Abstract
Poly(ethylene glycol) (PEG) modified thiolated gelatin nanoparticles (PEG-SHGel) were developed as a long-circulating passively-targeted delivery system that respond to intracellular glutathione concentrations to enhance DNA delivery and transfection. Reporter plasmid expressing enhanced green fluorescent protein (EGFP-N1) was encapsulated in the nanoparticles. DNA-containing gelatin (Gel) and thiolated gelatin (SHGel) nanoparticles were found to have a size range of 220–250 nm, whereas surface modification with PEG resulted in particles with slightly larger size range of 310–350 nm. PEG modification was confirmed by electron spectroscopy for chemical analysis (ESCA), where an increase in the ether peak intensities of the C1s spectra corresponds to the surface presence of ethylene oxide residues. In addition, the PEG-SHGel nanoparticles released encapsulate plasmid DNA in response to varying concentrations of glutathione (0 – 5.0 mM GSH in phosphate buffered saline). The stability of the encapsulated DNA was confirmed by agarose gel electrophoresis. Lastly, from the qualitative and quantitative results of in vitro transfection studies in murine fibroblast cells (NIH-3T3), PEG-Gel and PEG-SHGel nanoparticles afforded the highest transfection efficiency of the reporter plasmid. The results of these studies show that PEG-modified thiolated gelatin nanoparticles could serve as a very efficient nanoparticulate vector for systemic DNA delivery to solid tumors where the cells are known to have significantly higher intracellular glutathione concentrations.
Keywords: Thiolated gelatin nanoparticles, poly(ethylene glycol), surface modification, DNA delivery, transfection
INTRODUCTION
The objective of any gene therapy strategy is to efficiently and safely deliver therapeutic DNA to the cells at the target site. There are various limitations to the delivery of DNA, which include its stability during transit from the site of injection to the target tissue, cellular uptake by endocytosis, the endosomal/lysosomal escape of the DNA, and finally the nuclear import followed by transcription and translation of the protein of interest 1, 2. Further understanding these challenges encountered in the delivery of DNA would help in designing a vector that could overcome the barriers for gene delivery.
Gene delivery to target cells can be achieved by either viral or non-viral methods. Viral vectors such as the adenoviruses, retroviruses, adeno-associated viruses (AAVs), lentivirus and herpes simplex virus are continued to be used in many clinical protocols. Although efficient in transfection, the viral vectors are plagued by issues of integration with the host genome, self-replication, recombination potential, and immunogenicity. There are reported incidents of insertional mutagenesis during treatment in humans and their safety is an issue 3–5. As such, there is a tremendous need to develop non-viral gene delivery vectors that would be safe, easier to produce in large quantities, and would not have a packaging DNA size limitation. Over the last few years, there has been extensive ongoing research in the application of lipids and polymers to deliver DNA for local and systemic gene therapy applications. An ideal vector for gene therapy needs to be inert while in circulation and release its payload in the cell at the target site resulting in efficient transfection of the cells. The vector used should have sufficiently small size and stability, minimal aggregation in blood and have the ability to efficiently target cells, and disassemble and release the DNA intracellularly, and allow for the DNA to be imported into the nucleus. The design of an ideal vector system is still the major limiting step for effective non-viral gene therapy 6.
Several types of polymeric materials have been investigated on their physical, chemical, and biological properties for capability to be used as vectors for gene delivery applications. The polymer used should be biocompatible and biodegradable, as accumulation of non-metabolizable materials in vivo results in toxicity 7. In case of gene delivery applications, the polymeric material should also be non-immunogenic with a high efficiency to complex and/or encapsulate the payload. There has been a fair amount of success in reducing immunogenicity and cytotoxicity with the concomitant enhancement in efficiency of transfection using polymeric vectors.
Gelatin is one of the most versatile, naturally occurring biopolymers widely used in cosmetics, pharmaceutical formulations, as well as in many different types of food products. Gelatin is obtained by acid or base hydrolysis of collagen. The nanoparticulate carriers of gelatin have been used for efficient intracellular delivery of the encapsulated hydrophilic payload. Over the last few years, our group is engaged in exploring gelatin and modified gelatin-based nanoparticles for intracellular drug and gene delivery.9–13 From the results published so far, it is evident that thiolated gelatin nanoparticles can result in a rapid release of their contents in a highly reducing environment, such as one with high glutathione concentration. This could be attributed to the thiol content of gelatin, which would result in the formation of disulfide bonds within the polymer structure, thus strengthening the tertiary and quaternary protein structure in the case of gelatin. The disulfide bonds also stabilize the nanoparticles during systemic circulation. However, in the cell, where the glutathione concentrations are usually 1000 fold higher, these disulfide bonds are broken, the biopolymer unfolds releasing its contents (Figure 1). In addition, preliminary data indicate that the thiolated gelatin nanoparticles have better transfection efficiency over gelatin nanoparticles.
Figure 1.
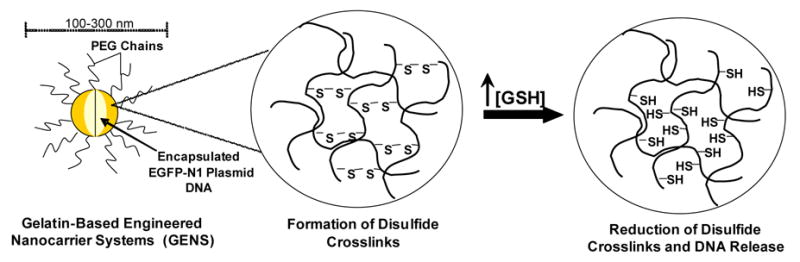
Schematic illustration for the mechanism of intracellular DNA delivery with thiolated gelatin nanoparticles in the presence of higher glutathione (GSH) concentrations.
Gelatin nanoparticles, like many other conventional nanoparticulate systems, are predominantly engulfed by the cells of the reticuloendothelial system (RES) upon systemic administration. Surface modification of gelatin nanoparticles with hydrophilic polymers, such as poly(ethylene glycol) (PEG), affords long circulation times of these nanoparticles in vivo. The hydrophilic nature of these surface modified particles reduces their binding capacity to serum proteins (e.g., opsonins) resulting in reduced uptake by the reticuloendothelial system (RES) through steric repulsion by surface bound PEG chains 8, 9. When administered in the systemic circulation, PEG-modified nanoparticles are preferentially distributed to solid tumor due to the hyperpermeability of the angiogenic blood vessels by the enhanced permeability and retention effect 10–12.
In the present study, we have modified the thiolated gelatin nanoparticle surface with PEG in order to prolong the circulation time through evasion of RES uptake. PEG-modified thiolated gelatin nanoparticles, when administered in human breast cancer-bearing nude mice, show enhanced circulation time in plasma (t1/2 of >15 hours) and preferential tumor localization by the enhanced permeability and retention effect 12. Here we report PEG modification, encapsulation of reporter plasmid DNA (EGFP-N1), DNA stability, and in vitro transfection studies in NIH-3T3 murine fibroblast cells.
