Abstract
A majority of cervical cancers are associated with HPV-16. A DNA vaccine (E7IR) was designed for prophylactic and therapeutic treatment of HPV-16+ tumors containing two repeats of the E7 gene to inactivate transformation and duplicate available epitopes. Mice were vaccinated then tumor challenged, or challenged and then immunized and monitored for tumor volume and survival. Splenocytes were utilized for in vivo CTL assays. The E7IR vaccine demonstrated decreased tumor volume and enhanced survival in prophylactic and therapeutic experiments and improved CTL mediated lysis. The E7IR vaccine shows promise in prevention of tumor formation and elimination of established tumors.
Keywords: Human Papillomavirus (HPV), DNA Vaccine, Cervical Cancer
1.Introduction
Cancer of the uterine cervix is the second most common cause of cancer-related deaths in women worldwide. Approximately 510,000 new cases and 288,000 deaths are reported annually [1]. The development of cervical cancer is associated with infection with high risk types of Human Papillomaviruses (HPV). HPV is detected in 99.7% of cervical carcinomas [2], with high risks types HPV-16 and 18 accounting for nearly 70% of cervical cancer cases [3]. In contrast to many other cancers, the fact that cervical cancer is highly associated with a viral infection creates a unique opportunity for therapeutic vaccination against HPV to either prevent cervical cancer once an individual is infected or after the virus has caused cervical cancer. Integration of the viral genome into the host cell genome, a necessary step in cellular transformation, leads to the loss of some genes necessary for a productive viral life cycle and to deregulated or overexpression of the E6 and E7 oncoproteins. Because E7 has been shown to be consistently expressed in HPV-16 positive tumors, it is a logical target in HPV-16 vaccination strategies.
Recent efforts in the field of HPV tumor immunotherapy have focused on the delivery of naked DNA vaccines. DNA based vaccines are attractive candidates because of simplicity of delivery, stability, and overall safety. DNA vaccines based on the E6 and or E7 oncoproteins can either stably integrate into the genome or are maintained in an episomal form allowing for extended expression of HPV antigens. Of course, in this instance mutations in the p53 and pRB binding sites are essential in order to prevent vaccination induced cellular transformation. Furthermore, administration of DNA based vaccines has been shown to induce CTL to HPV antigens [4].
Preclinical studies using DNA based vaccine approaches tend to focus on enhancing presentation of HPV antigens via the MHC class I or MHC class II pathways [5]. Several different approaches have been examined including the attachment of ubiquitin to DNA constructs [6, 7], targeting of tumor antigens to centrosomal compartments to enhance MHC class I presentation [8], the attachment to constructs of calreticulin to target HPV antigens to the MHC I pathway [9], optimization of codon usage for enhanced expression and presentation of HPV oncogenes [6, 10, 11] , utilization of modified genes to decrease the transformation capacity [12-14] or enhancement of intercellular spreading in order to increase the number of cells expressing antigen [15]. All of these approaches have demonstrated increased antigen presentation and superior tumor eradication in mouse models.
The current study was undertaken in order to assess whether insertional replication of the HPV-16 E7 gene could enhance immune responses compared to vaccination with the wild type gene, by increasing the overall number of T cell epitopes while simultaneously providing a safe construct. In this study, this approach did in fact enhance immunity compared to vaccination with the wild-type construct as evidenced by the ability to prevent tumor formation and therapeutically treat existing tumors in C57BL/6 mice and furthermore this effect appears to be CTL mediated as evidenced by increased killing seen in vivo CTL assays. Taken together these results suggest that insertional replication of the E7 gene provides a unique method to ablate the transforming activity of the E7 gene and simultaneously enhance its immunogenicity. This strategy provides an important basic DNA construct for superior vaccination against HPV-16 E7 especially when coupled with current techniques for enhancing vaccine potency.
