto-3-deoxygluconate even at alkaline pH and in the presence of an amine that stimulates its activity on methylglyoxal (unpublished results). Thus ketoaldehyde dehydrogenase appears to be very distinct from 3-DGDH/ALDH1A1, which is the main enzyme responsible for the oxidation of 3-deoxyglucosone.
ALDH1A1 is known to act on several substrates such as acetaldehyde, benzaldehyde, 4-hydroxynonenal, malondialdehyde, and retinaldehyde [12,17,18]. The latter, which is the precursor of retinoic acid, a compound playing a major role in cellular differentiation, is also a substrate for ALDH1A2 and ALDH1A3. Mouse gene knock out models have shown that ALDH1A1 deficiency does not lead to any detectable phenotype [11], although compound mutant mice revealed that ALDH1A1 does play a redundant role in retinoic acid synthesis needed for eye development [19]. This is in marked contrast with ALDH1A2 and ALDH1A3 deficiencies which lead to in utero death or lethal choanal atresia, respectively [21, 21]. Furthermore, comparison of the crystal structure of ALDH1A1 and ALDH1A2 indicate that ALDH1A1 has a larger catalytic cavity compared to ALDH1A2, consistent with the broader substrate specificity of this enzyme [22, 23]. These observations suggest that ALDH1A1 plays essentially a metabolic role, which is consistent with its high activity in liver.
3-Deoxyglucosone is likely to be quantitatively an important substrate for ALDH1A1. Assuming that the urinary excretion of 2-keto-3-deoxygluconate entirely derives from 3-deoxyglucosone, it can be calculated that this enzyme catalyses the oxidation of about 20 μmol 3-deoxyglucosone/day. By comparison, the daily intake of Vitamin A, the main source of retinaldehyde, represents about 3 mg [24], i.e., 10 μmol. This suggests that 3-deoxyglucosone is quantitatively a slightly more important substrate than retinaldehyde for ALDH1A1.
Acknowledgments
This work was supported by the Concerted research action program of the Communauté Française de Belgique, the Interuniversity Attraction Poles Program-Belgian Science Policy, by the Belgian Scientific Fund for Medical Research (FRSM), by the European Foundation for the study of Diabetes and by the Juvenile Diabetes Foundation International. JF is fellow of the Fonds pour l’Encouragement à la Recherche dans l’Industrie et dans l’Agriculture.
Abbreviation list
- ALDH1A1
aldehyde dehydrogenase 1A1
- 3-DGDH
3-deoxyglucosone dehydrogenase
- HEK
human embryonic kidney
- FN3K
fructosamine 3-kinase
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
François Collard, Université catholique de Louvain, Christian de Duve Institute of Cellular Pathology, Avenue Hippocrate 75, B-1200 Brussels, Belgium.
Didier Vertommen, Université catholique de Louvain, Christian de Duve Institute of Cellular Pathology, Avenue Hippocrate 75, B-1200 Brussels, Belgium.
Juliette Fortpied, Université catholique de Louvain, Christian de Duve Institute of Cellular Pathology, Avenue Hippocrate 75, B-1200 Brussels, Belgium.
Gregg Duester, Burnham Institute for Medical Research, 10901 North Torrey Pines Road, La Jolla, CA 92037, USA.
Emile Van Schaftingen, Université catholique de Louvain, Christian de Duve Institute of Cellular Pathology, Avenue Hippocrate 75, B-1200 Brussels, Belgium.
References
- 1.Delpierre G, Rider MH, Collard F, Stroobant V, Vanstapel F, Santos H, Van Schaftingen E. Identification, cloning, and heterologous expression of a mammalian fructosamine-3-kinase. Diabetes. 2000;49:1627–1634. doi: 10.2337/diabetes.49.10.1627. [DOI] [PubMed] [Google Scholar]
- 2.Delpierre G, Collard F, Fortpied J, Van Schaftingen E. Fructosamine-3-kinase is involved in an intracellular deglycation pathway. Biochem J. 2002;365:801–808. doi: 10.1042/BJ20020325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Szwergold BS, Howell S, Beisswenger PJ. Human fructosamine-3-kinase: purification, sequencing, substrate specificity, and evidence of activity in vivo. Diabetes. 2001;50:2139–2147. doi: 10.2337/diabetes.50.9.2139. [DOI] [PubMed] [Google Scholar]
- 4.Baynes JW, Watkins NG, Fisher CI, Hull CJ, Patrick JS, Ahmed MU, Dunn JA, Thorpe SR. The Amadori product on protein: structure and reactions. Prog Clin Biol Res. 1989;304:43–67. [PubMed] [Google Scholar]
- 5.Niwa T. 3-Deoxyglucosone: metabolism, analysis, biological activity, and clinical implication. J Chromatogr B Biomed Sci Appl. 1999;731:23–36. doi: 10.1016/s0378-4347(99)00113-9. [DOI] [PubMed] [Google Scholar]
- 6.Fujii E, Iwase H, Ishii-Karakasa I, Yajima Y, Hotta K. The presence of 2-keto-3-deoxygluconic acid and oxoaldehyde dehydrogenase activity in human erythrocytes. Biochem Biophys Res Commun. 1995;210:852–7. doi: 10.1006/bbrc.1995.1736. [DOI] [PubMed] [Google Scholar]