EXPERIMENTAL METHODS
Materials
Type-B gelatin (225 bloom strength) having 100–115 millimoles of carboxylic acid per 100 g of protein, an isoelectric point of 4.7–5.2 and an average molecular weight 40,000 – 50,000 daltons was purchased from Sigma-Aldrich (St. Louis, MO). Methoxy-PEG-succinimidyl glutarate of molecular weight of 2,000 Da was purchased from Sun-Bio, Inc. (Orinda, CA). Absolute ethyl alcohol (200 proof, 99.5% ACS reagent) was obtained from Acros Chemicals (Pittsburgh, PA). Glyoxal 40% (w/v) and glycine were purchased from Fisher Scientific (Fair Lawn, NJ). Protease, 2,4,6-trinitrobenzene sulfonic acid (TNBS) and salt of ethylene diamine tetraacetic acid were obtained from Sigma (St.Louis, MO). 0.8% agarose gels pre-stained with ethidium bromide from Invitrogen (Carlsbad, CA), Picogreen® from Molecular Probes (Eugene, OR) and DNAse-I from Takara Bio, Inc (Palo Alto, CA). The NIH3T3, mouse embryonic fibroblast cell line, was obtained from American Type Culture Collection (ATCC, Roockville, MD) and maintained in Dulbecco’s modified Eagle medium (DMEM) with 4 mM L-glutamine, 1.5 g/l sodium bicarbonate and 4.5 g/l glucose and supplemented with 10% fetal bovine serum and 1% each of penicillin-streptomycin, HEPES buffer, and sodium pyruvate. General purpose serum free medium (UltraCulture®) was obtained from BioWhittaker (Walkersville, MD). Lipofectin® reagent was obtained from Invitrogen (Carlsbad, CA). The plasmid DNA pEGFP-N1 was obtained from Clontech (Palo Alto, CA) All aqueous solutions and reagents were prepared in deionized distilled water from Nanopure II (Barnstead/Thermolyne, Dubuque, IA).
Synthesis of Thiolated Gelatin and Preparation of Nanoparticles
Thiolated gelatin was synthesized as reported previously 13. Briefly, thiolated gelatin was prepared by the covalent modification of the primary amino groups of type B gelatin by the addition of sulfahydryl moieties. For the synthesis, 1 g of gelatin was dissolved in 100 ml of deionized water and then incubated with varying amount of 2-iminothiolane hydrochloride, (up to 100 mg per gram) at room temperature for 15 hours. Any unreacted 2-iminothiolane was removed by repeated dialysis against 5 mM HCl, followed by 1 mM HCl solution for 24 hours each. The purified thiolated gelatin was dried in-vacuo and stored at −80°C for further use.
Based on initial cytotoxicity and transfection results 13, the nanoparticles for these studies were prepared with thiolated gelatin that was formed by reaction of 20 mg of 2-iminothiolane per gram of type-B gelatin, which has an average of 6.1 mM sulfahydryl groups equivalent per gram of the biopolymer. The nanoparticles were prepared with 1% (w/v) aqueous solution of thiolated gelatin in a temperature-controlled water bath at 37ºC. The pH of the resulting solution was adjusted to 7.0 with 0.2 M sodium hydroxide. The nanoparticles were formed when the solvent composition was changed from 100% water to 75% by volume of hydro-alcoholic solution upon gradual addition of absolute ethanol under continuous stirring conditions. The formed nanoparticles were further crosslinked with 0.1 ml of 40% (v/v) aqueous solution of glyoxal for the desired time interval and any unreacted aldehyde residues were quenched with 0.2 M glycine solution. The particles obtained were centrifuged at 16,000 rpm for 30 minutes and the pellet was washed twice with deionized distilled water. The purified nanoparticles were freeze-dried and stored at room temperature.
Surface Modification with PEG
The control gelatin (Gel) and thiolated gelatin (SHGel) nanoparticles collected after centrifugation were suspended in 0.1 M phosphate buffer (pH 7.4) and incubated with 5 times molar excess (2 mg of PEG per mg of gelatin or thiolated gelatin nanoparticles) of methoxy-PEG-succinimidyl glutarate (Mol. wt. 2,000 Da) for 2 hours at room temperature. At the end of the reaction, the nanoparticles were collected by centrifugation and assayed for the degree of PEG modification by using trinitrobenzene sulfonic acid (TNBS) assay 14. In the TNBS method, the number of free amino groups is estimated by a colorimetric reaction that results in the formation of a yellow-colored product, which shows maximum absorbance at 420 nm. The Gel and SHGel nanoparticles and their corresponding PEG conjugated analogs (i.e., PEG-Gel and (PEG-SHGel) were dispersed in pH 8.5 alkaline borate buffer and allowed to react with the TNBS reagent at room temperature. The reaction mixture was centrifuged at 5000 rpm for 5 min and the absorbance of the supernatant solution was measured at 420 nm using a Shimadzu UV160U spectrophotometer (Columbia, MD). Using this assay, the percentage of surface-accessible amine groups that could react with TNBS was calculated. Since methoxy PEG-succinimidyl glutarate reacts with the primary amine residues in Gel and SHGel nanoparticles, the degree of PEG modification was estimated indirectly based on differences between the absorbance values of the supernatant from the control and after PEG modification.
Characterization of PEG-Modified Nanoparticles
Particle Size Analysis
The mean particle sizes and size distribution of unmodified (i.e, Gel and SHGel) and the PEG-modified nanoparticles (i.e., PEG-Gel and PEG-SHGel) were analyzed using ZetaPALS, 90Plus (Brookhaven Instruments Corporation, Holtsville, NY). The colloidal suspension of the nanoparticles was diluted with deionized distilled water and the particle size analysis was carried out at a scattering angle of 90° and at 25°C.
Surface Charge Measurements
Zeta potential (ζ) measurements of the unmodified control and PEG modified gelatin-based nanoparticle systems in suspension in deionized water were performed using a ZetaPALS (Brookhaven Instruments Corporation, Holtsville, NY). The nanoparticles were dispersed in deionized water and the zeta potential values were measured at the default parameters of dielectric constant, refractive index, and viscosity of water. The average zeta potential values were computed based on Smoluchowski equation.
