Summary
E. coli DNA topoisomerase III belongs to the type IA family of DNA topoisomerases, which transiently cleave single-stranded DNA (ssDNA) via a 5′ phosphotyrosine intermediate. We have solved crystal structures of wild-type E. coli topoisomerase III bound to an 8-base ssDNA molecule in three different pH environments. The structures reveal the enzyme in three distinct conformational states while bound to DNA. One conformation resembles the one observed previously with a DNA-bound, catalytically inactive mutant of topoisomerase III where DNA binding realigns catalytic residues to form a functional active site. Another conformation represents a novel intermediate in which DNA is bound along the ssDNA-binding groove but does not enter the active site, which remains in a catalytically inactive, closed state. A third conformation shows an intermediate state where the enzyme is still in a closed state, but the ssDNA is starting to invade the active site. For the first time, the active site region in the presence of both the catalytic tyrosine and ssDNA substrate is revealed for a type IA DNA topoisomerase, although there is no evidence of ssDNA cleavage. Comparative analysis of the various conformational states suggests a sequence of domain movements undertaken by the enzyme upon substrate binding.
Introduction
DNA topoisomerases perform a fundamental role in the cell by regulating DNA topology during a number of metabolic processes, including DNA replication, transcription, recombination, and chromosome condensation 1–3. Topoisomerase-mediated transformation of DNA involves the transient cleavage of single-stranded (ssDNA) or double-stranded DNA (dsDNA), the passage of DNA through the resulting break, and the subsequent rejoining of the broken phosphodiester backbone. Topoisomerases use this basic reaction mechanism to catalyze a variety of complex DNA rearrangements, including the supercoiling and relaxation of DNA, and the catenation and decatenation of DNA molecules.
While all known DNA topoisomerases function through the formation of a covalent phosphotyrosine intermediate, several different subfamilies have evolved that differ in sequence, structure, and mechanism. Type I topoisomerases cleave a single strand of DNA while type II topoisomerases cleave both strands of a DNA duplex concurrently. Members of the type I family of enzymes are classified further as type IA topoisomerases, which form a 5′ phosphotyrosine linkage, or type IB topoisomerases, which function via a 3′ phosphotyrosine intermediate. Additionally, an archaeal type I enzyme, topoisomerase V, appears to define a new type I subfamily 4. Type IA topoisomerases, exemplified by Escherichia coli DNA topoisomerase I and III, differ structurally and mechanistically from type IB enzymes, such as human topoisomerase I 1. Type II enzymes bear no sequence similarity to type I topoisomerases, yet structural similarities observed between several type IA and type II topoisomerases suggest possible mechanistic similarities between the two families 1.
Crystal structures of several type IA enzymes in the absence of DNA have been determined, including a 67 kDa N-terminal fragment of E. coli topoisomerase I 5, E. coli topoisomerase III 6, Thermotoga maritima topoisomerase I 7, and Archaeoglobus fulgidus reverse gyrase 8. The structures reveal that the core of the enzyme, which participates in the cleavage and strand passage events, adopts an overall four-domain, toroidal fold enclosing a central hole large enough to accommodate DNA. The active site of the apo enzyme is buried at the interface between two domains and is closed off to DNA. In the absence of DNA, the catalytic tyrosine participates in a hydrogen bond network with conserved residues from an adjacent domain, thereby remaining inaccessible to DNA. The variable C-terminal region, shown to be involved in DNA binding 9,10, may be responsible in defining functional specificity of each enzyme.
There are only two structures of complexes of a type IA enzyme with DNA. The crystal structure of a catalytically inactive mutant of E. coli topoisomerase III (Y328F) complexed to an 8-base ssDNA molecule illustrated the conformational changes undertaken by the enzyme to accommodate its scissile ssDNA substrate 11. In the structure of the noncovalent complex, the oligonucleotide binds along a groove that leads into the active site and is specific for ssDNA. DNA binding is accompanied by a domain reorientation resulting in the assembly of a catalytically competent active site where the Y328F substitution is ideally poised above the scissile phosphodiester bond and other conserved residues have realigned in preparation for catalysis. The proposed structure-based mechanism suggests that a conserved arginine (Arg330) and lysine (Lys8) serve to stabilize the phosphotyrosine transition state while an invariant glutamate (Glu7) is positioned to donate a proton to the 3′ leaving oxygen to facilitate cleavage. The second structure, a structure of a complex of wild-type E. coli topoisomerase I and ssDNA 12 reveals a different conformational stage. In this structure, the enzyme remains closed and the active site is not formed, but part of the oligonucleotide is bound in the groove leading to the active site. A single helix forming part of this groove is displaced yet the rest of the enzyme maintains an overall apo-like conformation. The conformational stage observed appears to be an early intermediate just before the enzyme undergoes large domain rearrangements to open the active site and fully accommodate the ssDNA substrate.
While the structures of the E. coli topoisomerase III (Y328F)+ssDNA complex and E. coli topoisomerase I+ssDNA complex provided valuable insights into the pre-cleavage state of the enzyme, several questions remain to be addressed. The mechanism underlying activation of the catalytic tyrosine remains unclear. It is possible that the active site configuration seen in the structure of the DNA-bound topoisomerase III mutant does not accurately reflect the reorganization that occurs in the case of the active enzyme when the catalytic tyrosine is present. Furthermore, since the type IA topoisomerase reaction cycle proceeds through several different protein-DNA conformational intermediates, the capture of other enzyme-substrate intermediates, including a covalent enzyme-substrate complex containing the phosphotyrosine intermediate, is needed for a comprehensive understanding of the topoisomerase mechanism. We report here several crystal structures, ranging from 2.35 to 2.5 Å resolution, of the catalytically active form of E. coli topoisomerase III in complex with an 8-base ssDNA molecule at pH values ranging from 5.5 to 8. The structures reveal new enzyme+ssDNA conformational intermediates while further establishing key mechanistic aspects of the current type IA topoisomerase model for catalysis.
Results
Structure determination and overall structure
Initial crystals were obtained of wild-type E. coli topoisomerase III complexed to the same 8-base oligonucleotide used previously in structural studies with the E. coli topoisomerase III (Y328F) mutant 11. The DNA sequence used for crystallization (5′-CGCAACT↑T-3′, where the arrow denotes the preferred site of cleavage) contains an asymmetric cleavage site recognized by E. coli topoisomerase III 13. Surprisingly, the wild type enzyme+ssDNA complex crystallized under the same conditions that produced crystals of the E. coli topoisomerase III (Y328F)+ssDNA complex yet the new crystals grow in a different crystal form. In the new crystals, there are two molecules in the asymmetric unit almost related by a two-fold axis (~178°).
Interestingly, we discovered that the conformational state of each complex in the asymmetric unit depended on the cryoprotectant used prior to data collection. For crystals that had been cryoprotected in 25% glucose (Form I), the resulting structure shows both complexes in the same conformation. In this case, each enzyme molecule adopts a closed conformation with the domain I–III interface found in an apo-like state such that the active site remains inaccessible to DNA (Figure 1A). The ssDNA occupies the binding groove with 5′ to 3′ polarity towards the active site, but only 6 out of the 8 nucleotides are bound. This conformation reflects a novel pre-cleavage intermediate in which the ssDNA molecule binds along the groove but does not yet enter the active site and the active site is still not fully formed. However, when glycerol is used as the cryoprotectant (Form II) at a pH at or below 7, both enzyme molecules in the asymmetric unit are found in different conformations, with each one deviating from the apo-form of E. coli topoisomerase III. Whereas one complex remains in the closed conformation, the other switches to an open conformation where DNA binding results in the assembly of a functional active site (Figure 1B). As seen in the topoisomerase III (Y328F)+ssDNA complex, conformational changes induced by substrate binding in the open complex allow the oligonucleotide to fully occupy the ssDNA binding cleft and enter the realigned active site. Finally, crystals soaked in 25% glycerol and at pH 8.0 resulted in a third structure revealing one molecule in the open conformation with the ssDNA fully bound and the other molecule in an intermediate conformation, where the enzyme is mostly closed, the ssDNA is also in an intermediate conformation between the closed and open forms, and a helix forming part of the groove has shifted. This conformation appears to be an intermediate between the closed and the open conformations (Figure 1C).
Figure 1. Overall structure of the open and closed complexes.
