Abstract
Complete genomes of HPV101 and HPV103 were PCR amplified and cloned from cervicovaginal cells of a 34-year-old female with cervical intraepithelial neoplasia grade 3 (CIN 3) and a 30-year-old female with a normal Pap test, respectively. HPV101 and HPV103 contain 4 early genes (E7, E1, E2 and E4) and 2 late genes (L2 and L1), but both lack the canonical E6 ORF. Pairwise alignment similarity of the L1 ORF nucleotide sequences of HPV101 and HPV103 indicated that they are at least 30 % dissimilar to each other and all known PVs. However, similarities of the other ORFs (E7, E1, E2, and L2) indicated that HPV101 and HPV103 are most related to each other. Phylogenetic analyses revealed that these two types form a monophyletic clade, clustering together with the gamma- and pi-PV groups. These data demonstrated that HPV genomes closely related to papillomaviruses identified from cutaneous epithelia can be isolated from the genital mucosal region. Moreover, this is the first report of HPVs lacking an E6 ORF and phylogenetic evidence suggests this occurred subsequent to their emergence from the gamma-/pi-PVs.
Keywords: Human papillomavirus, novel type, complete genome, phylogeny, molecular clock
Introduction
Papillomaviruses (PVs) are a heterogeneous group of DNA viruses with circular double-stranded DNA genomes about 8 kb in size that contain three general regions. An upstream regulatory region (URR) contains sequences that control transcription and replication, an early region contains genes (e.g., E6, E7, E1, E2, E4 and E5) involved primarily in enzymatic activities, and a structural region consisting of the L1 capsid protein and L2 that facilitates packaging of viral DNA. PVs are highly species-specific pathogens that cause proliferation of predominantly epithelial cells in a wide range of host species. Currently, over one hundred types of human papillomaviruses (HPVs) and at least twenty-eight types of nonhuman PVs have been fully characterized. For a papillomavirus to be recognized as a distinct type, the DNA sequence of the L1 open reading frame (ORF) of the cloned genome should be no more than 90% similar to previously typed PVs (de Villiers, 2001; Delius et al., 1998). Historically, PV phylogenies based on tissue tropism and genomic sequence have typically been subdivided into clades infecting mucosal / genital lesions, cutaneous / epidermodysplasia verruciformis (EV) regions and three main animal hosts: the artiodactyls ruminant, the distant avian and a group containing canine, feline, rabbit and rodent (Chan et al., 1995; de Villiers, 2001; de Villiers et al., 2004). Based on the L1 ORF, papillomaviruses have recently been classified into alpha-PV, beta-PV and gamma-PV, and at least 13 other genera, however, analyses based on the early region of HPV genomes indicates that the phylogeny of the alpha-PVs is more complex than initially suspected (Narechania et al., 2005).
Most PVs target the basal cells of dermal or mucosal epithelia, and infection has been implicated in both benign and malignant lesions of epithelia in a broad range of animals and humans. Some papillomatous proliferations induced by specific types of PVs have a high risk for malignant progression (Clifford et al., 2003; Munoz et al., 2003). Nevertheless, the majority of PVs appear to be nonpathogenic, since a wide variety of different types can be detected at different sites of healthy skin of human and animals (Antonsson et al., 2003; Antonsson and Hansson, 2002), and over 40 types have been detected in the mucosal epithelia of human with only a minority associated with high-risk disease (Munoz et al., 2003). PVs seem to have coexisted with their host over long periods of time. With the exception of certain bovine PVs (BPVs), each PV type has been found only in a single host species. In some host species, particularly in human beings, numerous PV types have been found. The available data suggests associations between PV types, pathological properties, and tissue tropism; however, it is unclear what molecular properties link specific PV types to particular hosts and tissues, and, in particular, how numerous types that have similar tissues tropisms (e.g., mucosal or cutaneous) and pathologic manifestation evolved in the same host.
This report describes the characterization of two novel HPV types, HPV101 and HPV103, isolated from the cervicovaginal cells using an overlapping PCR method. The viruses were initially identified by PCR amplification of a 480-bp fragment using FAP primers (Forslund et al., 1999). Nucleotide sequence similarity by pairwise alignment of 5 ORFs (E7, E1, E2, L2 and L1) indicated that they are closely related to the gamma-PV genus (usually associated with cutaneous lesions) and not alpha-PVs. Furthermore, both HPV genomes lack an E6 ORF. Phylogenetic analyses revealed that HPV101 and HPV103 form a monophyletic group at the root of the gamma- and pi-PV genera.
Results and discussion
Determination of the complete genome sequences
The complete nucleotide sequences of HPV101 and HPV103 contain 7259 bp and 7263 bp, with a GC-content of 42.8 % and 41.6 %, respectively. The complete nucleotide sequences of HPV101 and HPV103 L1 ORFs share 60.9 % and 65.3 % similarity to HPV65 L1 (gamma-PV), 62.6 % and 59.5 % to HPV19 L1 (beta-PV), 59.4 % and 65.2 % to HaOPV (pi-PV); and 64.7 % similarity to each other (Table 1). HPV101 and HPV103 are at least 30 % dissimilar to all known PVs and satisfy the criteria for novel types.