Electron Spectroscopy for Chemical Analysis (ESCA)
In order to determine the surface presence of PEG chains on the modified nanoparticles, freeze-dried formulations of the unmodified control and the PEG modified nanoparticles were analyzed by ESCA. ESCA (or X-ray photoelectron spectroscopy) determines the elemental and chemical composition of the surface at up to 10 nm distance. ESCA was performed at the National ESCA and Surface Analysis Center for Biomedical Problems (NESAC/BIO), University of Washington (Seattle, WA). The samples were placed in an ultrahigh vacuum environment and exposed to a low energy X-rays beam resulting in an emission of secondary photoelectrons from the surface of the nanoparticles. The energy of the electrons emitted is characteristic of the element and the molecular orbital from which it is emitted. Only those electrons that escape from the surface of the sample without loss of energy contribute to the signal peak. For each surface chemical element, a spectrum was obtained by plotting the number of detected electrons as a function of the binding energy (in millivolts). The surface elemental composition was determined using standard Scofield photoemission cross sections. High resolution analysis of the C1S spectra was performed to determine the exact chemical composition from the hydrocarbon (C-C or C-H at 285 mV), ether (C-O at 286.4 mV), and the carbonyl (C=O at 288.1 mV) envelops and the relative composition of each functionality was determined from the area under the peak. In PEG modified nanoparticles, the surface presence of ethylene oxide residues would translate into an increase in the ether signal in the C1S spectra 15.
Plasmid DNA (EGFP-N1) Encapsulation
The EGFP-N1 plasmid DNA upon transfection results in an expression of enhanced green fluorescent protein (GFP) in cells. The DNA encapsulated nanoparticles were prepared by using a 1% (w/v) solution of the gelatin or thiolated gelatin adjusted to pH 7.0, followed by the addition of plasmid DNA (EGFP-N1) at a final concentration of 0.5% (w/w) of the polymer and formed into nanoparticles by the desolvation process as described previously. The plasmid DNA-loaded nanoparticles were separated from the free plasmid DNA by a series of centrifugation and washing steps. The nanoparticles were re-dispered, diluted appropriately and the surface charge measured using ZetaPALS, 90Plus (Brookhaven Instruments Corporation, Holtsville, NY). The loading efficiency of plasmid DNA in the nanoparticle formulations was determined by dissolving the nanoparticle sample in protease (0.2 mg/ml)-containing PBS at 37°C. The plasmid DNA concentrations was estimated in solution using the PicoGreen® dsDNA reagent (Molecular Probes, Eugene, OR), a one step, sensitive, fluorescent, quantitation kit for double-stranded DNA. The reagent was incubated with the samples for 5 min and the fluorescence intensity was measured at 480 nm excitation and 520 nm emission wavelengths, using a Bio-Tek plate reader.
Glutathione-Responsive In Vitro DNA Release
The in vitro release of plasmid DNA from the unmodified control and PEG modified gelatin-based nanoparticles was performed in PBS containing different concentrations (i.e., 0 mM, 0.1 mM, and 5.0 mM) glutathione at 37°C. Twenty-mg of the EGFP-N1 plasmid DNA loaded nanoparticulate samples were weighed into microcentrifuge tubes and incubated with 1.5 ml of the respective buffer solutions. The samples were placed in a temperature controlled shaker 10,000 rpm for 5 minutes. Sink conditions were maintained by replacing an equal volume of the release medium each time. The plasmid DNA concentration in the release medium was measured using the Picogreen® assay described above. Percentage of encapsulated plasmid DNA released as a function of time was calculated from these data.
Stability of the Encapsulated DNA
The stability of the plasmid DNA encapsulated in the control and PEG-modified nanoparticles was performed in presence of polymer matrix degrading enzyme protease and an endonuclease DNAse-I that catalyzes the degradation of both single and double stranded DNA. The nanoparticles were treated with 0.2 mg/ml protease and 0.2 U (per batch of nanoparticles weighing 200mg) DNAse-I, for a period of 30 min and 20 min respectively at 37°C. Additionally, sequential addition of protease followed by DNAse and DNAse followed by protease treatments to confirm that the plasmid was physically encapsulated in the nanoparticles and not adsorbed to the surface. The activity of the DNAse was quenched by using a solution of EDTA which chelates the Ca+2 and Mg+2 ions that are required for the functioning of the enzyme. The resulting nanoparticulate solutions were diluted appropriately and mixed with 5-fold diluted gel loading buffer.
The samples were then loaded onto a 0.8% (w/v) agarose gel (E-Gel®, Invitrogen, Carlsbad, CA) pre-stained with ethidium bromide at an equivalent DNA concentration of 100 ng/well in a 15 μl volume per well. The gels were run at 65 V for 15 minutes using a Biorad™ model 200/2 power supply and the bands are visualized using Kodak Gel logic® 100 imaging system (Scientific Imaging, New Haven, CT).
In Vitro Transfection Studies
Qualitative Analysis of Transfection
NIH-3T3 murine fibroblast cells were grown in 6-well cell culture plates containing a Corning’s circular glass cover-slip with 2 X 105 cells seeded per well. The cells were grown to semi-confluence in an incubator at 37°C and 5% CO2 atmosphere. The plasmid DNA encapsulated, unmodified control and PEG-modified gelatin-based nanoparticle systems were dispersed in serum free medium. Twenty-μg of EGFP-N1 plasmid DNA complexed with 20 μl of Lipofectin®, a cationic lipid transfection reagent, was used as a positive control and untreated cells were used as a negative control. The DNA-containing nanoparticle suspension was added at a concentration equivalent to 20 μg of plasmid DNA per well and incubated with the cells at 37°C for a period of 6 hours. The serum free media was removed from each well and replaced with the DMEM. The cover slips were washed three times with sterile PBS at time intervals of 12, 24, 48 and 72 hours post-incubation and mounted on to microscopic slides containing a drop of fluorescence-free mounting medium (Fluoromount-G®, Southern Biotechnology Associates, Birmingham, AL). The expression of GFP in the cells was observed by a fluorescence microscope. Differential interference contrast (DIC) and fluorescence images were acquired using Olympus BX61 microscope and the digital images were processed with Adobe Photoshop software.
Quantitative Analysis of Transfection
NIH-3T3 cells were grown to semi-confluence in a 6 well cell culture plate, transfected with EGFP-N1 plasmid DNA encapsulated nanoparticulate systems of unmodified control and after PEG modification and compared with the Lipofectin®-plasmid DNA transfected cells and cells treated with serum free media alone. The nanoparticles carrying plasmid DNA and the Lipofectin®-pDNA mixture were dispersed in serum free media and added to each of the 6 wells at a concentration of 20 μg of plasmid DNA per well. The cells are allowed to incubate for a period of 6 hours at 37 °C in a 5 % CO2 incubator and then replaced by fresh growth medium. The adherent cells were trypsinized at 12, 24, and 48 and 96 hours post-incubation, centrifuged, and re-suspended in 0.5 ml of DMEM before fixing with 100 μl of 4% formalin buffer solution. The fluorescence of the GFP produced in the transfected cells was detected by a Beckman Coulter (Epics/Altra) (FACS) equipped with an argon 488 laser. The FL1 channel was used to detect the cells expressing GFP fluorescence and the results obtained were analyzed using Expo 32 software.