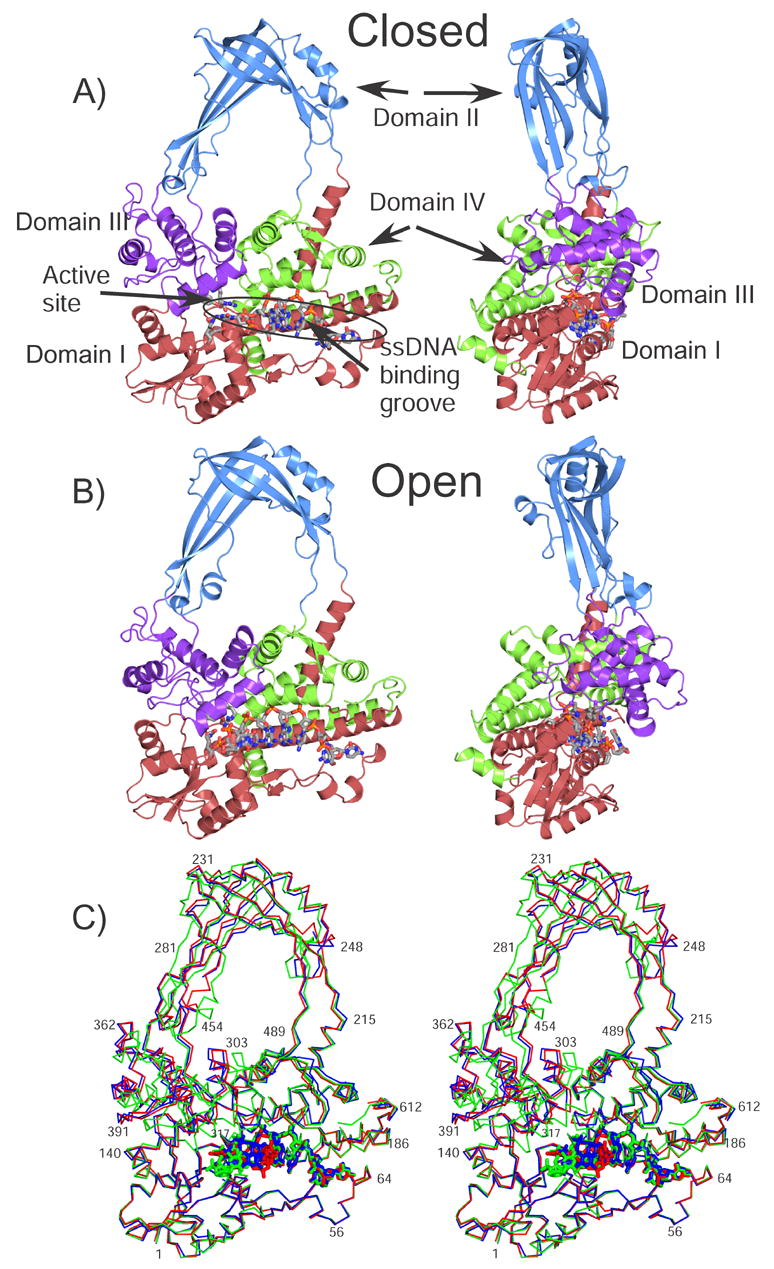
A) The diagram shows a schematic representation of the closed complex (Form I, pH 5.5). The four major domains of the protein are colored red, blue, purple, and green for Domain I, II, III, and IV. The active site is found at the intersection of Domains I and III. The ssDNA binding groove extends from Domain IV to the active site. The ssDNA in the complex is shown in a ball and stick representation. B) Schematic diagram of the open complex (Form II, pH 5.5), colored as in the previous diagram. C) Stereo view showing the superposition of the closed (red), intermediate (blue), and open (green) complexes. The structures were aligned by superposing Domain I only. The three structures correspond to Form I pH5.5, Form II pH 8.0, and Form II, pH 5.5.
The use of glycerol or glucose as a cryoprotectant leads to a small change in the c axis, which increases from ~444 Å to ~453 Å. Analysis of the crystal packing shows that Domain II, forming the top of the molecule, forms few crystal packing contacts and is in a position to move. For this reason, it is not surprising that the molecules in the crystal can change conformation in order to form the open conformation. The quality of the electron density in Domain II is significantly worse than in the rest of the molecule, suggesting that this domain is more mobile. In particular, the temperature factors in Domain II in the A monomers (closed monomers) are much larger than for the entire molecule (average value of 74.9 Å2 vs. 47.3 Å2, 76.7 Å2 vs. 55.8 Å2, 92.3 Å2 vs. 69.8 Å2, 83.9 Å2 vs. 52.0 Å2, and 97.5 Å2 vs. 64.4 Å2 for the glycerol forms at pH 5.5, 7.0 and 8.0 and glucose forms at pH 5.5 and 7.0 respectively). This monomer is also the one that can change conformation from the closed to the intermediate form at pH8.0. In contrast, electron density around the active site and most of the DNA molecule was clear and allowed for unambiguous placing of the side chains (Figure 2).
Figure 2. Difference map around the ssDNA in the intermediate form.
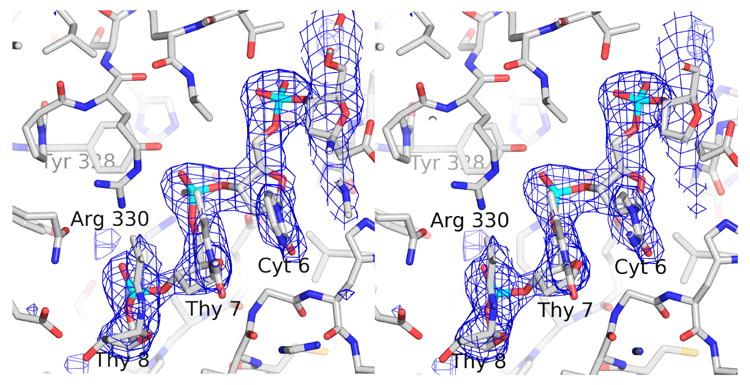
The difference map was calculated in the absence of the ssDNA. The coordinates of the protein in the final, refined Form I, pH 8 closed form structure were randomly changed by 0.3 Å r.m.s.d. and then refined in the absence of the ssDNA using Refmac5 24. The difference map was calculated using Refmac5 and it is shown at the 3.0 σ level.
In the open complex, the catalytic tyrosine (Y328) is found perfectly poised for cleavage with the tyrosine OH only ~3.2 Å away from the scissile phosphate. Nonetheless, well-defined electron density in the active site region in all the different experiments indicates that the catalytic tyrosine is not involved in a covalent linkage with its substrate in any of the complexes. No evidence was found to suggest that DNA cleavage has occurred. A probable explanation is that the low pH of the crystallization environment (pH 5.5) inhibits the wild-type enzyme from cleaving its substrate. For this reason, crystals were soaked in high pH solutions that could be more conducive to cleavage in an attempt at inducing DNA cleavage within the crystalline state. Nevertheless, DNA cleavage was not observed in any of the structures. Type IA topoisomerases require magnesium for activity 14,15. Based on structural 5 and biochemical 16 observations in E. coli topoisomerase I, the magnesium binding site has been suggested to include three highly conserved acidic residues, Asp111, Asp113, and Glu115 that are found in the vicinity of the active site. The equivalent residues are Asp103, Asp105, and Glu107 in topoisomerase III. Despite the presence of magnesium in the crystal soaking solution, there is no evidence of a metal ion in the resulting structures near the active site or at any other location. At pH 7.0, the overall conformation of each molecule in the asymmetric unit remains the same as that observed at pH 5.5. In the pH 8 structure, one of the molecules is in the open conformation and the other in the intermediate conformation, but no cleavage is observed. It is likely that the conditions used for crystallization shift the equilibrium towards the religation reaction and not towards the cleavage reaction. Alternatively, cleavage must be concomitant with an even larger conformational change that cannot be accommodated in the crystalline lattice.