Table 1.
Nucleotide sequence pairwise similarity (%) of HPV101 and HPV103 ORFs with representative PVs.
| HPV101 | Pi-PV
|
Gamma-PV
|
Beta-PV
|
Mu-PV
|
Alpha-PV
|
Eta-PV
|
||||
|---|---|---|---|---|---|---|---|---|---|---|
| HPV103 | HPV103 | HaOPV | HPV65 | HPV60 | HPV48 | HPV4 | HPV19 | HPV1 | HPV16 | FPV |
| E7 | 69.7 | 42.1 | 57.0 | 56.4 | 58.8 | 57.4 | 55.9 | 52.9 | 54.5 | 33.1 |
| ---- | 43.1 | 57.5 | 51.3 | 55.1 | 60.6 | 56.1 | 53.0 | 54.3 | 34.1 | |
| E1 | 73.3 | 60.2 | 62.4 | 62.4 | 58.8 | 63.3 | 60.5 | 60.9 | 61.1 | 54.9 |
| ---- | 60.9 | 62.9 | 63.6 | 61.7 | 63.8 | 61.4 | 61.5 | 60.0 | 54.0 | |
| E2 | 68.8 | 58.6 | 58.5 | 61.5 | 58.6 | 59.0 | 51.4 | 57.7 | 56.8 | 53.2 |
| ---- | 59.5 | 59.7 | 61.4 | 59.4 | 60.3 | 50.8 | 57.3 | 57.5 | 52.4 | |
| L2 | 67.1 | 56.4 | 57.7 | 58.0 | 56.3 | 58.2 | 56.5 | 55.6 | 53.4 | 51.3 |
| ---- | 55.5 | 58.8 | 57.3 | 57.0 | 59.1 | 57.6 | 56.2 | 55.9 | 51.9 | |
| L1 | 64.7 | 59.4 | 60.9 | 58.1 | 58.8 | 62.0 | 62.6 | 58.3 | 61.2 | 54.2 |
| ---- | 65.2 | 65.3 | 61.6 | 64.2 | 64.8 | 59.5 | 60.8 | 59.9 | 57.1 | |
The genomes of HPV101 and HPV103 contain six ORFs, potentially encoding four early genes (E7, E1, E2 and E4) and two late genes (L2 and L1) (Supplement 1).
The putative E7 ORFs of HPV101 and HPV103 contain one conserved zinc binding domain, CxxC(x)29CxxC, and a motif (LxCxE) for binding to the pRB protein. The ATP-binding site of the ATP-dependent helicase (GPPDTGKS for HPV101, GPSDTGKS for HPV103) is conserved in the carboxy-terminal region of E1. The E4 ORF contains a start codon and overlaps the E2 ORF. A polyadenylation consensus sequence (AATAAA) for processing of early viral mRNA transcripts is present at the beginning of the L2 gene (nucleotide [nt] 3524 to 3529 for HPV101, nt 3453 to 3458 for HPV103). The major (L1) and minor (L2) capsid proteins of both types show a nuclear localization signal at their 3′ end.
Both HPV101 and HPV103 lack a canonical E6 ORF. Although there is a novel ORF downstream of the L1 ORF in HPV101 (nt 6734 to 7225, 6995 for the first ATG) calculated to produce an 8.9 kDa protein (Supplement 1), it does not contain a zinc binding domain and is embedded in the long control region (LCR). This is the first characterization of human PVs lacking an E6 ORF. Interestingly, bovine papillomavirus type 3 (BPV3), BPV4 and BPV6, bird PVs Fringilla coelebs papillomavirus (FPV) and Psittacus erithacus timneh papillomavirus (PePV) also lack a definable E6 ORF (Jackson et al., 1991; Terai, DeSalle, and Burk, 2002), while Phocoena spinipinnis papillomavirus (PsPV) that causes genital warts in small cetaceans, has an E6 ORF but lacks an identifiable E7. It appears that E6 and E7 functions are either not required by these viruses or are performed by another viral (or host) protein. One hypothesis is that ancient genomic rearrangement may have contributed to the evolution of PV types lacking E6 or E7 ORFs. The E6 and E7 proteins, the major oncogenes in PVs, may play a central function in adapting PV genome to various hosts and tissues. In contrast, the E1, E2, L2, and L1 ORFs, are well conserved in all PVs and their products are essential proteins for the PV life cycle (Terai, DeSalle, and Burk, 2002). In addition, HPV101 and HPV103 lack a distinct E5 ORF. The region between the early and late genes (ELR) is about 100 bp in length.