RESULTS AND DISCUSSION
Surface Modification with PEG
Methoxy PEG-succinimidyl glutarate is an amine reactive derivative of PEG which readily reacts with primary amino groups of gelatin and thiolated gelatin at room temperature and alkaline pH. There was an increase in PEG modification of the surface amino groups of the gelatin and thiolated gelatin nanoparticles with an increase in incubation time of methoxy PEG-succinimidyl glutarate for up to 2 hours. At the end of this time period, about 90% of the surface accessible amine groups were modified upon incubation with 5-times molar excess (i.e., 2 mg of PEG derivative per mg of gelatin or thiolated gelatin nanoparticles) of methoxy PEG-succinimidyl glutarate. This technique of post-PEG modification carried out after preparation of nanoparticles has been used to successfully modify the surface with PEG chains to achieve maximum stearic hindrance.
Characterization of PEG-Modified Nanoparticles
The mean particle diameters and surface charge values of the Gel, SHGel, PEG-Gel, and PEG-SHGel nanoparticles are shown in Table 1. For the controls Gel and SHGel nanoparticles, the average size was found to be in the range of 220 – 250 nm. However, owing to the surface modification, both of the PEG-modified nanoparticles were found to have a larger particle size ranging from 290 – 350 nm. The increase in size of the nanoparticles from an average of 200–250 nm to about 290–350 nm clearly indicates the addition of PEG molecules on the surface of the nanoparticles, which results in the change in hydrodynamic diameter of the system in aqueous environment. Even with PEG modification, the nanoparticles still have the size range below 400–600 nm that is necessary for accumulation in solid tumor by the enhanced permeability and retention effect 16–19.
Table 1.
Particle Size and Surface Charge of the Control and Poly(Ethylene Glycol)-Modified Nanoparticles*
| Nanoparticle Type | Empty Nanoparticles | DNA Encapsulated Nanoparticles | ||
|---|---|---|---|---|
| Size (nm) | Zeta Potential (mV) | Zeta Potential (mV) | ||
| Gelatin (Gel) | 230 ± 11.5 | − 8.45 ± 0.45 | − 9.01 ± 0.84 | |
| PEG- Modified Gelatin (PEG-Gel) | 329 ± 17.7 | − 7.72 ± 0.88 | − 6.89 ± 0.26 | |
| Thiolated Gelatin (SHGel) | 244 ± 6.8 | − 9.36 ± 0.64 | − 8.98 ± 0.52 | |
| PEG-Modified Thiolated Gelatin (PEG-SHGel) | 302 ± 9.2 | − 6.67 ± 0.77 | − 6.84 ± 0.14 | |
Mean ± S.D. (n = 5)
The controls Gel and SHGel nanoparticles were found to have average zeta potential values of −.8.45 mV and −9.36 mV, respectively. In comparison, the PEG-Gel and PEG-SHGel nanoparticles were found to have a surface charge of −7.72 and −6.67 mV respectively. Upon measurement of surface charge there was a slight decrease in the negative charge on the surface of PEG-modified nanoparticles, which could be due to the modification of the surface amino groups on the surface of gelatin and thiolated gelatin nanoparticles.
ESCA is a surface sensitive technique that exploits the photoelectric effect to obtain information about the chemical composition and structure of the surface layer up to 10 nm depth. It utilizes a source of X-rays to eject valence band electrons or photo electrons from the surface of the sample. The energy of the electrons emitted is characteristic of the element and the molecular orbital from which it is emitted. Only those electrons that escape from the surface of the sample without loss of energy contribute to the signal peak and will be analyzed for carbon-1s (C1s) envelopes. The results in Figure 2 shows the peak intensities of the species C-H (hydrocarbon), -C-O (ether), and C=O (carbonyl) obtained from C1S scan of the control and PEG-modified nanoparticles at their characteristic binding energies. The surface presence of PEG chains in the PEG-modified nanoparticles was confirmed by the increase in the relative peak intensities of ether linkage from the high resolution spectra. The peak intensities of ether linkage in case of PEG-modified gelatin (PEG-Gel) were found to be 79.2% and PEG-modified thiolated gelatin (PEG-SHGel) nanoparticles resulted in 82.6%. In comparison, the gelatin and thiolated gelatin nanoparticles have shown ether peak intensities of 36% and 28.4% respectively.
Figure 2.
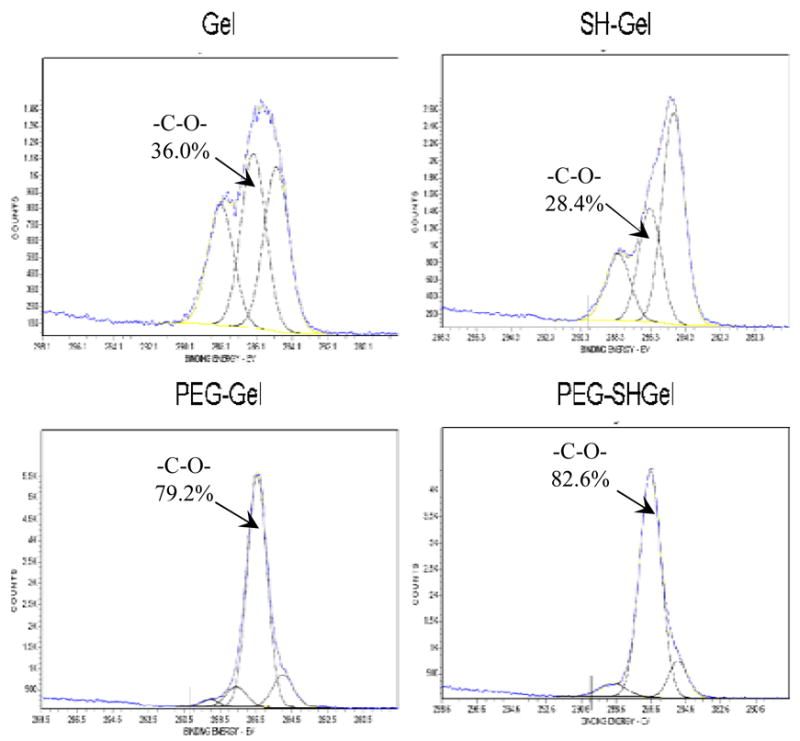
High resolution C1s scans of electron spectroscopy for chemical analysis for the surfaces of gelatin (Gel), thiolated gelatin (SHGel), PEG-modified gelatin (PEG-Gel) and PEG-modified thiolated gelatin nanoparticles (PEG-SHGel). The scans show the peak intensities of the species C-H (hydrocarbon), C-O (ether), and C=O (carbonyl) groups at 285.0, 286.3 and 288.1 eV respectively. The percentage of C-O peak area for each spectra is provided.
The loading capacity and efficiency of plasmid DNA in the nanoparticle formulations was determined from a standard curve of plasmid DNA in protease-containing PBS. The results of the plasmid DNA capacity and efficiency are been reported in Figure 3. The control Gel and SHGel nanoparticles were found to have a DNA loading efficiency (at 0.5% w/w) of 99.5% and 94.4%, respectively. In comparison, upon PEG-modification, the DNA loading efficiency was 98.3% for PEG-Gel nanoparticles and 91.7% for PEG-SHGel nanoparticles.