Topoisomerase III+ssDNA complex: closed conformation
The relative domain arrangement of topoisomerase III in the closed complex resembles that seen in the apo form of the enzyme. In all, there are seven different closed structures: one of the molecules in the asymmetric unit in each of the five different conditions (Form I-pH 5.5 and 7.0 and Form II-pH 5.5, 7.0, and 8.0), and two additional closed complexes in the glucose or Form I crystals. Superposition of the closed structures with the structure of the apo enzyme yields a root mean square deviation (r.m.s.d.) for all main chain atoms (residues 1–617) ranging between 1.5 to 2.4 Å. Superposition of each domain in the apo form and in the closed complexes results in r.m.s.d. values ranging from 0.87 to 1.0 Å, 0.66 to 0.93 Å, 0.42 to 0.45 Å, and 1.74 to 3.1 Å for domains I, II, III, and IV, respectively. Thus, rearrangements within domain IV have occurred in order to accommodate the ssDNA molecule along the binding groove (Figures 1C and 3). There is a slight rotation of domains II and III away from domain I (~5°), but this shift is not sufficient to allow DNA to enter the active site. Unambiguous electron density is seen for six nucleotides, which are positioned along the DNA binding cleft leading up to the active site. A seventh nucleotide was built at the 3′ end, but the density is not as well-defined as for the other 6 ones. Cyt1 at the 5′ end is found interacting with the 5′ end of its symmetry related counterpart. In addition to base interactions, two chloride ions found at the interface between the 5′ ends of the ssDNA molecules contribute to crystal packing. Five nucleotides are positioned within the DNA binding groove such that the phosphate groups are buried within the protein and the bases are either oriented towards the bottom of the groove or outwards from the protein. Cyt3, Ade4, and Ade5 are involved in stacking interactions with one another. Cyt6 at the 3′ end of the oligonucleotide is found in a flipped conformation with respect to the fully formed complex. There is disconnected electron density extending from the 3′ end of the oligonucleotide, which seems to correspond to the final two nucleotides. Strong difference density indicative of a planar group found adjacent to the base of Cyt6 suggests that the 3′ end of the oligonucleotide Thy7 stacks against Cyt6. It is possible that the enzyme cleaved the oligonucleotide, thus explaining why only 6 nucleotides can be seen clearly. However, this does not explain the residual density that extends from the 3′ end, and furthermore, the electron density agrees well with the sequence of the intact oligonucleotide and no other density is present that could correspond to a cleaved dinucleotide product. In general, many of the conserved residues involved in DNA interactions in the topoisomerase III-Y328F+ssDNA structure are also observed coordinating the DNA in the closed conformation. The enzyme only makes direct contacts to four phosphates of the oligonucleotide compared with contacts with all seven phosphates in the fully formed complex. Protein-DNA interactions at the 5′ end of the oligonucleotide are nearly identical in the intermediate complex and the topoisomerase III-Y328F-DNA complex. Interactions at the 3′ end involve the same residues in both complexes but are shifted slightly since the DNA is not fully bound along the groove and within the active site in the closed complex.
Figure 3. Domain IV rearranges to accommodate the ssDNA.
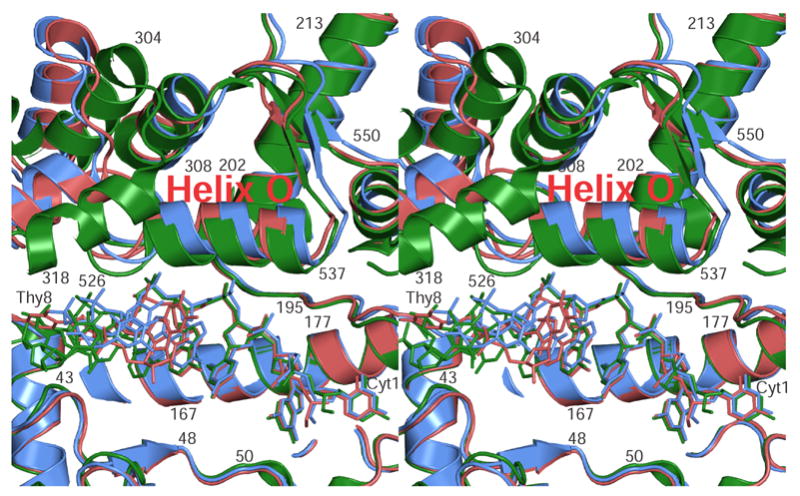
The stereo diagram shows a superposition of domain IV in the closed, intermediate, and open forms (red, blue, and green respectively). The ssDNA for each form is shown in the same coloring scheme. The diagram illustrates that helix O, lying just above the ssDNA, is found in a different conformation in each of the different forms. This helix appears to be responsible for the triggering of the larger domain changes that lead from the closed to the open forms. As the helix moves, the ssDNA moves as well and starts entering the active site. The helices on the top left part of the diagram form part of Domain III, which has already moved in the open form (green). Overall, the only major change observed in Domains I and IV among the three forms is the movement of helix O.
The active site environment of the closed conformation is very similar to that observed in the apo-form (Figure 4B). The phosphate group belonging to Cyt6 is the closest one to Tyr328 but it is still ~7.7 Å away from the hydroxyl group of the catalytic tyrosine. The hydrogen bonding network creating the domain I–III interface, as seen in the apo-structure, is maintained in this complex. In comparison to the orientation of active site residues in the apo structure, Arg330 and Lys8 have adjusted their conformations in order to interact with Cyt6. Arg330 coordinates the phosphate group of Cyt6. Additionally, the guanidinium group of Arg330 lies parallel against the cytosine ring of Cyt6 while Lys8 and Asp130 interact with the cytosine ring thus serving to stabilize the base in a flipped conformation.
Figure 4. Assembly of the active site and entry of the ssDNA.
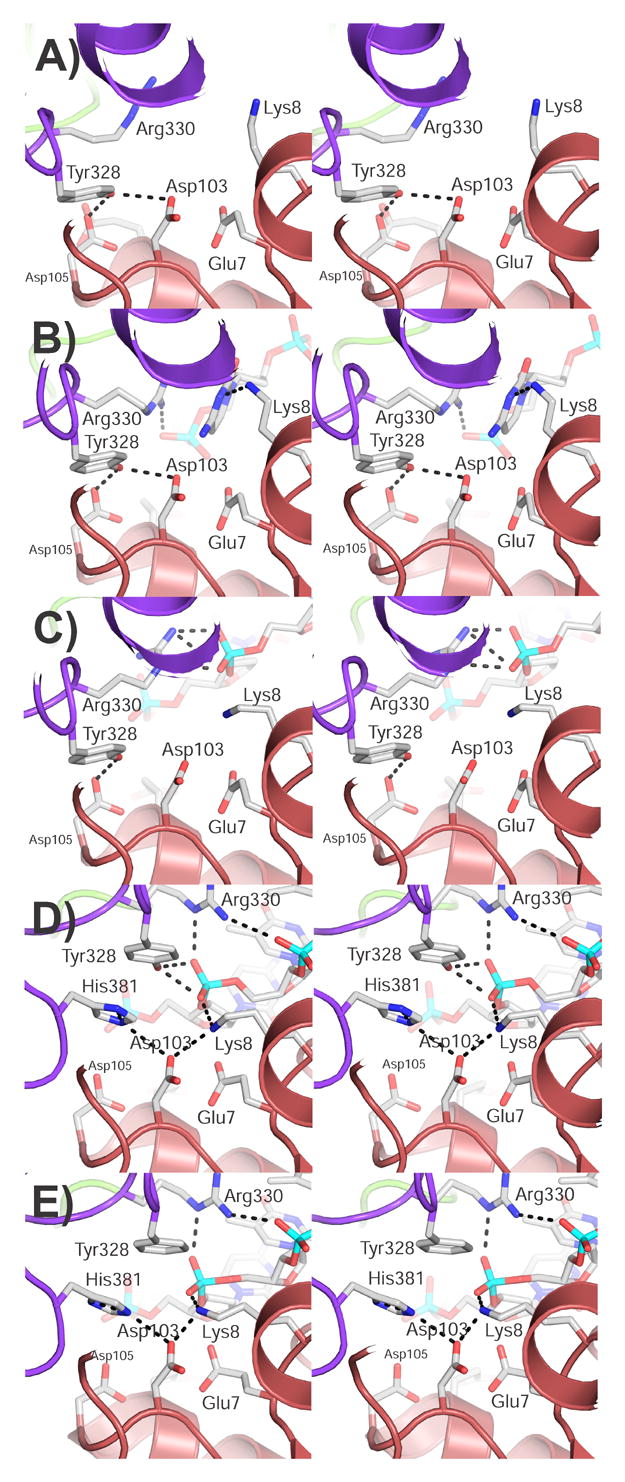
The stereo diagrams show the active site region in the different forms whose structure is known. A) Apo structure, B) closed structure, C) intermediate structure, D), wild-type open form, and E) Y328F mutant open form. The diagrams illustrate the way the active site is assembled and the interactions with ssDNA. As the protein changes conformation from the closed to the open form, the ssDNA approaches the active site and makes different interactions until it is in its final position. Domain I is shown in red and domain III in purple. Some of the side chains forming the active site are shown as well as some possible hydrogen bonds. The ssDNA is shown as sticks.