The sequence between the stop codon of L1 and the first ATG of E6 is known as the long control region (LCR) or the upstream regulatory region (URR), a noncoding region (NCR) containing many of the cis-acting regulating regulatory sequences that control viral transcription and replication. Because of the lack of a definable E6 ORF for HPV101 and HPV103, the LCR sequence was defined as the region between the L1 stop codon and the first start codon of the E7 ORF. The LCR is 678 bp in HPV101 (nt 6582 to 7259) and 771 bp in HPV103 (nt 6493 to 7263) (Supplement 2). Analysis of the HPV101 LCR revealed three typical palindromic E2-binding sites [ACC(N)6GGT] at nt 6806 to 6817, 6922 to 6933, 6999 to 7010, and one modified putative E2-binding sites [AAC(N)6GGT] at nt 6893 to 6904. The LCR also contains a second overlapping polyadenylation site (AATAAA; nt 6642 to 6647 vs 6646 to 6651) at the 5′ end. Multiple binding sites for transcriptional regulatory factors such as AP-1 (Chan et al., 1990), NF-1 (Apt et al., 1993), SP-1 (Gloss and Bernard, 1990), transcriptional enhancer factors (TEF)-1 (Ishiji et al., 1992), and YY-1 (Dong et al., 1994) are also present within the LCR region. Analysis of HPV103 LCR revealed two typical palindromic E2-binding sites (nt 6775 to 6786 and 6968 to 6979), and similar to HPV101, it also contains a polyadenylation site (nt 6753 to 6758) and multiple transcription factor binding sites.
DNA diversity and phylogenetic analyses of HPV101 and HPV103
The nucleotide sequence similarity of HPV101 and HPV103 to HPV1, HPV4, HPV48, HPV60 and HPV65 (gamma-PVs), HPV19 (a beta-PV), HPV16 (a high-risk alpha-PV) and chaffinch (Fringilla coelebs) PV (FPV) were investigated by pairwise alignment of each ORF (Table 1). The selected types represent the main PV clades. The nucleotide sequence similarity of HPV101 and HPV103 L1 ORFs was 64.7 % when compared to each other; whereas they were approximately 60–65 % similar to the L1 of PVs in other genera. However, all other ORFs similarities (E7, E1, E2 and L2) indicated these two novel types are most closely related to each other. Furthermore, HPV101 and HPV103 are more closely related to the gamma-PVs (e.g., HPV4, HPV65) than to the alpha-PVs (e.g., HPV16).
In order to examine the phylogenetic relationship among HPVs and other animal PVs, trees using multiple algorithms were constructed. The Bayesian tree inferred from the concatenated amino acids and nucleotide sequences of six ORFs (E1, E2, E6, E7, L1 and L2) clusters all characterized PVs into genera consistent with one phylogeny, similar to that proposed by de Villiers et al. (Fig. 1) (de Villiers et al., 2004). The incongruence of the branching order of some major genera between these two trees could be the result of an early recombination event, an ecological niche change, and / or asymmetric genome convergence driven by intense selection across different PV genes (Narechania et al., 2005). As shown in the Fig. 1, large evolutionary distances significantly separate PVs of hosts belonging to different mammalian orders (e.g., human and bovine). There is also a significant distance between mammalian PVs and avian PVs (FPV and PePV) that infect bird and not mammals. This distribution supports the notion of an ancient virus-host association implicit in a coevolution history.
Fig. 1.
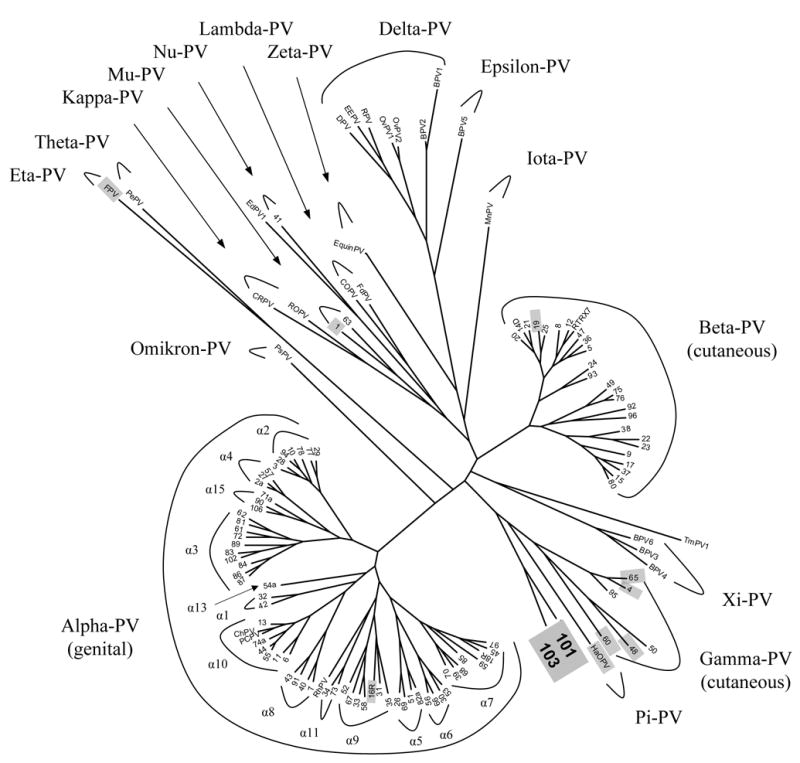