Figure 3.
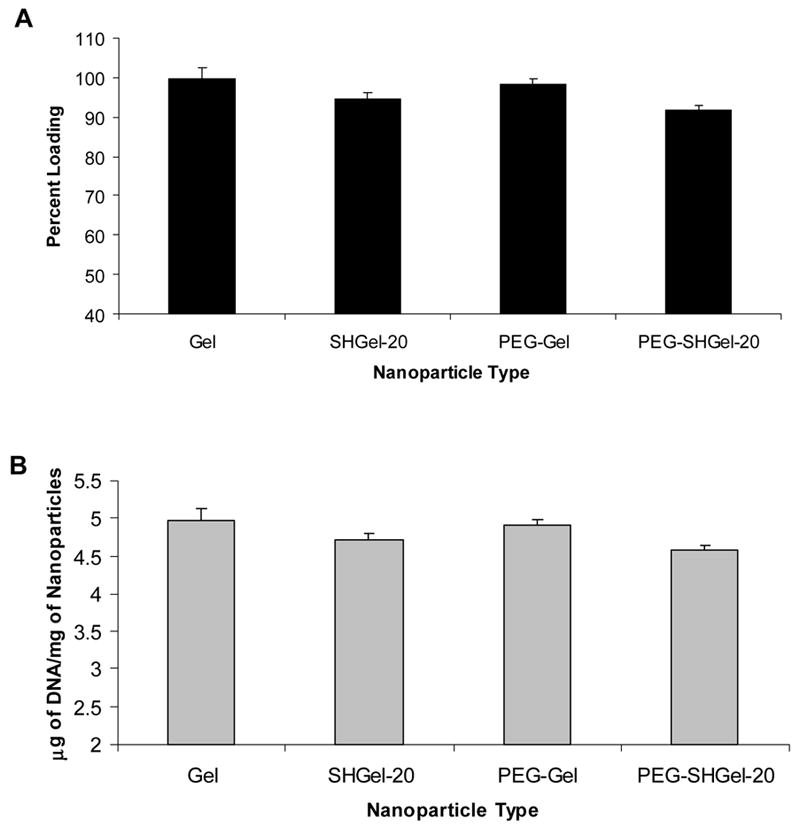
The encapsulation efficiency (A) and capacity (B) of EGFP-N1 plasmid DNA in gelatin (Gel), thiolated gelatin (SHGel-20), poly(ethylene glycol)-modified gelatin (PEG-Gel), and poly(ethylene glycol)-modified thiolated gelatin (PEG-SHGel) nanoparticles. Each of the plasmid DNA containing formulation was prepared with 0.5% (w/w) DNA. The loading efficiency and capacity was measured by treating each of the DNA-containing nanoparticle formulations in 0.2 mg/ml protease-containing phosphate-buffered saline (pH 7.4) to degrade the biopolymer matrix.
With almost 91 – 99 % loading efficiency of the added DNA, both the control and the PEG-modified nanoparticles were proved to have good encapsulation efficiency. The slight decrease in DNA loading in SHGel and PEG-SHGel nanoparticles could be due to the presence of the free sulfahydryl residues or formed disulfide bonds that were found to affect DNA encapsulation. The results also indicate that PEG modification of gelatin and thiolated gelatin had resulted in a slight decrease in the loading capacity of the nanoparticulate systems, which could be attributed to the leaching of DNA from the nanoparticles after repeated processing steps involved in post modification of the of the nanoparticulate systems with PEG derivative. In our earlier studies, almost 100% of the DNA was encapsulated when we formed the PEG-modified gelatin nanoparticles using pre-modified gelatin derivative20, 21.
Glutathione-Responsive In Vitro DNA Release Studies
The release of plasmid DNA from the control and PEG-modified nanoparticles in presence of different glutathione concentrations is plotted as a function of time and is shown in Figure 4. The in vitro release profiles in PBS at 37°C show a greater fraction of the DNA being released from PEG-modified nanoparticles (40–45%) when compared to the unmodified nanoparticles (25–30%). In the presence of glutathione (0.1 mM and 5 mM) the release of plasmid DNA from the thiolated nanoparticles, both modified and unmodified has been enhanced with almost 60% of the encapsulated payload being released over a period of 5h in case of PEG-modified thiolated gelatin nanoparticles. On increasing the GSH concentration from 0 to 5mM in the release medium, it resulted in an enhanced release of the DNA from the thiolated nanoparticles indicating the reducing action of the enzyme on the disulfide bonds formed within the nanoparticles.
Figure 4.
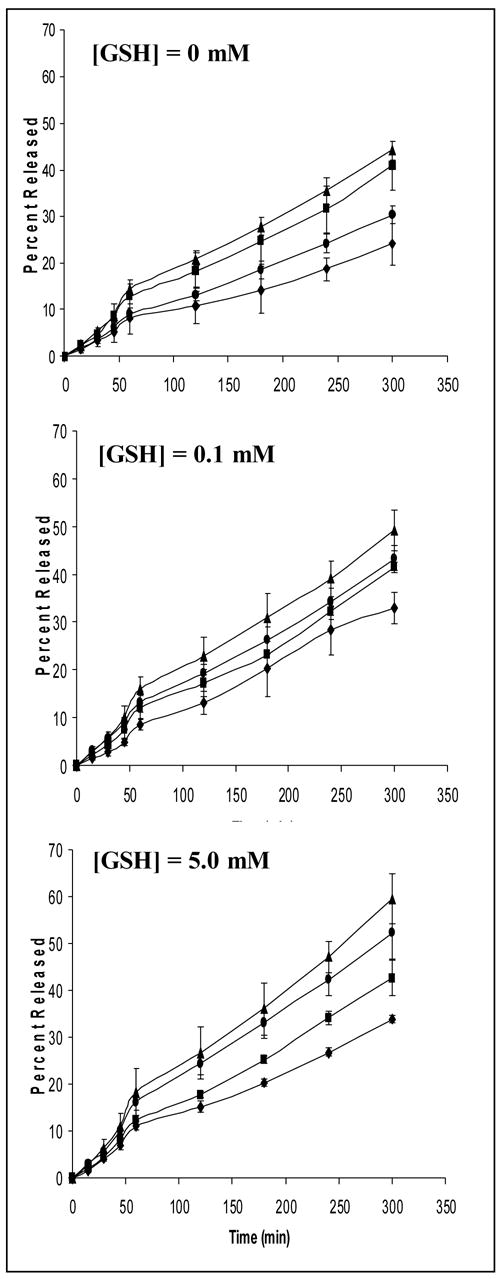
In vitro release profiles of EGFP-N1 plasmid DNA from gelatin (Gel) (◆), thiolated gelatin (SHGel) (●),poly(ethylene glycol)-modified gelatin (PEG-Gel) (■), and poly(ethylene glycol)-modified thiolated gelatin (PEG-SHGel) (▲) nanoparticles in phosphate-buffered saline (PBS, pH 7.4) at 37°C containing 0 mM (PBS control), 0.1 mM glutathione, and 5.0 mM glutathione (GSH) concentrations. The plasmid DNA concentration in the release medium at each time point was measured by fluorescent PicoGreen® dsDNA assay.