- 7.Feather MS, Flynn TG, Munro KA, Kubiseski TJ, Walton DJ. Catalysis of reduction of carbohydrate 2-oxoaldehydes (osones) by mammalian aldose reductase and aldehyde reductase. Biochim Biophys Acta. 1995;1244:10–6. doi: 10.1016/0304-4165(94)00156-r. [DOI] [PubMed] [Google Scholar]
- 8.Takahashi M, Fujii J, Teshima T, Suzuki K, Shiba T, Taniguchi N. Identity of a major 3-deoxyglucosone-reducing enzyme with aldehyde reductase in rat liver established by amino acid sequencing and cDNA expression. Gene. 1993;127:249–53. doi: 10.1016/0378-1119(93)90728-l. [DOI] [PubMed] [Google Scholar]
- 9.Sato K, Inazu A, Yamaguchi S, Nakayama T, Deyashiki Y, Sawada H, Hara A. Monkey 3-deoxyglucosone reductase: tissue distribution and purification of three multiple forms of the kidney enzyme that are identical with dihydrodiol dehydrogenase, aldehyde reductase, and aldose reductase. Arch Biochem Biophys. 1993;307:286–94. doi: 10.1006/abbi.1993.1591. [DOI] [PubMed] [Google Scholar]
- 10.Achouri Y, Noel G, Vertommen D, Rider MH, Veiga-Da-Cunha M, Van Schaftingen E. Identification of a dehydrogenase acting on D-2-hydroxyglutarate. Biochem J. 2004;381:35–42. doi: 10.1042/BJ20031933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fan X, Molotkov A, Manabe S, Donmoyer CM, Deltour L, Foglio MH, Cuenca AE, Blaner WS, Lipton SA, Duester G. Targeted disruption of Aldh1a1 (Raldh1) provides evidence for a complex mechanism of retinoic acid synthesis in the developing retina. Mol Cell Biol. 2003;13:4637–48. doi: 10.1128/MCB.23.13.4637-4648.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vasiliou V, Pappa A, Petersen DR. Role of aldehyde dehydrogenases in endogenous and xenobiotic metabolism. Chem Biol Interact. 2000;129:1–19. doi: 10.1016/s0009-2797(00)00211-8. [DOI] [PubMed] [Google Scholar]
- 13.Hsu LC, Chang WC, Hoffmann I, Duester G. Molecular analysis of two closely related mouse aldehyde dehydrogenase genes: identification of a role for Aldh1, but not Aldh-pb, in the biosynthesis of retinoic acid. Biochem J. 1999;339:387–95. [PMC free article] [PubMed] [Google Scholar]
- 14.Monder C. Alpha-keto aldehyde dehydrogenase, an enzyme that catalyses the enzymic oxidation of methylglyoxal to pyruvate. J Biol Chem. 1967;242:4603–4609. [PubMed] [Google Scholar]
- 15.Dunkerton J, James SP. Purification of 2-oxoaldehyde dehydrogenase and its dependence on unusual amines. Biochem J. 1975;149:609–17. doi: 10.1042/bj1490609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vander Jagt DL, Hunsaker LA. Methylglyoxal metabolism and diabetic complications: roles of aldose reductase, glyoxalase-I, betaine aldehyde dehydrogenase and 2-oxoaldehyde dehydrogenase. Chem Biol Interact. 2003;143–144:341–51. doi: 10.1016/s0009-2797(02)00212-0. [DOI] [PubMed] [Google Scholar]
- 17.Gagnon I, Duester G, Bhat PV. Kinetic analysis of mouse retinal dehydrogenase type-2 (RALDH2) for retinal substrates. Biochim Biophys Acta. 2002;1596:156–62. doi: 10.1016/s0167-4838(02)00213-3. [DOI] [PubMed] [Google Scholar]
- 18.Vasiliou V, Pappa A, Estey T. Role of human aldehyde dehydrogenases in endobiotic and xenobiotic metabolism. Drug Metab Rev. 2004;36:279–99. doi: 10.1081/dmr-120034001. [DOI] [PubMed] [Google Scholar]
- 19.Molotkov A, Molotkova N, Duester G. Retinoic acid guides eye morphogenetic movements via paracrine signaling but is unnecessary for retinal dorsoventral patterning. Development. 2006;133:1901–1910. doi: 10.1242/dev.02328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Niederreither K, Subbarayan V, Dolle P, Chambon P. Embryonic retinoic acid synthesis is essential for early mouse post-implantation development. Nat Genet. 1999;21:444–8. doi: 10.1038/7788. [DOI] [PubMed] [Google Scholar]
- 21.Dupe V, Matt N, Garnier JM, Chambon P, Mark M, Ghyselinck NB. A newborn lethal defect due to inactivation of retinaldehyde dehydrogenase type 3 is prevented by maternal retinoic acid treatment. Proc Natl Acad Sci U S A. 2003;100:14036–41. doi: 10.1073/pnas.2336223100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moore SA, Baker HM, Blythe TJ, Kitson KE, Kitson TM, Baker EN. Sheep liver cytosolic aldehyde dehydrogenase: the structure reveals the basis for the retinal specificity of class 1 aldehyde dehydrogenases. Structure. 1998;6:1541–51. doi: 10.1016/s0969-2126(98)00152-x. [DOI] [PubMed] [Google Scholar]
- 23.Lamb AL, Newcomer ME. The structure of retinal dehydrogenase type II at 2.7 A resolution: implications for retinal specificity. Biochemistry. 1999;38:6003–11. doi: 10.1021/bi9900471. [DOI] [PubMed] [Google Scholar]
- 24.Stitt KR. Nutritive value of diets today and fifty years ago. Nutr Rev. 1963;21:257–9. doi: 10.1111/j.1753-4887.1963.tb04812.x. [DOI] [PubMed] [Google Scholar]