Bayesian tree of 125 papillomavirus types inferred from the concatenated amino acids and nucleotide sequences of six ORFs (E6, E7, E1, E2, L2 and L1). The ORF sequence was left blank if unavailable. Classification at the “genus” level is according to de Villiers (10). The number at the ends of each of the branches identifies an HPV type. All other abbreviations refer to animal PVs but the RTRX7. Two novel HPV types in this work (HPV101 and HPV103) form a monophyletic clade at the root of the gamma-PV genus. PV genomes representing main clades of papillomaviruses that were used in Table 1 are were highlighted in gray. All complete genome sequences of PVs were obtained from NCBI / GenBank database (with their accession numbers): HPV01a (NC_001356), HPV02a (NC_001352), HPV03 (NC_001588), HPV04 (NC_001457), HPV05 (NC_001531), HPV06 (NC_000904), HPV07 (NC_001595), HPV08 (NC_001532), HPV09 (NC_001596), HPV10 (NC_001576), HPV11 (NC_001525), HPV12 (NC_001577), HPV13 (NC_001349), HPV14D (NC_001578), HPV15 (NC_001579), HPV16R (AY686581), HPV17 (NC_001580), HPV18R (AY262282), HPV19 (NC_001581), HPV20 (NC_001679), HPV21 (NC_001680), HPV22 (NC_001681), HPV23 (NC_001682), HPV24 (NC_001683), HPV25 (NC_001582), HPV26 (NC_001583), HPV27 (NC_001584), HPV28 (NC_001684), HPV29 (NC_001685), HPV30 (NC_001585), HPV31 (NC_001527), HPV32 (NC_001586), HPV33 (NC_001528), HPV34 (NC_001587), HPV35 (NC_001529), HPV36 (NC_001686), HPV37 (NC_001687), HPV38 (NC_001688), HPV39 (NC_001535), HPV40 (NC_001589), HPV41 (NC_001354), HPV42 (NC_001534), HPV43 (NC_005349), HPV44 (NC_001689), HPV45 (NC_001590), HPV47 (NC_001530), HPV48 (NC_001690), HPV49 (NC_001591), HPV50 (NC_001691), HPV51 (NC_001533), HPV52 (NC_001592), HPV53 (NC_001593), HPV54 (NC_001676), HPV55 (NC_001692), HPV56 (NC_001594), HPV57 (NC_001353), HPV58 (NC_001443), HPV59 (NC_001635), HPV60 (NC_001693), HPV61 (NC_001694), HPV62 (NC_006357), HPV63 (NC_001458), HPV65 (NC_001459), HPV66 (NC_001695), HPV67 (NC_004710), HPV68 (DQ080079), HPV69 (NC_002171), HPV70 (NC_001711), HPV71 (NC_002644), HPV72 (NC_006164), HPV73 (NC_006165), HPV74 (NC_004501), HPV75 (NC_006166), HPV76 (NC_006167), HPV77 (NC_006168), HPV78 (unpublished), HPV80 (NC_006169), HPV81 (NC_005351), HPV82 (NC_002172), HPV83 (NC_000856), HPV84 (NC_002676), HPV85 (NC_004762), HPV86 (NC_003115), HPV87 (NC_002627), HPV89 (NC_004103), HPV90 (NC_004104), HPV91 (NC_004085), HPV92 (NC_004500), HPV93 (NC_005133), HPV94 (NC_005352), HPV95 (AJ620210), HPV96 (NC_005134), HPV101 (DQ080081 ), HPV103 (DQ080078), bovine BPV1 (X02346), BPV2 (NC_001521), BPV3 (NC_004197), BPV4 (NC_004711), BPV5 (NC_004195), BPV6 (NC_005350), canine oral CoPV (NC_001619), commom chimpanzee ChPV (NC_001838), cottontail rabbit CRPV (NC_001541), deer DPV (NC_001523), Erethizon dorsatum EdPV-1(AY684126), Equinus caballus EquiPV (NC_004194), European elk EEPV (NC_001524), feline FdPV (NC_004765), Fringilla coelebs FPV (NC_004068), Hamster oral HaOPV (E15111), multimammate rat MnPV (NC_001605), ovine OvPV1 (NC_001789), ovine OvPV2 (NC_001790), Procyon lotor PIPV-1 (AY763115), Phocoena spinipinnis PsPV (NC_003348), Psittacus erithacus PePV (NC_003973), pygmy chimpanzee PcPV (NC_006163), Trichechus manatus latirostris TmPV1 (NC_006563), rabbit oral ROPV (NC_002232), reindeer RPV (NC_004196), and rhesus monkey RhPV (NC_001678).
Except for HPV1, 41 and 63, all characterized HPVs are subdivided into three genera: one is predominantly associated with genital mucosal infection (alpha-PVs) and two predominantly infect cutaneous epithelia (beta- and gamma-PVs). Molecular phylogeny is consistent with the classification by tissue tropism. This suggests that selection pressure, e.g., species-specific immunoresponses from the host, may have shaped the evolution of PVs. HPV101 and HPV103 diverged near the root of the gamma-PV genus, and are joined in a monophyletic group by the pi-PV (represented by hamster oral PV, HaOPV). The L1 ORF of HPV103 has a higher similarity to the L1 nucleotide sequence of HaOPV (Table 1), however, it is difficult to localize HPV101 and HPV103 in a phylogenetic tree by placing them into the gamma-PV genus or the pi-PV genus, as they potentially form a novel genus inferred from the nucleotide sequence similarities. HPV101 and HPV103 seem to have common features with HaOPV and are close to papillomaviruses involved in cutaneous neoplasia; whereas, the lack of an E6 ORF and the mucosal tissue tropism may separate them from the pi-PV group.