In addition, we have also examined the release of plasmid DNA in the absence and presence of protease at 2 mg/ml concentration in PBS to show the effect of PEG surface modification on the degradation and release of gelatin and thiolated gelatin matrix (results not shown). These results showed the release of entire payload within 2 h in case gelatin and thiolated gelatin nanoparticles, while only 80% of the encapsulated pDNA has been released from the PEG-modified nanoparticles. The decrease in the release of the plasmid DNA from the PEG-modified nanoparticles was probably due to steric repulsion of the PEG chains which prevents the access of the enzyme to the nanoparticles.
Stability of Encapsulated DNA
The results of agarose gel electrophoresis (Figure 5) show DNA ladder in lane 1 and intact EGFP-N1 plasmid DNA in lane 2. The plasmid DNA when treated with protease is not affected and the intact bands of the DNA are shown in lane 3. This was followed by plasmid DNA treated with DNAse-I in lane 4 that resulted in extensive fragmentation of DNA and similar results were seen in case of plasmid DNA treated with protease and DNAse-I in lane 5. Plasmid DNA encapsulated nanoparticulate formulations subjected to different treatments were loaded in lanes 6–9. The nanoparticles treated with protease were loaded in lane 6 and shows intact plasmid DNA that has been released upon degradation of the biopolymer by the proteolytic activity of the enzyme. This was followed by formulations that were treated by DNAse-I as shown in lane 7, which did not affect the DNA as it was completely encapsulated in the polymer matrix. Nanoparticles treated with protease followed by DNAse-I resulted in extensive fragmentation of the DNA (lane 8). However, upon changing the order of treatment (i.e., DNAse-I followed by protease), it resulted in the release of intact plasmid DNA and is shown in lane 9. Exactly the same gel eletrophoresis profiles were observed for all four types of nanoparticulate systems: Gel, SHGel, PEG-Gel, and PEG-SHGel systems.
Figure 5.
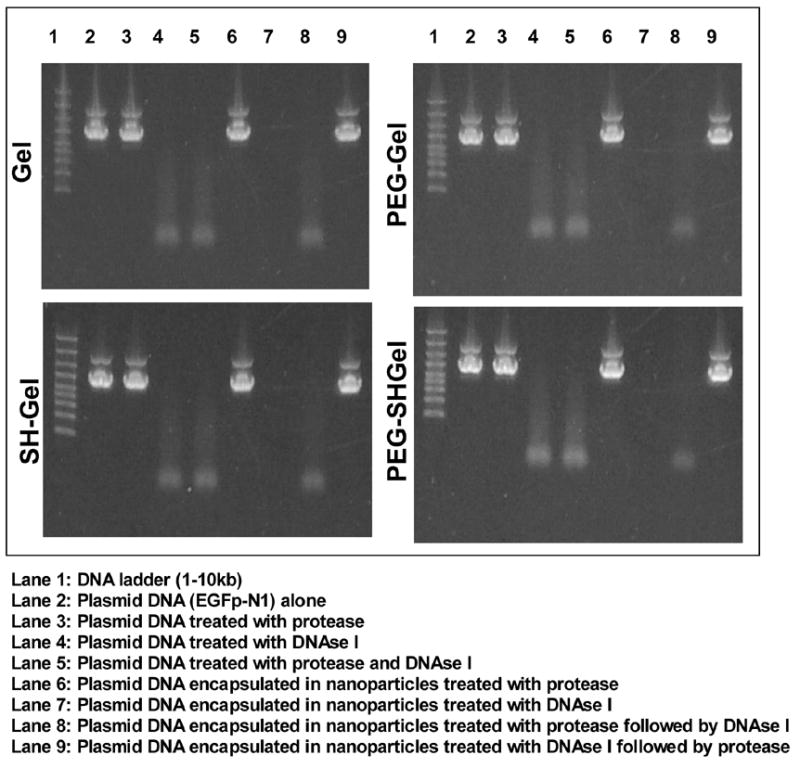
Agarose gel eletrophoresis to show stability of EGFP-N1 plasmid DNA encapsulated in gelatin (Gel), thiolated gelatin (SH-Gel), poly(ethylene glycol)-modified gelatin (PEG-Gel), and poly(ethylene glycol)-modified thiolated gelatin (PEG-SHGel) nanoparticles. The nanoparticles were treated with 0.2 mg/ml of protease or 0.2 units (per batch of nanoparticles) of DNAse-I. The sequence of enzyme addition was either protease followed by DNAse to degrade the matrix and release the plasmid or DNAse followed by protease to show that the plasmid was physically encapsulated in the matrix and did not undergo degradation.
These results unequivocally indicate that the method of preparation, nanoparticle crosslinking, surface PEG modification, and freeze-drying did not have any detectable negative effect on the DNA encapsulated in the control and PEG-modified nanoparticles. Throughout these experiments, the plasmid DNA was seen running at two molecular weight ranges indicating the super coiled state of the DNA and the possibility of some of the plasmid DNA being nicked. More importantly, the fraction of the supercoiled DNA remained the same in the different nanoparticle system.
In Vitro Transfection Studies
Qualitative analysis of transfection of NIH3T3 cells by plasmid encapsulated nanoparticles was carried out by fluorescence microscopy. The Figure 6 shows the DIC and fluorescence images of cells transfected with Lipofectin®-DNA complexes, PEG-Gel, and PEG-SHGel nanoparticles at 48 hours post-transfection. While the DIC images show intact cells, the fluorescent images show that the cells transfected with nanoparticles encapsulated with plasmid DNA resulted in stable expression of the green fluorescent protein. These results clearly show that PEG-modification has resulted in stabilization of the thiolated gelatin nanoparticle at the same time without any compromise on the transfection ability of the nanoparticles. The use of PEG-chains of lower molecular weight (2,000 Da) and post-modification technique on the surface of the nanoparticles has in fact resulted in an enhanced stability of the particle and ultimately an effective escape of DNA from the endosome. The consequence of which has resulted in improved transfection efficiency of the PEG-SHGel nanoparticles in cell culture in vitro.
Figure 6.