Topoisomerase III+ssDNA: open conformation
In the open complex, the enzyme has undergone a significant conformational change to allow complete binding of the oligonucleotide resulting in a structure similar to the one of the topoisomerase III-Y328F+ssDNA complex. There are three different open complex structures, all in glycerol conditions. The r.m.s.d. for all main chain atoms between the open complex and the topoisomerase III-Y328F+ssDNA complex range from 1.3–1.4 Å, emphasizing that all structures are basically identical. Individual superposition of each domain in the open complex with the apo structure results in r.m.s.d. values ranging from 0.91 to 0.94 Å, 0.56 to 0.63 Å, 0.49 to 0.51 Å, and 2.99 Å for domains I, II, III, and IV, respectively. Again, a rearrangement of domain IV relative to the apo-form has occurred in order to accommodate ssDNA along the DNA binding groove (Figure 1C and 3). The r.m.s.d. difference for domain IV goes down to 0.39 – 0.45 Å when the comparison is made between the open and topoisomerase III-Y328F+ssDNA complex structures. In contrast to the closed complex, domains II and III in the open complex have also rotated ~20° thereby making the active site at the interface of domains I and III accessible to the 3′ end of the oligonucleotide. The relative orientation of the four domains in the complex is comparable to that observed in the topoisomerase III-Y328F+ssDNA complex.
As a result of these conformational differences, the ssDNA is not as deeply buried in the active site of the open complexes as it is in the topoisomerase III-Y328F+ssDNA complex. The slight displacement of active site residues observed in the open complex relative to that in the topoisomerase III-Y328F+ssDNA complex could also be caused in part by the presence of the hydroxyl group of the catalytic tyrosine in the wild type structure. Nevertheless, the active site configuration in the fully bound wild type topoisomerase III+ssDNA complex is nearly identical to the active site environment in the topoisomerase III-Y328F+ssDNA complex (Figure 4D,E). Tyr328 is poised for nucleophilic attack on the phosphate moiety of Thy7 with the hydroxyl group of Tyr328 located ~3.2 Å away from the phosphorous atom of Thy7. Arg330 and Lys8 coordinate the phosphate of the putative scissile bond thus likely serving to stabilize the negative charge developing on the transition state intermediate during cleavage. Glu7, which forms a clear, ~2.8 Å hydrogen bond in the topoisomerase III-Y328F+ssDNA complex, forms what appears to be a long hydrogen bond to the bridging 3′ oxygen of the scissile bond (~3.5 Å) and is also part of a hydrogen bonding network with Asp103 and His381.
Superposition of the bound ssDNA molecules in the open conformation and the mutant topoisomerase III complexes yields an average r.m.s.d. of 0.62 Å (all atoms) thus showing that the overall DNA conformation does not change (Figure 5). Enzyme-DNA interactions along the binding groove in the fully bound wild type-DNA complex are analogous to those seen in the topoisomerase III-Y328F-DNA structure. In general, the structural similarities between the wild type and catalytically inactive forms of the DNA-bound enzyme in two different crystal forms corroborate the functional relevance of these conformational states, which are unlikely to be dictated exclusively by crystal packing interactions.
Figure 5. Conformation of the ssDNA in the closed, intermediate, and closed forms.
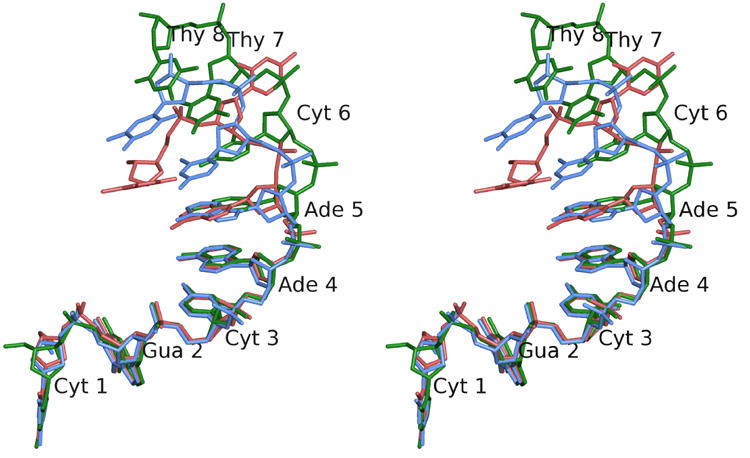
Stereo diagram of the ssDNA in the closed (red), intermediate (blue), and open (green) conformations. The diagram illustrates the differences in conformation of the ssDNA as it enters the active site. The first five nucleotides remain essentially unchanged, while nucleotides 6 and 7 change conformation to approach the active site. Nucleotide 8 is ordered only in the open conformation. Note the base of Thy 7, which is in a completely different conformation in the closed form. As the structure changes from the closed to the open forms, the phosphate of Thy 7, the scissile phosphate, approaches the active site. Nucleotides are numbered from the 5′ to the 3′ end starting at Cyt 1.
Topoisomerase III+ssDNA: intermediate conformation
At a pH below 8, crystals cryo-protected with glycerol show one monomer in the closed conformation and one in the open conformation. When the crystals are moved to pH 8, there is a change in the structure of the closed complex. The structure of the open complex remains the same as at lower pH, but the closed one shows two major changes: i) the ssDNA moves closer to the active site while the base seen flipped in the closed complex changes to a conformation more similar to the one in the fully-formed complex, and ii) a helix (helix O) forming part of the ssDNA binding groove shifts position and is found in a conformation intermediate between the closed and fully-formed conformations. In the intermediate complex the relative domain arrangement is also in an intermediate state between the apo form and the fully formed complex. Superposition of the intermediate structure with the structure of the apo enzyme yields an r.m.s.d. of 2.4 Å for all main chain atoms, which is the largest difference for all the structures in the closed conformation. Pairwise superposition of each domain in the apo form and in the intermediate complex results in r.m.s.d. values of 1.0, 0.92, 0.45, and 3.0 Å for domains I, II, III, and IV, respectively. When domains I and IV from the Y328F and intermediate complexes are superposed simultaneously, the r.m.s.d. is only 0.8 Å, showing that these domains are oriented in the same manner. In contrast, when domains I and IV in the apo form and the intermediate complex are compared, the r.m.s.d. is 2.8 Å. These observations indicate that domains I and IV rearrange in the intermediate complex to a relative orientation closer to the one observed in the Y328F complex. A careful comparison shows that the one major difference between the closed and intermediate conformations is found in the ssDNA binding groove. Helix O (residues 525 to 539) shifts in the intermediate form to a position in between the one observed in the open and closed forms (Figure 3). Thus, this helix appears to adopt an intermediate conformation as well. No other major changes are seen in the structure, aside from a difference in the overall position of Domain II, which appears to be directly related to the changes in the helix. Interestingly, the corresponding helix in E. coli topoisomerase I was also observed to change conformation upon DNA binding, although in this case the helix did not adopt a single conformation 12.
In the intermediate complex, electron density is seen for 7 nucleotides. The DNA is almost in the same conformation as in the closed conformation and the five nucleotides positioned in the ssDNA binding groove (nucleotides 1–5) are essentially identical in the topoisomerase III-Y328F+ssDNA complex and in the intermediate conformation (Figure 5). Near the active site there are significant changes. The phosphodiester backbone of the last two ordered nucleotides moves towards the protein and the base of Cyt6 is not flipped out, but it faces the protein and is stacked in between the adjacent bases. While the phosphate adjacent to the scissile bond is only 3.5 Å away from the OH of Y328 in the open complexes and 7.7 Å away in the closed form, in the intermediate conformation it is 5.5 Å away. Thus, this complex appears to place this region of the DNA also in an intermediate conformation, not quite ready for cleavage but much nearer than in the closed form.
Overall, the active site resembles the one in the apo form, although there are notable differences (Figure 4C). The hydrogen bonding network creating the domain I–III interface, as seen in the apo-structure, is largely maintained in this complex. In comparison to the orientation of active site residues in the apo structure, Arg330 and Lys8 have adjusted their conformation. Arg330 forms an ionic interaction with the phosphate group of Thy7 and Lys8 faces this phosphate as well.