Separate Bayesian trees for early genes (E1 and E2) and late genes (L1 and L2) were compared to determine whether there is incongruence of the phylogenetic signals emanating from different genes, which would potentially reflect different evolutionary histories or different evolutionary pressures (Narechania et al., 2005). Representative types of the beta- and gamma-PV genera were utilized for tree construction. As shown in Fig. 2, the phylogeny indicated stable and consistent patterns by the early and late gene regions, which was confirmed by the congruence test of ILD in PAUP (data not shown). Therefore, HPV101 and HPV103 genomes have evolved as a unit rather than certain genes having different evolutionary histories. HaOPV, however, showed variable phylogenetic positions between early and late genes.
Fig. 2.
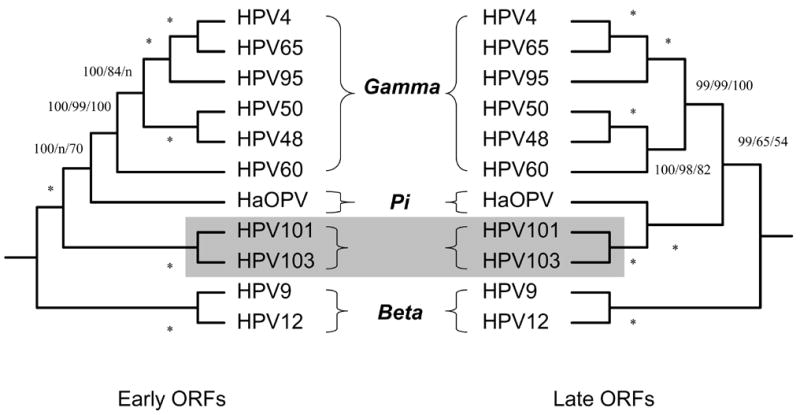
Bayesian trees inferred from the concatenated amino acids and nucleotide sequences of early (E6, E7, E1 and E2) and late (L2 and L1) ORFs. The ORF sequence was left blank if unavailable. Numbers on or near branches indicate support indices from multiple algorithms in the following order: Bayesian credibility value using MrBayes; maximum parsimony (MP) bootstrap percentage and neighbor joining (NJ) bootstrap percentage using PAUP. An asterisk (*) indicates complete agreement between methods. “n” reflects disagreement between a method and the reference Bayesian tree at a given node. HPV101 and HPV103 genomes are highlighted in gray.
Molecular clock prediction
The divergence times of the most recent common ancestor (MRCA) of representative PVs were predicted and were shown in Fig. 3. The MRCA of HPV was predicted to occur around 30–50 million years ago (Mya), and subsequently split into three potential ancestors of HPV1 (also including HPV41 and HPV63), genital HPVs (e.g., HPV16), and cutaneous HPVs (e.g., HPV19 and HPV4). The MRCA of present-day Gamma HPVs was predicted to have occurred approximately 15–30 Mya ago.
Fig. 3.
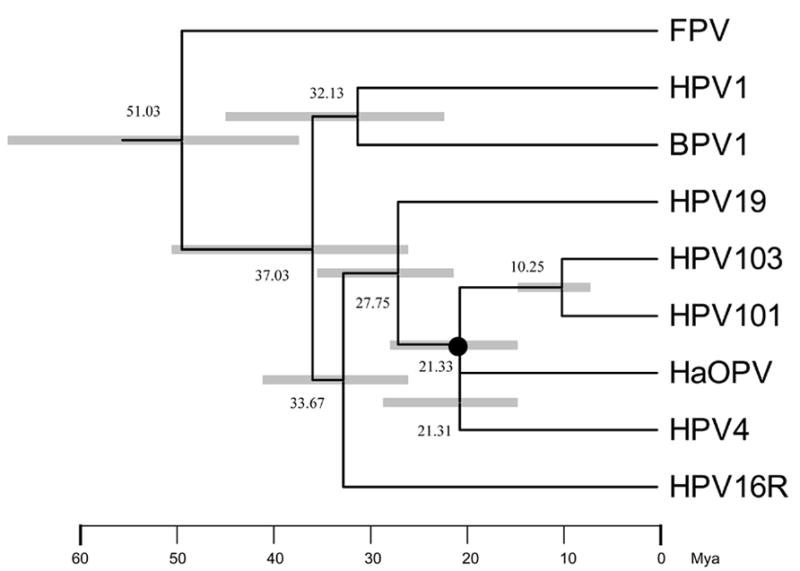
Molecular tree topology of representative papillomaviruses combined with dating of the phylogenetic nodes. Branch lengths are proportional to divergence times. Numbers above the nodes are the mean estimated divergence time (in million years, Mya). The bars in grey represent the 95% highest posterior density (HPD) interval for the divergence times. A Bayesian Markov chain Monte Carlo (MCMC) method was used to calculate divergence times in the BEAST program by selecting the uncorrelated lognormal distribution (UCLD) model. The node of the most recent common ancestor (MRCA) between the clade of HPV101/103 and the Gamma HPVs is denoted with a black circle.