2.Materials and Methods
2.1 Mouse strains and cell lines
Specific pathogen free 6-10 week old female C57BL/6 mice were purchased from Taconic Farms (Germantown, NY. USA) and housed under SPF conditions, conforming to institutional animal use guidelines at the animal facilities at the University of Southern California. Tumor challenge studies were performed utilizing the HPV-16 E6/E7 expressing TC-1 tumor cell line and cultured as described [16]. Phoenix cells (Orbigen, Inc. San Diego, CA) and NIH 3T3 cells (ATCC number CRL-1658, American Type Culture Collection Manassas, VA) were cultured in DMEM (Invitrogen) containing 10% heat inactivated fetal bovine serum (Omega), 100 U/ml Penicillin, 100 U/ml Streptomycin (Invitrogen), and 2 mM L-Glutamine (Invitrogen).
2.2 Construction of the E7IR vaccine
The native 98 amino acid HPV16 E7 protein contains three regions that were arbitrarily designated as a, b, or c. The E7IR DNA construct was designed containing two repeats of the first, second, and third regions of the E7 gene. The nucleotide sequence was synthesized by Entelechon (Entelechon GmbH, St. Veit-Weg 2, 93051 Regensburg) and was cloned into a PCR4 topo vector and sub-cloned into the APL023 expression vector [7]. In the a region, amino acids 21, 24, and 26 were modified from DLYCYEQ to GLYGYGQ to reduce transformation activity and enhance antigenicity [17, 18]. In the b region, a split was introduced between two cystine residues at position 31 and 32 to reduce transformation activity. In the c region two zinc finger binding motifs were disrupted by a split between amino acids 60 and 61 and modifications of C61 to G61 and C94 to G94. All three regions, a, b, c were replicated to enhance expression, and H-2Db and HLA-A*0201 epitopes were added downstream of the aabbcc construct with Ala-Ala-Tyr (AAY) as an optimal spacer for proteosomal cleavage [7]. In addition to further enhancing expression, addition of these epitopes provided unique primer sites at the 3’ end of the construct that were necessary for cloning. The addition of a Kozak sequence on the 5’ end of the construct, also served to create a unique primer site as well as enhance translation of the construct in eukaryotic cells [19]. Lastly, the codons in the E7IR construct were optimized for expression in mice. The construct was cloned into the multiple cloning site of the APL023 expression vector [7] utilizing Bam H I and Not I restriction enzymes.
2.3 Retroviral Expression of the E7IR gene
PCR of the E7IR gene was performed utilizing the following primers sets 5’ CGT GGA TCC GCC GCC ACC ATG CAT GGA GAC ACA CCA ACC CTG 3’ and 5’ GCT CAG CAC GCC GGC GTT AAT CGA TCT CGA TCT CGA GGG 3’, 5’ CGT GGA TCC GCC GCC ACC ATG TAC CCA TAC GAT GTT CCA GAT TAC GCT ATG CAT GGA GAC ACA CCA ACC CTG 3’ containing a Bam HI restriction site in the forward primer and a Not I restriction site in the reverse primer. The last primer listed was utilized as the forward primer in a separate reaction to generate a HA-tag encoding sequence for use in western blotting analysis. The resultant PCR products were cloned into the LZRSpBMN-Z retroviral expression vector (a kind gift of Dr. Gary Nolan, Stanford University, Palo Alto, CA.). Retroviral infection of Phoenix cells with LZRSpBMN-Z constructs was conducted utilizing the reagents and protocol from Orbigen Inc. San Diego, CA. USA. Briefly, Phoenix cells were transfected by the CaCl2 precipitation method and after 48 hours, 3 ml of harvested retroviral supernatants were utilized to infect 5×105 NIH 3T3 cells. Forty-eight hours later, NIH 3T3 cells were screened for retroviral infectivity by x-gal staining and lac z negative cells were selected by adding 2μg/ml puromycin (EMD Biosciences, Inc., San Diego, CA.).