Discussion
Domain changes triggered by ssDNA binding
There are now structures for four distinct complexes between topoisomerase III and DNA: the wild type enzyme in the closed, intermediate and open forms, and the topoisomerase III-Y328F+ssDNA complex. In addition, the structure of the apo form shows the conformation in the absence of DNA. Analysis of the different conformational states of the enzyme indicates that the domain movements are complex and do not originate at a single hinge point. The crystal structure of Topoisomerase III-Y328F complexed with ssDNA suggested that protein-DNA interactions at the base of a helix (residues 198 – 215 ) leading from domain I into domain II (helix G following the E. coli topoisomerase I nomenclature 5 and corresponding to residues 196 – 213 in topoisomerase I) help trigger the large-scale movement of domains II and III. In the closed complex, protein-DNA interactions formed at the base of helix G are identical to those seen in the topoisomerase III-Y328F+ssDNA complex. However, the resulting displacement of helix G does not produce a complete rotation of domains II and III. Further movement of domains II and III in the complex is hindered partly due to crystal packing.
Structural comparison of all available topoisomerase III models highlights the movement of another helix spanning residues 525 – 539 (helix O whose equivalent in E. coli topoisomerase I spans residues 497 – 509) in domain IV that may be a contributing factor in eliciting the overall conformational change in the enzyme. Helix O forms part of a helix-turn-helix motif that has been identified in type IA and type II topoisomerases and is proposed to be involved in binding DNA 17. In the presence of ssDNA along the binding groove, helix O aligns itself in a parallel manner along the phosphodiester backbone. Several residues in helix O, such as Thr524, Thr527, and Arg538 (Thr496, Ser499, and Arg507 in E. coli topoisomerase I), coordinate phosphate groups. Positioning of helix O along the bound ssDNA molecule likely promotes a more stable association of DNA within the binding cleft. Comparison of the closed, intermediate, and fully bound forms of wild type topoisomerase III reveals a progressive displacement of helix O upon ssDNA binding. There is also evidence showing movement of the equivalent helix in topoisomerase I when ssDNA is present in the binding cleft 12. In topoisomerase III, the progressive movement of helix O seems to be coupled to the rotation of domains II and III away from the base of the enzyme. Moreover, the shift in helix O effects other changes within domain IV, including a displacement of the decatenation loop. Rearrangement of structural elements within domain IV are linked to the concerted movement of domains II and III away from domain I. Taken together, these observations suggest that protein-DNA interactions along the binding groove result in local changes within domain IV, which lead into the more global movement of domains II and III required to expose the active site.
Active site assembly
The available structures of topoisomerase III present snapshots of a type IA topoisomerase along the catalytic reaction cycle and illustrate how the DNA enters the protein and starts forming the catalytically competent active site (Figure 4). As the DNA approaches the active site, there are several rearrangements of the protein side chains. In addition, as domain III moves to form the active site, side chains that were distant are now positioned to interact with other conserved residues for catalysis. The initial and final stages of the sequence are the apo-form, with no DNA around, and the final complex with the DNA positioned in the fully formed active site. The closed and intermediate complexes appear to correspond to states where the DNA is entering the active site and changing conformation. The open form is basically identical to the fully formed complex.
In the apo form, the active site tyrosine, Tyr328 (Tyr319 in E. coli topoisomerase I), forms hydrogen bonds to two highly conserved acidic residues, Asp103 and Asp105 (Asp 111 and Asp 113 in E. coli topoisomerase I) (Figure 4A). A third completely conserved acidic residue, Glu7 (Glu9 in E. coli topoisomerase I), is too distant to interact with the tyrosine. Arg330 and Lys8 (Arg321 and Lys13 in E. coli topoisomerase I), two basic residues that interact with DNA, flank the tyrosine. In the apo conformation, domain III is positioned in a manner that allows the interaction of the catalytic tyrosine with the acidic residues. In the fully formed complex, domain III has moved relative to domain I and the interaction between the two acidic residues and Tyr328 (Tyr319 in E. coli topoisomerase I) is broken. One of the acidic residues, Asp103 (Asp111 in E. coli topoisomerase I), interacts with His381 (His365 in E. coli topoisomerase I), which was too distant before and is brought in close proximity by the movement of the domains. The equivalent histidine has been studied extensively in E. coli topoisomerase I 18 and is known to be responsible for the pH dependence of the topoisomerization reaction. The active site tyrosine is positioned close to the scissile phosphate and interacting with the non-bridging oxygens. The two conserved basic residues form clear ionic interactions with the phosphate. Arg330 (Arg 321 in E. coli topoisomerase I) forms a salt bridge to the scissile phosphate and to the phosphate adjacent to it. Lys8 (Lys13 in E. coli topoisomerase I) also interacts with the scissile phosphate. Finally, Glu7 (Glu9 in E. coli topoisomerase I), which in the apo form is far from the active site tyrosine, interacts with the 3′ bridging oxygen. Thus, the phosphate and the 3′ bridging oxygen that form the scissile bond are in contact with protein residues.
The closed form shows an intermediate where the protein has not undergone any major conformational changes, but the presence of the DNA near the active site drives the rearrangement of some side chains. The phosphate backbone of the DNA is too far to interact directly with most of the residues forming the active site except for Arg330 (Arg321 in E. coli topoisomerase I). This residue stretches out to form a ionic pair with the phosphate group adjacent to the scissile phosphate. Interestingly, the base of this nucleotide is flipped and enters the active site. The flipping has a marked consequence: while the scissile phosphate is far too distant to interact with any region of the protein, the base can approach the protein and form contacts with Lys8 and Asp103 (Lys13 and Asp111 in E. coli topoisomerase I). These contacts are clearly base-specific and may not be formed when a different base is at that position. Given that type IA topoisomerases have a very weak sequence preference 13, it is unlikely that these interactions are conserved.
The intermediate form is more similar in overall structure to the apo-form but, as was mentioned before, one important helix has changed to an intermediate conformation. The active site is still not fully formed, but the DNA has changed conformation as it begins to approach the active site. The base that was flipped in the open form is not flipped in this conformation and this allows the phosphate backbone to come closer to the active site residues. The scissile phosphate is now making contacts with Arg330 (Arg321 in E. coli topoisomerase I), which changes conformation again. The arginine now forms an ionic interaction with the scissile phosphate. The other important basic residue, Lys8 (Lys13 in E. coli topoisomerase I), is oriented in a manner appropriate for interaction with the phosphate, but the DNA is not close enough to allow this. Finally, the tyrosine is still too distant to interact with the DNA as domain III has not moved to bring the tyrosine closer to the DNA. It is interesting to note that Arg330 (Arg321 in E. coli topoisomerase I) already contacts the group it has to interact in the active form, but that the movement of domain III will bring it to a position where it can approach two phosphates and not just one.
The open form is almost identical to the one observed in the topoisomerase III-Y328F mutant. Nevertheless, the presence of the catalytic tyrosine instead of the phenylalanine has two consequences. The scissile phosphate is ideally positioned for attack by the tyrosine with the two basic residues which may be involved in transition state charge stabilization already in position. The only group that is not forming a close enough contact appears to be Glu7 (Glu9 in E. coli topoisomerase I), which in the wild-type complex is at ~3.5 Å from the 3′ bridging oxygen. In the topoisomerase III-Y328F+ssDNA complex, this glutamate is clearly interacting with the 3′ bridging oxygen (~2.8 Å). The longer distance may simply reflect the inherent ambiguities in a 2.5 Å structure, but the quality of the map in this region suggests that this difference is real. It is more likely to reflect the impossibility of forming the final complex without leading to cleavage and the expected concomitant conformational change. Thus, the open complex may reflect a conformation that is not the final conformation, but rather a conformation that is just one step away from the chemically active one.
Comparison of the complexes provides structural evidence in support of a minimal sequence length of 7 bases required for cleavage, as previously determined biochemically 13. In the closed complex, the enzyme forms direct contacts with only 4 phosphates whereas the enzyme in the open complex directly interacts with 6 phosphates. The structures suggest that the enzyme must directly contact at least 6 phosphates (7 bases) of an ssDNA region to produce the domain realignment required to position the scissile bond for catalysis.