Molecular epidemiology of variants of several HPVs suggests that these viruses already existed at the time humans originated as a species, a few hundred thousand years ago (Ho et al., 1993; Ong et al., 1993). Since between 10 and 20 million years ago a variety of hominoid primates lived in Africa, Europe and Asia (Andrews, 1992), the genomic heterogeneity and organization typical of HPVs must have existed prior to that time. Our MRCA prediction confirmed that all HPV types should have existed before the origin of humans. HPV101 and HPV103, however, are the only two HPVs that lack an E6 gene. A gene loss in this lineage seems to be the most parsimonious explanation, rather than a number of independent acquisitions of E6 assuming that the common ancestor of HPVs did not have an E6 gene. Hence, the loss of E6 might have happened during the development of the evolutionary lineage from the MRCA of the gamma- and pi-PVs to the emergence of HPV101 and HPV103 (≈15–30 Mya). However, genomic rearrangement or lack of selective pressure may be responsible for PV types lacking an E6 ORF.
In this work, we report the characterization of two novel HPVs obtained from cervicovaginal cells. Based on the genomic and phylogenetic analyses, these two novel types are related to gamma-PVs although they were isolated from cervicovaginal cells, since (i) the region between the early and late genes (ELR) is about 100 bp in length and lacks a distinct E5 ORF, similar to beta- and gamma-PVs (de Villiers et al., 2004); (ii) HPV101 and HPV103 are more closely related to the gamma-PV genus (e.g., HPV65) than to the alpha-PV genus (e.g., HPV16) inferred from sequence comparisons of 5 ORFs; (iii) they phylogenetically form a clade clustering together with the gamma- and pi-PV genera.
Materials and methods
Specimens and HPV cloning
HPV101 was isolated from the cervicovaginal (CV) cells from a 34-year-old female with cervical intraepithelial neoplasia grade 3 (CIN 3) that also contained HPV33 and HPV35. HPV103 was isolated from the CV cells of a 30-year-old female with a normal cytological screening test. Both females were participants of a population-based HPV study in Costa Rica (Herrero et al., 2000). HPV DNA was initially identified by PCR amplification. The 480 bp fragments amplified by the FAP primers within the L1 ORF were sequenced (Forslund et al., 1999). A BLAST search against GenBank revealed that sequences of both fragments were less than 70% related to other PV genomes. Hence, type-specific primer sets were thereafter designed based on the sequence of the FAP product and a consensus region of E1 ORF to allow amplification of the complete genome in two fragments (3249 bp and 4105 bp for HPV101, 3253 bp and 4161 bp for HPV103). Oligonucleotide primer sequences used in this study are available from the authors.
For overlapping PCR, an equal mixture of the AmpliTaq Gold Taq DNA polymerase (Applied Biosystems, USA) and Pwo Platinum Taq DNA Polymerase (High Fidelity, Invitrogen, USA) were utilized as previously described (Terai and Burk, 2001a; Terai and Burk, 2001b). PCR products were separated by electrophoresis in agarose gels, stained with ethidium bromide, and visualized under ultraviolet illumination.
After confirmation of appropriate product size, each PCR product was purified (Qiagen Gel Extraction Kit, Qiagen, Valencia, CA), ligated into the pGEM-T easy vector (Promega, Madison, WI) and sequenced by the Einstein Sequencing Facility. Subsequent sequencing was performed using primer walking. The complete genomic sequences were determined from a combination of sequences from the two overlapping amplification products. DNA clones were submitted to the Human Papillomavirus Reference Laboratory (Heidelberg, Germany) for official designation. The genome sequences of HPV101 and HPV103 were submitted to NCBI / GenBank database (Access numbers: HPV101, DQ080081; HPV103, DQ080078).
Phylogenetic analysis and tree construction
Phylogenetic trees were inferred from alignments of concatenated amino acids and nucleotide sequences of six ORFs (E6, E7, E1, E2, L2 and L1) of papillomaviruses. The amino acid sequence of each ORF was aligned using ClustalX software (Thompson et al., 1997). Codon Align (ver 1.0) (available from website http://www.sinauer.com/hall/) was implemented to align the nucleotide sequence of each coding region corresponding to the aligned proteins.
The computer program, MrBayes v3.0b4 (Huelsenbeck and Ronquist, 2001) was used for Bayesian tree construction, with 100,000 cycles for the Markov chain Monte Carlo (MCMC) algorithm. The gamma model was set for among-site rate variation, and allowed all substitution rates of aligned sequence to be different. Maximum parsimony (MP) and neighbor joining (NJ) trees were calculated by a heuristic search in PAUP* v4.0b10 (Swofford, 1998). For maximum parsimony analyses, amino acid and nucleotide sequences were reduced to phylogenetically informative sites. Data were bootstrap resampled 1,000 times. Sequences of related PVs analyzed in this work are available from NCBI database.
A computational algorithm to test the incongruence length difference (ILD) was used to assess the relative degrees of incongruence across different node/partition pairs (Narechania et al., 2005). The partition homogeneity test employed in the PAUP* v4.0b10 (Swofford, 1998) was performed by treating early and late genes as separate partitions. The ILD values were calculated.