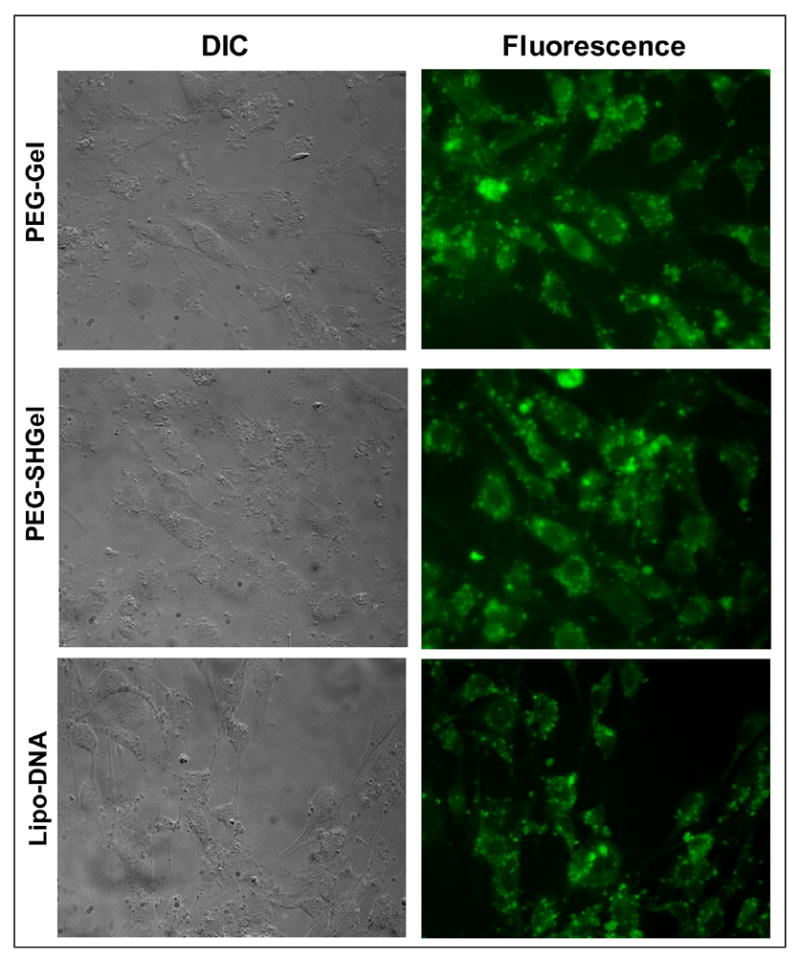
Qualitative evaluation of enhanced green fluorescent protein expression following transfection with the EGFP-N1 plasmid DNA encapsulated nanoparticles in NIH-3T3 murine fibroblast cells. Differential interference contrast (DIC) and fluorescence microscopy images of cells at 48 hours post-transfection with DNA encapsulated poly(ethylene glycol)-modified gelatin (PEG-Gel) and poly(ethylene glycol)-modified thiolated gelatin (PEG-SHGel) nanoparticles along with Lipofectin®-plasmid DNA complexes used as a positive control.
The quantitative determination of transfection efficiency of the plasmid encapsulated nanoparticles in NIH3T3 cells was done by FACS analysis. The Figure 7 shows transfection results at various time points for gelatin (Gel), thiolated gelatin (SH-Gel) and their respective PEG-modified nanoparticulate systems (PEG-Gel and PEG-SHGel) encapsulated with plasmid DNA. The nanoparticles were compared with Lipofectin-Plasmid DNA complexes by plotting percentage transfected cells as a function of time. The scatter plots of the transfected and normal cells represented in the form of pixels in regions A1 to A4 and transfected cells expressed as a percentage of the total cells present in quadrants A3 and A4. When compared to Lipofectin®-Plasmid DNA complexed and other nanoparticulate formulations the PEG-SHGel nanoparticles resulted in highest expression of the GFP protein with almost 33% of the cells being transfected at the end of 72 hours. As observed in the previous study, thiolation has resulted in rapid release of the DNA payload in the cells with reducing environment such as that of glutathione. In addition the PEG-modification on the surface of the nanoparticles has resulted in greater stability of the particle during the process of endocytosis, resulting in enhanced protection of the plasmid during intracellular transport. Lastly, the control and PEG-modified gelatin-based nanoparticulate system maintains the supercoiled structure of the plasmid. We believe that the supercoiled structure of the plasmid is essential for nuclear import in non-dividing cells. The lower transfection efficiency observed with cationic lipids and polymers may be due to the decomplexation that needs to occur at the peri-nuclear region, which can change the plasmid structure from supercoiled to open circular. Open circular plasmids have a size of about 100 nm, while the nuclear pores in non-dividing cells are only 9–10 nm in diameter.
Figure 7.
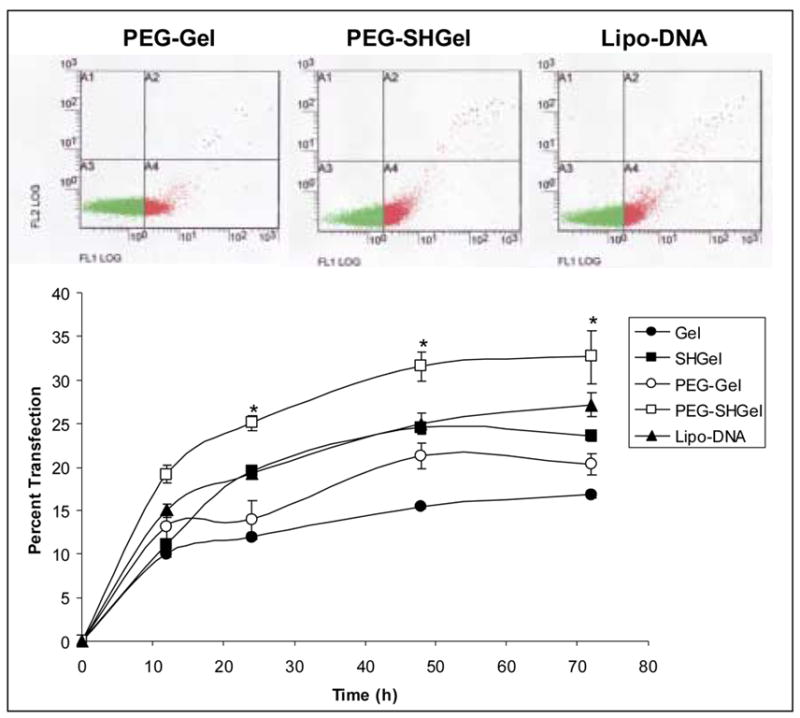
Quantitative transfection profiles of nanoparticles encapsulated with EGFP-N1 plasmid DNA in NIH-3T3 murine fibroblast cells. The percentage of cells transfected was plotted as a function of time post-administration of the plasmid in the gelatin (Gel), thiolated gelatin (SHGel), poly(ethylene glycol)-modified gelatin (PEG-Gel) and poly(ethylene glycol)-modified thiolated gelatin (PEG-SHGel) nanoparticle formulations. Lipofectin®-complexed DNA and naked DNA were used as controls. The top panels show the scatter plot results from flow cytometry, with the cells expressing the fluorescent protein following transfection represented in red and the non-expressing cells in green. (* Statisically significant difference (p<0.05) between the transfection efficiency with PEG-SHGel as compared to Gel and SH-Gel nanoparticles).