The structures so far appear to show a sequence of steps in the catalytic cycle (Figure 6). Starting from the apo structure, the DNA enters the binding groove and approaches the active site, but is unable to interact with the active site residues as Domain III is still blocking the active site. A further change in Domain IV, the movement of helix O, allows the DNA to approach the active site and align the DNA with some of the key basic side chains (Arg330 and Lys8; Arg321 and Lys13 in E. coli topoisomerase I). A further change in conformation, which dislocates Domain III from Domain I, exposes the active site and allows the DNA to fully enter and form the final catalytically competent complex. It is expected that once the DNA is properly positioned, cleavage can occur and a further, much larger conformational change ensues. In the structures, the crystal lattice probably impedes the final conformational change and hence precludes cleavage. The intermediate conformation may reflect this by capturing a complex that is slightly too “loose” and is not yet poised for cleavage.
Figure 6. Diagram illustrating the proposed mechanism of DNA relaxation catalyzed by type IA topoisomerases.
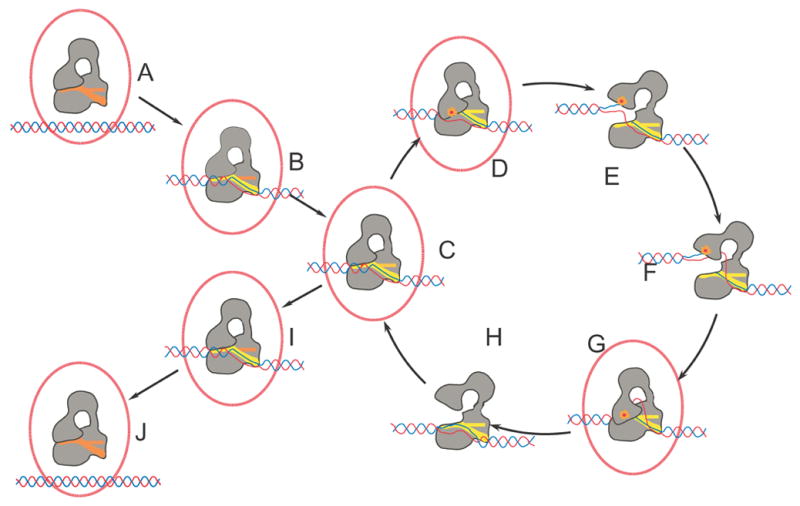
The proposed mechanism involves several transient conformational intermediates both of the protein and the DNA. The sequence of the steps is hypothetical. It is very likely that more states are involved in the cycle. Processivity by the enzyme requires that after one relaxation event the protein continues to another relaxation cycle without releasing the DNA. The selection of states B and I as outside the main cycle is arbitrary. In the diagram, the protein is shown in grey and the DNA in red/blue. The proposed intermediates whose structures are known are surrounded by a red ellipse. The orange dot represents the presence of the covalent protein/DNA complex, which at this stage is hypothetical and has not been observed. The ssDNA binding groove is shown in red or yellow. Helix O, which is found in Domain IV and is part of the binding groove, is shown as a horizontal bar. The color of the binding groove and helix O reflects its conformation. It is red in the closed form, orange in the intermediate form, and yellow in the open form.
Entrance to the cycle
A. Enzyme and supercoiled DNA prior to catalysis. The enzyme is in the closed conformation as found in the apo structure of E. coli DNA topoisomerase III 6.
B. The ssDNA starts invading the ssDNA binding groove and Domain IV in the protein changes conformation to accept the ssDNA. This state is illustrated by the closed form described here.
Processive cycle
C. The ssDNA approaches the active site concomitant with the movement of helix O. The overall structure of the protein resembles the closed form, but the ssDNA and helix O have changed conformation and the scissile phosphate approaches the active site. This state is illustrated by the intermediate form described here.
D. Entrance of the ssDNA into the protein triggers a conformational change to create a catalytically competent active site, as observed in the structure of the complex of topoisomerase III-Y328F with ssDNA 11 and the open form described here.
E. The enzyme opens, bridges the gap between the broken ends of the cleaved DNA, and allows passage of the other strand between the separated ends and into the central hole of the protein. No structures are known of this state, but the structure of a 30 kDa fragment of E. coli topoisomerase I comprising Domains II and III 29 may illustrate the extent of the conformational changes.
F. Enzyme-catalyzed strand passage of the second strand of DNA into the central hole of the enzyme. No structure is known of this intermediate.
G. Following strand passage, the enzyme closes, trapping the passing DNA strand inside the protein. Once the gate is closed, the cleaved strand is religated by the enzyme. The complex of topoisomerase III-Y328F with ssDNA 11 and the open form described here illustrate this state.
H. The cycle is completed by the enzyme opening to release both the religated strand and the one that was passed through the gap. No structure is known of this intermediate.
During the cycle, one DNA strand has passed through the other to change the topology of the DNA and alter the linking number by 1. At this point, the enzyme can continue in the cycle and perform another relaxation event or exit the cycle and release the DNA. The reaction always proceeds toward relaxation of the DNA.
Exit of the cycle
I. The ssDNA starts exiting the ssDNA binding groove and Domain IV in the protein changes conformation to expel the ssDNA. This state is illustrated by the closed form described here.
J. Upon complete release of the DNA, the enzyme returns to the apo conformation 6. The DNA has been relaxed by 1 during each round of the cycle.
Overall, all the structures of topoisomerase I and III alone and in complex with DNA are starting to present a detailed picture of the catalytic cycle of this family of enzymes. A major difference among members of the type IA family is the presence of different types of DNA binding domains at the C-terminus 9,10. Although the role of this domain is still not completely defined, it is clear that it does not play a central role in the reaction, as evidenced by the fact that it can be removed in some instances without total loss of activity and without affecting the requirements for positioning the ssDNA in the active site 9. The structures described here show that some of the movements are subtle and involve relatively small rearrangements of the domains of the enzyme or even of particular secondary structure elements. Sensing the negatively supercoiled state of DNA is thought to involve recognizing single stranded DNA regions. The structures show how the sensing and subsequent changes occur in steps: first the protein recognizes the presence of ssDNA as it enters the single stranded DNA binding region and once ssDNA is accommodated the active site is formed. This ensures that the active site is only formed when the correct type of DNA has been recognized. The synergy between ssDNA binding and domain rearrangements show how a binding event away from the active site triggers a large movement of domains that lead to the formation of the catalytically competent active site and positioning of the ssDNA in it. Surprisingly, the structures suggest that domain rearrangements occur not in one large step, but instead through a series of small and discrete changes that lead from one conformation to another. This ordered and well-defined sequence of events may not only occur in type IA topoisomerases, but also in other complex molecular machines.
Materials and Methods
Crystallization and data collection
Intact wild-type E. coli topoisomerase III (residues 1-653) containing a C-terminal hexa-histidine tag was expressed in BL21 cells and purified as described previously 11. The oligonucleotide used for crystallization (5′-CGCAACTT-3′) was synthesized by Integrated DNA Technologies and purified by reverse phase HPLC 19. The topoisomerase III-DNA complex was prepared by incubating the wild-type enzyme (3.5–5 mg/ml) and ssDNA (15–30 mg/ml) at a 1:2 molar ratio of protein:DNA at 4°C for 30 minutes in 50 mM Tris-HCl (pH 8.0), 0.2 M NaCl, 1 mM DTT, and 1 mM EDTA. Co-crystals appeared at 22°C in hanging drops equilibrated against 1.5–1.6 M (NH4)2SO4, 0.1 M sodium citrate (pH 5.5), and 0.5 M NaCl. Crystals with typical dimensions of 200 × 200 × 50 Å3 appeared within 7–10 days and grew to maximum size within two months. These crystals correspond to the pH 5.5 conditions.
For the pH 7.0 studies, crystals were soaked for 6–18 hours at 22°C in 1.5 M (NH4)2SO4, 0.1 M sodium citrate (pH 7), 0.5 M NaCl, and 25 mM magnesium acetate (adjusted to a final pH 7.0). Additionally, 5% glycerol was included in the pH 7.0 crystal soaking solution for crystals that were to be cryoprotected in glycerol. For the pH 8.0 studies, the crystals were transferred to pH 8.0 during the cryoprotection transfer process as described below. The final conditions were the same as for the pH 7.0 crystals but with the final pH adjusted to 8.
Prior to data collection, pH 5.5 and 7 crystals were transferred either in 5% steps (5 min/step) or in a single step (1–2 minutes) to mother liquor supplemented with 25% glucose or 25% glycerol before being flash frozen in liquid nitrogen. For the pH 8 studies, crystals were transferred stepwise to pH 7.0 mother liquor supplemented with 20% glycerol followed by a 30 sec. transfer to pH 8.0 mother liquor supplemented with 25% glycerol. The fast transfer of the pH 8 crystals was necessary as the crystals stop diffracting if they are incubated for longer periods of time at pH 8.