Molecular divergence estimates
In order to predict the potential divergence times of papillomaviruses, a Bayesian Markov chain Monte Carlo (MCMC) method was used in the BEAST 1.3 program (Drummond and Rambaut, 2003). The complete L1 nucleotide sequences of representative types were selected for prediction. Gblocks (Castresana, 2000) was used to refine ambiguous alignment. We assumed a general time-reversible model of nucleotide substitution with gamma-distributed rate heterogeneity among sites and a proportion of invariant sites. In addition, we assumed an uncorrected lognormal distribution (UCLN) molecular clock model of rate variation among branches in the tree. A mean substitution rate was set to 1.84 × 10−08 (Lower, 1.27 × 10−08; Upper, 2.35 × 10−08) substitutions per site per year. The MCMC analysis was run for 10,000,000 steps. Results were determined by Tracer 1.2 (Rambaut and Drummond, 2003).
Supplementary Material
Supplement 1. ORF map. Location of the predicted ORFs of (a) HPV101, (b) HPV103, and (c) HPV4. Each ORF is represented as a rectangle occupying either the first, second, or third reading frames. Numbers show the nucleotide positions of the start and stop codons.
Supplement 2. Comparison HPV101 and HPV103 LCRs with other PV types (HaOPV, HPV4, and HPV16). LCR length and position of multiple binding sites are shown as indicated. AP-1, activator protein 1; TEF-1, transcriptional enhancer factor -1; Poly(A), poly(A) signal; E1, E1 binding domain; E2, E2 binding domain. A novel ORF (X) embedded in the LCR of HPV101 is indicated with an arrow.
Acknowledgments
This work was supported in part by CA78527 from the National Cancer Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Andrews P. Evolution and environment in the Hominoidea. Nature. 1992;360(6405):641–6. doi: 10.1038/360641a0. [DOI] [PubMed] [Google Scholar]
- Antonsson A, Erfurt C, Hazard K, Holmgren V, Simon M, Kataoka A, Hossain S, Hakangard C, Hansson BG. Prevalence and type spectrum of human papillomaviruses in healthy skin samples collected in three continents. J Gen Virol. 2003;84(Pt 7):1881–6. doi: 10.1099/vir.0.18836-0. [DOI] [PubMed] [Google Scholar]
- Antonsson A, Hansson BG. Healthy skin of many animal species harbors papillomaviruses which are closely related to their human counterparts. J Virol. 2002;76(24):12537–42. doi: 10.1128/JVI.76.24.12537-12542.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apt D, Chong T, Liu Y, Bernard HU. Nuclear factor I and epithelial cell-specific transcription of human papillomavirus type 16. J Virol. 1993;67(8):4455–63. doi: 10.1128/jvi.67.8.4455-4463.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 2000;17(4):540–52. doi: 10.1093/oxfordjournals.molbev.a026334. [DOI] [PubMed] [Google Scholar]
- Chan S-Y, Delius H, Halpern AL, Bernard H-U. Analysis of genomic sequences of 95 papillomavirus types: uniting typing, phylogeny, and taxonomy. J Virol. 1995;69:3074–3083. doi: 10.1128/jvi.69.5.3074-3083.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan WK, Chong T, Bernard HU, Klock G. Transcription of the transforming genes of the oncogenic human papillomavirus-16 is stimulated by tumor promotors through AP1 binding sites. Nucleic Acids Res. 1990;18(4):763–9. doi: 10.1093/nar/18.4.763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clifford GM, Smith JS, Plummer M, Munoz N, Franceschi S. Human papillomavirus types in invasive cervical cancer worldwide: a meta-analysis. Br J Cancer. 2003;88(1):63–73. doi: 10.1038/sj.bjc.6600688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Villiers E-M. Taxonomic classification of papillomaviruses. Papillomavirus Rep. 2001;12(3):57–63. [Google Scholar]
- de Villiers EM, Fauquet C, Broker TR, Bernard HU, zur Hausen H. Classification of papillomaviruses. Virology. 2004;324(1):17–27. doi: 10.1016/j.virol.2004.03.033. [DOI] [PubMed] [Google Scholar]
- Delius H, Saegling B, Bergmann K, Shamanin V, de Villiers EM. The genomes of three of four novel HPV types, defined by differences of their L1 genes, show high conservation of the E7 gene and the URR. Virology. 1998;240(2):359–65. doi: 10.1006/viro.1997.8943. [DOI] [PubMed] [Google Scholar]
- Dong XP, Stubenrauch F, Beyer-Finkler E, Pfister H. Prevalence of deletions of YY1-binding sites in episomal HPV 16 DNA from cervical cancers. Int J Cancer. 1994;58(6):803–8. doi: 10.1002/ijc.2910580609. [DOI] [PubMed] [Google Scholar]
- Drummond AJ, Rambaut A. BEAST version 1.3 [computer program] [Accessed 31 January 2006];2003 Available: http://evole.zoo.ox.ac.uk/beast.