CONCLUSIONS
The thiolated gelatin nanoparticles have been successfully modified with the hydrophilic polymer PEG and the nanoparticles had a mean size of 290–350 nm. The PEG-modification on the surface of the nanoparticles was confirmed by increase in relative peak intensity of the ether peaks by ESCA. The presence of PEG chains on the surface of the nanoparticles was found to resist digestion by proteolytic enzymes. The absence of degradation of the encapsulated DNA when treated with protease alone or DNase-I or DNase-I followed by protease as seen by gel electrophoresis indicate the enhanced stability of the plasmid DNA encapsulated in the control and PEG-modified nanoparticles. In addition, the in vitro transfection studies prove the ability of PEG-SHGel nanoparticles to transfect cells in vitro and could potentially be used as vectors for gene delivery in vivo.
Acknowledgments
This study was supported by a grant RO1-CA095522 from the National Cancer Institute of the National Institutes of Health. The flow cytometry studies were performed at the Forsyth Dental Institute (Boston, MA). We deeply appreciate the assistance of Dr. Jean Eastcott with the flow cytometric experiments and results interpretation. Additionally, Dr. Lara Gamble’s help with the ESCA investigations at the NESAC/BIO (Seattle, WA) is gratefully acknowledged. NESAC/BIO is supported by the National Institutes of Health grant EB-002027.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Labhasetwar V. Nanotechnology for drug and gene therapy: the importance of understanding molecular mechanisms of delivery. Curr Opin Biotechnol. 2005;16(6):674–80. doi: 10.1016/j.copbio.2005.10.009. [DOI] [PubMed] [Google Scholar]
- 2.Read ML, Logan A, Seymour LW. Barriers to gene delivery using synthetic vectors. Adv Genet. 2005;53:19–46. [PubMed] [Google Scholar]
- 3.Lehrman S. Virus treatment questioned after gene therapy death. Nature. 1999;401(6753):517–8. doi: 10.1038/43977. [DOI] [PubMed] [Google Scholar]
- 4.Marshall E. Gene therapy. Second child in French trial is found to have leukemia. Science. 2003;299(5605):320. doi: 10.1126/science.299.5605.320. [DOI] [PubMed] [Google Scholar]
- 5.Thomas CE, Ehrhardt A, Kay MA. Progress and problems with the use of viral vectors for gene therapy. Nat Rev Genet. 2003;4(5):346–58. doi: 10.1038/nrg1066. [DOI] [PubMed] [Google Scholar]
- 6.Blessing T, Remy JS, Behr JP. Monomolecular collapse of plasmid DNA into stable virus-like particles. Proc Natl Acad Sci U S A. 1998;95(4):1427–31. doi: 10.1073/pnas.95.4.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Han S, Mahato RI, Sung YK, Kim SW. Development of biomaterials for gene therapy. Mol Ther. 2000;2(4):302–17. doi: 10.1006/mthe.2000.0142. [DOI] [PubMed] [Google Scholar]
- 8.Naper DH. Polymeric Stabilization of Colloidal Dispersions. Academic Press; New York: 1983. [Google Scholar]
- 9.Kommareddy S, Tiwari SB, Amiji MM. Long-circulating polymeric nanovectors for tumor-selective gene delivery. Technol Cancer Res Treat. 2005;4(6):615–26. doi: 10.1177/153303460500400605. [DOI] [PubMed] [Google Scholar]
- 10.Kaul G, Amiji M. Biodistribution and targeting potential of poly(ethylene glycol)-modified gelatin nanoparticles in subcutaneous murine tumor model. J Drug Target. 2004;12(9–10):585–91. doi: 10.1080/10611860400013451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kaul G, Amiji M. Tumor-targeted gene delivery using poly(ethylene glycol)-modified gelatin nanoparticles: in vitro and in vivo studies. Pharm Res. 2005;22(6):951–61. doi: 10.1007/s11095-005-4590-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kommareddy S, Amiji M. Biodistribution and pharmacokinetic analysis of long-circulating thiolated gelatin nanoparticles following systemic administration in breast cancer-bearing mice. J Pharm Sci. 2006 doi: 10.1002/jps.20813. (In press) [DOI] [PubMed] [Google Scholar]
- 13.Kommareddy S, Amiji M. Preparation and evaluation of thiol-modified gelatin nanoparticles for intracellular DNA delivery in response to glutathione. Bioconjug Chem. 2005;16(6):1423–32. doi: 10.1021/bc050146t. [DOI] [PubMed] [Google Scholar]
- 14.Snyder SL, Sobocinski PZ. An improved 2,4,6-trinitrobenzenesulfonic acid method for the determination of amines. Anal Biochem. 1975;64(1):284–8. doi: 10.1016/0003-2697(75)90431-5. [DOI] [PubMed] [Google Scholar]
- 15.Sherwood MA. Data analysis in X-ray photoelectron spectroscopy. In: Briggs D, Seah MP, editors. Practical surface analysis. Vol. 2. John Wiley and Sons; New York: 1990. pp. 555–586. [Google Scholar]
- 16.Yuan F, Leunig M, Huang SK, Berk DA, Papahadjopoulos D, Jain RK. Microvascular permeability and interstitial penetration of sterically stabilized (stealth) liposomes in a human tumor xenograft. Cancer Res. 1994;54(13):3352–6. [PubMed] [Google Scholar]
- 17.Yuan F, Salehi HA, Boucher Y, Vasthare US, Tuma RF, Jain RK. Vascular permeability and microcirculation of gliomas and mammary carcinomas transplanted in rat and mouse cranial windows. Cancer Res. 1994;54(17):4564–8. [PubMed] [Google Scholar]
- 18.Jain RK. Physiological barriers to delivery of monoclonal antibodies and other macromolecules in tumors. Cancer Res. 1990;50(3 Suppl):814s–819s. [PubMed] [Google Scholar]
- 19.Jang SH, Wientjes MG, Lu D, Au JL. Drug delivery and transport to solid tumors. Pharm Res. 2003;20(9):1337–50. doi: 10.1023/a:1025785505977. [DOI] [PubMed] [Google Scholar]
- 20.Kaul G, Amiji M. Long-circulating poly(ethylene glycol)-modified gelatin nanoparticles for intracellular delivery. Pharm Res. 2002;19(7):1062–8. doi: 10.1023/a:1016486910719. [DOI] [PubMed] [Google Scholar]
- 21.Kaul G, Amiji M. Cellular interactions and in vitro DNA transfection studies with poly(ethylene glycol)-modified gelatin nanoparticles. J Pharm Sci. 2004;94(1):184–198. doi: 10.1002/jps.20216. [DOI] [PubMed] [Google Scholar]