All diffraction data were collected at 100 °K. Although the crystals diffract to better than 2.1 Å, they often exhibited a high degree of mosaicity and poor spot quality. The combination of high mosaicity, poor spot quality, and the long unit cell dimension along the c-axis limited high resolution data collection. Macromolecular crystal annealing improved mosaicity and spot quality, but was accompanied by a ~0.1–0.2 Å loss in resolution. Nevertheless, annealing was necessary in many instances as otherwise it was not possible to collect the data. To anneal the crystals, they were removed from the liquid nitrogen stream and submerged in the appropriate cryoprotectant for 3 minutes before being remounted in the cryostream 20.
An initial data set for Form I, pH 5.5 crystals was collected to 2.7 Å (0.5° oscillations) at the Stanford Synchrotron Radiation Laboratory, beamline 9-1, using a Mar345 imaging plate. All the high-resolution data sets ranging from 2.35–2.5 Å were collected with a 3Kx3K Marmosaic CCD detector at the Advanced Photon Source, beamline 5ID, using 0.1° oscillations. All diffraction data were processed with XDS 21, and scaled with SCALA 22. The crystals belong to the tetragonal space group P43212 with unit cell dimensions of a=b=102 Å and c ranging between 444 Å and 454 Å, depending on the freezing conditions, and contain two protein-DNA complexes in the asymmetric unit. Data collection statistics are presented in Table I.
Table I.
Data and refinement statistics
| Form I (pH 5.5) | Form I (pH 7.0) | Form II f (pH 5.5) | Form II (pH 7.0) | Form II (pH 8.0) | |
|---|---|---|---|---|---|
| Data collection | |||||
|
| |||||
| Wavelength (Å) | 0.9479 | 0.9479 | 0.9479 | 0.9756 | 1.0 |
| Cell dimensions (Å) (P43212 ) | a,b=102.2, c=443.8 | a,b=102.3, c=445.6 | a,b=102.2, c=451.7 | a,b=101.8, c=452.4 | a=b=101.6, c=453.6 |
| Resolution (Å) | 2.35 (2.41-2.35) | 2.50 (2.56-2.50) | 2.45 (2.51-2.45) | 2.48 (2.54-2.48) | 2.50 (2.56 – 2.50) |
| Measured reflections | 346,182 | 319,237 | 346,541 | 398,926 | 440,999 |
| Unique reflections | 95,028 | 79,675 | 87,792 | 82,671 | 81,345 |
| Completeness (%)a | 96.1 (74.2) | 96.5 (93.0) | 98.3 (94.1) | 96.8 (75.9) | 98.1 (89.6) |
| Rsym (%)a,b | 5.0 (21.5) | 6.0 (32.8) | 9.1 (30.2) | 7.2 (26.2) | 5.0 (26.9) |
| Rmeas (%)a,c | 5.8 (28.4) | 6.7 (40.8) | 10.3 (34.9) | 8.0 (31.4) | 6.2 (37.3) |
| Redundancy | 3.6 (1.7) | 4.0 (2.5) | 3.9 (3.6) | 4.8 (2.7) | 5.2 (3.0) |
| I/σ(I) | 10.4 (3.6) | 8.7 (2.3) | 6.8 (2.6) | 8.4 (2.9) | 10.8 (2.8) |
|
| |||||
| Refinement | |||||
|
| |||||
| Resolution (Å) | 15.0-2.35 (2.41-2.35) | 15.0-2.50 (2.56-2.50) | 15.0-2.45 (2.51-2.45) | 15.0-2.50 (2.56-2.50) | 15.0-2.50 (2.57 –2.50) |
| Number of reflections: | |||||
| working set/test set | 89879/4739 | 75333/3976 | 82938/4367 | 77357/4089 | 76707/4026 |
| R-factord | 20.7 (26.3) | 21.7 (33.3) | 21.2 (29.5) | 22.2 (30.5) | 22.1 (25.8) |
| Rfreee | 24.5 (30.3) | 26.0 (35.7) | 24.6 (31.6) | 26.9 (35.5) | 30.9 (35.3) |
| Number of protein atoms | 10,024 | 10,024 | 9993 | 9993 | 9,849 |
| Number of water molecules | 358 | 148 | 220 | 208 | 143 |
| Number of DNA atoms | 285 | 285 | 296 | 296 | 296 |
| Other | 2 | 2 | 6 | 6 | 6 |
| Residues observed | |||||
| A monomer | 1:617, 646:658 | 1:617, 646:658 | 1:620, 646:659 | 1:620, 646:659 | 1:243,246:453, 459:617,650:658 |
| B monomer | 1:620, 648:658 | 1:620, 648-658 | 1:618, 646:658 | 1:618, 646:658 | 1:620, 651:656 |
| R.m.s.d. from target values | |||||
| Bond lengths (Å) | 0.010 | 0.010 | 0.010 | 0.010 | 0.010 |
| Bond angles (°) | 1.16 | 1.25 | 1.22 | 1.27 | 1.24 |
| Average B-factor (Å2)h: | |||||
| Main chain (A/B) | 51.4/54.6 | 63.9/64.7 | 46.8/42.0 | 55.5/40.6 | 69.1/56.3 |
| Side chain (A/B) | 52.6/55.2 | 65.0/65.4 | 48.0/43.2 | 56.1/41.1 | 70.5/57.7 |
| Solvent | 38.0 | 46.6 | 30.8 | 36.0 | 45.0 |
| DNA (A/B) | 33.3/39.6 | 46.6/51.8 | 35.8/32.7 | 40.2/26.2 | 56.9/43.5 |
| Ramachandran favored (%)i | 98.0 | 97.1 | 97.5 | 96.2 | 96.6 |
| Ramachandran outliers (%)i | 0.2 | 0.2 | 0.1 | 0.2 | 0.2 |
| Rotamers outliers (%)i | 1.3 | 2.7 | 3.6 | 3.9 | 4.3 |
Numbers in parenthesis represent values in the highest resolution shell.
Rsym=Σ|I−〈I〉|/ΣI, where I=observed intensity, and 〈I〉=average intensity obtained from multiple measurements.
Rmeas as defined by 28.
R-factor=Σ||Fo|−|Fc||/Σ|Fo|, where |Fo|=observed structure factor amplitude and |Fc|=calculated structure factor amplitude.
Rfree: R-factor based on 5% of the data excluded from refinement.
Form I crystals were cryoprotected in 25% glucose and Form II crystals were in 25% glycerol.
The two numbers correspond to the A and B monomers respectively.
Calculated with MolProbity27.
Structure determination and refinement
The first structure (Form I pH 5.5) was solved by Molecular Replacement with the program AMoRe 23 using the intact apo E. coli topoisomerase III structure 6 as a search model. A clear solution was found that yielded a correlation coefficient of 57.8 and R-factor of 49.8%. Preliminary electron density maps showed evidence of a second molecule in the asymmetric unit. Therefore, the first solution was fixed in AMoRe and a second solution was found using the structure of the Y328F mutant of E. coli topoisomerase III bound to ssDNA with the DNA omitted from the search model 11. Initial refinement produced electron density maps that clearly showed Fo-Fc difference density for DNA bound to each monomer. With two protein-DNA complexes in the asymmetric unit, the solvent content of the crystals is ~68%. The first Form II structure was solved in a similar manner and showed the presence of two molecules in the same conformation. Once it was realized that different cryoprotectants resulted in different unit cell contents, it was easy to solve the other structures by Molecular Replacement.