- Forslund O, Antonsson A, Nordin P, Stenquist B, Hansson BG. A broad range of human papillomavirus types detected with a general PCR method suitable for analysis of cutaneous tumours and normal skin. J Gen Virol. 1999;80 ( Pt 9):2437–43. doi: 10.1099/0022-1317-80-9-2437. [DOI] [PubMed] [Google Scholar]
- Gloss B, Bernard HU. The E6/E7 promoter of human papillomavirus type 16 is activated in the absence of E2 proteins by a sequence-aberrant Sp1 distal element. J Virol. 1990;64(11):5577–84. doi: 10.1128/jvi.64.11.5577-5584.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrero R, Hildesheim A, Bratti C, Sherman ME, Hutchinson M, Morales J, Balmaceda I, Greenberg MD, Alfaro M, Burk RD, Wacholder S, Plummer M, Schiffman M. Population-based study of human papillomavirus infection and cervical neoplasia in rural Costa Rica. J Natl Cancer Inst. 2000;92(6):464–474. doi: 10.1093/jnci/92.6.464. [DOI] [PubMed] [Google Scholar]
- Ho L, Chan SY, Burk RD, Das BC, Fujinaga K, Icenogle JP, Kahn T, Kiviat N, Lancaster W, Mavromara-Nazos P, et al. The genetic drift of human papillomavirus type 16 is a means of reconstructing prehistoric viral spread and the movement of ancient human populations. J Virol. 1993;67(11):6413–23. doi: 10.1128/jvi.67.11.6413-6423.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huelsenbeck JP, Ronquist F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17(8):754–5. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- Ishiji T, Lace MJ, Parkkinen S, Anderson RD, Haugen TH, Cripe TP, Xiao JH, Davidson I, Chambon P, Turek LP. Transcriptional enhancer factor (TEF)-1 and its cell-specific co-activator activate human papillomavirus-16 E6 and E7 oncogene transcription in keratinocytes and cervical carcinoma cells. Embo J. 1992;11(6):2271–81. doi: 10.1002/j.1460-2075.1992.tb05286.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson ME, Pennie WD, McCaffery RE, Smith KT, Grindlay GJ, Campo MS. The B subgroup bovine papillomaviruses lack an identifiable E6 open reading frame. Mol Carcinog. 1991;4(5):382–7. doi: 10.1002/mc.2940040510. [DOI] [PubMed] [Google Scholar]
- Munoz N, Bosch FX, de Sanjose S, Herrero R, Castellsague X, Shah KV, Snijders PJ, Meijer CJ. Epidemiologic classification of human papillomavirus types associated with cervical cancer. N Engl J Med. 2003;348(6):518–27. doi: 10.1056/NEJMoa021641. [DOI] [PubMed] [Google Scholar]
- Narechania A, Chen Z, Desalle R, Burk RD. Phylogenetic Incongruence among Oncogenic Genital Alpha Human Papillomaviruses. J Virol. 2005;79(24):15503–15510. doi: 10.1128/JVI.79.24.15503-15510.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong CK, Chan SY, Campo MS, Fujinaga K, Mavromara-Nazos P, Labropoulou V, Pfister H, Tay SK, ter Meulen J, Villa LL, et al. Evolution of human papillomavirus type 18: an ancient phylogenetic root in Africa and intratype diversity reflect coevolution with human ethnic groups. J Virol. 1993;67(11):6424–31. doi: 10.1128/jvi.67.11.6424-6431.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambaut A, Drummond AJ. Tracer version 1.2 [computer program] [Accessed 31 January 2006];2003 aVailable: http://evole.zoo.ox.ac.uk.
- Swofford DL. “PAUP*Phylogenetic analysis Using Parsimony (*and other methods).” version 4 Sinauer. Sunderland, MA: 1998. [Google Scholar]
- Terai M, Burk RD. Characterization of a novel genital human papillomavirus by overlapping PCR: candHPV86 identified in cervicovaginal cells of a woman with cervical neoplasia. J Gen Virol. 2001a;82(Pt 9):2035–40. doi: 10.1099/0022-1317-82-9-2035. [DOI] [PubMed] [Google Scholar]
- Terai M, Burk RD. Complete nucleotide sequence and analysis of a novel human papillomavirus (HPV 84) genome cloned by an overlapping PCR method. Virology. 2001b;279(1):109–15. doi: 10.1006/viro.2000.0716. [DOI] [PubMed] [Google Scholar]
- Terai M, DeSalle R, Burk RD. Lack of canonical E6 and E7 open reading frames in bird papillomaviruses: Fringilla coelebs papillomavirus and Psittacus erithacus timneh papillomavirus. J Virol. 2002;76(19):10020–3. doi: 10.1128/JVI.76.19.10020-10023.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25(24):4876–82. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplement 1. ORF map. Location of the predicted ORFs of (a) HPV101, (b) HPV103, and (c) HPV4. Each ORF is represented as a rectangle occupying either the first, second, or third reading frames. Numbers show the nucleotide positions of the start and stop codons.
Supplement 2. Comparison HPV101 and HPV103 LCRs with other PV types (HaOPV, HPV4, and HPV16). LCR length and position of multiple binding sites are shown as indicated. AP-1, activator protein 1; TEF-1, transcriptional enhancer factor -1; Poly(A), poly(A) signal; E1, E1 binding domain; E2, E2 binding domain. A novel ORF (X) embedded in the LCR of HPV101 is indicated with an arrow.