The maps calculated from the Molecular Replacement solution show regions that had changed with respect to the initial model. Rebuilding of the model was followed by energy minimization and B-factor refinement. All refinements were performed with the program Refmac5 24 and model rebuilding was done using the programs O 25 and Coot 26. In all cases, unambiguous electron density can be seen for all 8 nucleotides bound to one monomer (open form). In the other monomer, 6 nucleotides of the bound DNA molecule are well-defined while there is partial density for the last 1 base at the 3′ end. The DNA was built in and refined at full occupancy. A large portion of domain IV also needed to be rebuilt for the closed and intermediate form molecules in the asymmetric unit. Two strong difference density peaks (>8σ) located at the interface between the two complexes were assigned as chloride ions. The packing of the molecules is such that Domain II of the molecules, especially in the closed monomer, makes few crystal packing contacts. For this reason, density for Domain II is much worse than for the rest of the molecule. To take into account the independent movement of the domains, TLS refinement of the molecules broke down into domains was also included in Refmac5. TLS refinement improved the overall Rfree and R-factors of the structures. Figure 2 shows a difference map around the active site region of the refined Form II, pH 8 closed form protein to illustrate the quality of the maps and models. Electron density is present for most residues in the main body of the protein (Table I). There is also electron density for 5 extra C-terminal residues encoded by the expression vector. The structures have been refined to R-factors ranging from 22 to 22.1 % and Rfree ranging from 24.1 to 26.7 % (Table I) with most residues in the most favored regions of the Ramachandran plot and with good rotamer distributions according to Molprobity 27.
For comparisons, the structures were superposed using the program LSQKAB of the CCP4 suite 22.
The coordinates and structure factors for all molecules have been deposited in the PDB with accession numbers 2O5E, 2O5C, 2O59, 2O19 and 2O54.
Acknowledgments
We thank A. Patel for technical assistance and Michael Blum for assistance with using the MARMosaic detector. Research was supported by NIH GM51350 to A.M. and an NRSA Institutional Training Grant in Molecular Biophysics (NIH GM08382) to A.C. We acknowledge the use of instruments in the Keck Biophysics Facility at Northwestern University. Support from the R.H. Lurie Cancer Center of Northwestern University to the Structural Biology Center is acknowledged. Portions of this work were performed at the DuPont-Northwestern-Dow Collaborative Access Team (DND-CAT) Synchrotron Research Center at the Advanced Photon Source (APS) and at the Stanford Synchrotron Radiation Laboratory (SSRL). DND-CAT is supported by DuPont, Dow, and the NSF and use of the APS is supported by the DOE. SSRL is operated by the DOE, Office of Basic Energy Sciences. The SSRL Biotechnology Program is supported by the NIH and the DOE.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Corbett KD, Berger JM. Structure, molecular mechanisms, and evolutionary relationships in DNA topoisomerases. Annu Rev Biophys Biomol Struct. 2004;33:95–118. doi: 10.1146/annurev.biophys.33.110502.140357. [DOI] [PubMed] [Google Scholar]
- 2.Champoux JJ. DNA Topoisomerases: Structure, Function, and Mechanism. Annu Rev Biochem. 2001;70:369–413. doi: 10.1146/annurev.biochem.70.1.369. [DOI] [PubMed] [Google Scholar]
- 3.Wang JC. DNA Topoisomerases. Annu Rev Biochem. 1996;65:635–692. doi: 10.1146/annurev.bi.65.070196.003223. [DOI] [PubMed] [Google Scholar]
- 4.Taneja B, Patel A, Slesarev A, Mondragon A. Structure of the N-terminal fragment of topoisomerase V reveals a new family of topoisomerases. EMBO J. 2006;25:398–408. doi: 10.1038/sj.emboj.7600922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lima CD, Wang JC, Mondragon A. Three-dimensional structure of the 67K N-terminal fragment of E. coli DNA topoisomerase I. Nature. 1994;367:138–146. doi: 10.1038/367138a0. [DOI] [PubMed] [Google Scholar]
- 6.Mondragon A, DiGate R. Structure of E. coli DNA topoisomerase III. Structure. 1999;7:1373–1383. doi: 10.1016/s0969-2126(00)80027-1. [DOI] [PubMed] [Google Scholar]
- 7.Hansen G, Harrenga A, Wieland B, Schomburg D, Reinemer P. Crystal structure of full length topoisomerase I from Thermotoga maritima. J Mol Biol. 2006;358:1328–1340. doi: 10.1016/j.jmb.2006.03.012. [DOI] [PubMed] [Google Scholar]
- 8.Rodriguez AC, Stock D. Crystal structure of reverse gyrase: insights into the positive supercoiling of DNA. EMBO J. 2002;21:418–426. doi: 10.1093/emboj/21.3.418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang HL, DiGate RJ. The carboxyl-terminal residues of Escherichia coli DNA topoisomerase III are involved in substrate binding. J Biol Chem. 1994;269:9052–9059. [PubMed] [Google Scholar]
- 10.Beran-Steed RK, Tse-Dinh YC. The carboxy terminal domain of Escherichia coli DNA topoisomerase I confers higher affinity to DNA. Proteins. 1989;6:249–258. doi: 10.1002/prot.340060307. [DOI] [PubMed] [Google Scholar]
- 11.Changela A, DiGate RJ, Mondragon A. Crystal structure of a complex of a type IA DNA topoisomerase with a single-stranded DNA molecule. Nature. 2001;411:1077–1081. doi: 10.1038/35082615. [DOI] [PubMed] [Google Scholar]
- 12.Perry K, Mondragon A. Structure of a complex between E. coli DNA topoisomerase I and single-stranded DNA. Structure. 2003;11:1349–1358. doi: 10.1016/j.str.2003.09.013. [DOI] [PubMed] [Google Scholar]
- 13.Zhang HL, Malpure S, DiGate RJ. Escherichia coli DNA topoisomerase III is a site-specific DNA binding protein that binds asymmetrically to its cleavage site. J Biol Chem. 1995;270:23700–23705. doi: 10.1074/jbc.270.40.23700. [DOI] [PubMed] [Google Scholar]
- 14.Wang JC. Interaction between DNA and an Escherichia coli Protein w. J Mol Biol. 1971;55:523–533. doi: 10.1016/0022-2836(71)90334-2. [DOI] [PubMed] [Google Scholar]
- 15.Domanico PL, Tse-Dinh YC. Mechanistic studies on E. coli DNA topoisomerase I: Divalent ion effects. J Inorg Biochem. 1991;42:87–96. doi: 10.1016/0162-0134(91)80035-g. [DOI] [PubMed] [Google Scholar]
- 16.Zhu CX, Tse-Dinh YC. The acidic triad conserved in type IA DNA topoisomerases is required for binding of Mg(II) and subsequent conformational change. J Biol Chem. 2000;275:5318–5322. doi: 10.1074/jbc.275.8.5318. [DOI] [PubMed] [Google Scholar]
- 17.Berger JM, Fass D, Wang JC, Harrison SC. Structural similarities between topoisomerases that cleave one or both DNA strands. Proc Natl Acad Sci USA. 1998;95:7876–7881. doi: 10.1073/pnas.95.14.7876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Perry K, Mondragon A. Biochemical characterization of an invariant histidine involved in Escherichia coli DNA topoisomerase I catalysis. J Biol Chem. 2002;277:13237–13245. doi: 10.1074/jbc.M112019200. [DOI] [PubMed] [Google Scholar]
- 19.Aggarwal AK. Crystallization of DNA binding proteins with oligodeoxynucleotides. METHODS: A Companion to Meth Enzym. 1990;1:83–90. doi: 10.1016/S1046-2023(05)80150-1. [DOI] [PubMed] [Google Scholar]
- 20.Harp JM, Timm DE, Bunick GJ. Macromolecular crystal annealing: overcoming increased mosaicity associated with cryocrystallography. Acta Crystallogr. 1998;D54:622–628. doi: 10.1107/s0907444997019008. [DOI] [PubMed] [Google Scholar]
- 21.Kabsch W. Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J Appl Crystallogr. 1993;26:795–800. [Google Scholar]
- 22.Collaborative-Computational-Project-4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. 1994;D50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 23.Navaza J. Amore - an Automated Package for Molecular Replacement. Acta Crystallogr. 1994;A50:157–163. [Google Scholar]
- 24.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. 1997;D53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 25.Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. 1991;A47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 26.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. 2004;D60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 27.Davis IW, Murray LW, Richardson JS, Richardson DC. MOLPROBITY: structure validation and all-atom contact analysis for nucleic acids and their complexes. Nucleic Acids Res. 2004;32:W615–619. doi: 10.1093/nar/gkh398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Diederichs K, Karplus PA. Improved R-factors for diffraction data analysis in macromolecular crystallography. Nat Struct Biol. 1997;4:269–275. doi: 10.1038/nsb0497-269. [DOI] [PubMed] [Google Scholar]
- 29.Feinberg H, Lima CD, Mondragon A. Conformational changes in E. coli DNA topoisomerase I. Nature Struct Biol. 1999;6:918–922. doi: 10.1038/13283. [DOI] [PubMed] [Google Scholar]