2.4 Western blots
Cellular protein extracts from transfected NIH 3T3 cells were obtained using the M-Per Mammalian Protein Extraction Kit (Pierce, Rockford, IL.) Thirty μg of protein extract was separated by SDS-PAGE using a 12% Bis-Tris gel (Invitrogen) and transferred onto a PVDF membrane (Bio Rad, Hercules, CA) using a semi dry blotting system (Invitrogen). HA-tagged E7IR was detected with a HA-specific polyclonal antibody (Santa Cruz Biotechnology, Santa Cruz, CA) and the WT E7 and the E7IR proteins were detected with an HPV-16 E7 specific mouse monoclonal antibody (Zymed clone 8C9, Invitrogen). The GAPDH-specific antibody (Chemicon International, Temecula, CA) was used to verify that equal amounts of protein were loaded. Bands were visualized by enhanced chemiluminescence for HRP detection (Pierce).
2.5 Soft Agar Assay
Anchorage-independent growth capability was determined by assessing the colony formation efficiency of cells suspended in soft agar. Transformed and control cells were plated in triplicate dilutions (1×104, 2×104, and 4×104) in 2 ml of 0.3% overlay agar and added to 6 well plates coated with 3 ml of 0.6% underlay agar. Plates were allowed to dry and were incubated at 37°C. Two weeks after the cells were plated; colonies were stained with p-iodonitrotetrazolium violet (Sigma) and counted. The mean transformation efficiency of the E7IR was calculated based on the triplicate dilution series and compared to the mean foci numbers of the E7 wild type.
2.6 In Vivo CTL Assay
On day one of the assay, spleens were harvested from naïve C57BL/6 mice and single cell suspensions were prepared for use as peptide loaded targets. Red blood cells were then lysed by treatment with RBC lysis buffer (Sigma) for 3 minutes and washed twice with IMDM media. Cells were then counted and split into two tubes; one containing the E7 (49-57) H-2Db specific peptide RAHYNIVTF [20] and the other containing prostate specific antigen 5 (PSA 5), VTWIGAAPL, a prostate specific antigen based peptide that binds to Kb and Db [32] as a non-specific peptide at a concentration of 0.5 μg/ml. Peptide labeled cells were then incubated at 37°C for 90 minutes. After incubation, the cells were washed three times in IMDM media, and once in 0.1% BSA in PBS before labeling with CFSE (Molecular Probes, Eugene, OR). Cells were labeled at a concentration of 5 μM/ml for the relevant peptide and 0.25μM/ml for the irrelevant peptide for 10 minutes at 37°C. The reaction was stopped by adding 20 ml ice cold IMDM (CamBrex, Walkersville, MD). Labeled cells were then washed twice in PBS, resuspended at a concentration of 50×106 cells/ml and relevant and irrelevant peptides were mixed in a 1:1 ratio and the peptide coated cells were injected i.v. into vaccinated or control mice. Eighteen hours later, vaccinated and control mice were sacrificed and spleens were harvested. Vaccinated and control spleens were resuspended to 3×106 total cells per sample and run on a Beckman Coulter Cytomics FC 500 Flow Cytometer (Beckman Coulter, Miami, FL). Data was analyzed using CXP software (Beckman Coulter) and percent lysis was calculated based on the equation: (Q-R/Q) × 100 where Q equals the CFSE high labeled relevant peptide and R equals the CFSE low labeled irrelevant peptide.
2.7 DNA-gold bullet preparation and gene gun-mediated delivery
The Helios gene gun system (Bio-Rad, Hercules, CA) was used for intradermal delivery of vaccine and control genes as previously described [7]. Bullets containing 2 μg of DNA/shot were generated according the manufacturer’s protocols. Briefly, 100 μg of DNA was precipitated on 25 mg of 1-mm gold particles in the presence of 100 μl of 0.05 M spermidine (Sigma, St. Louis, MO) using 100 μl of 1 M CaCl2/preparation. The gold was washed three times with 1 ml of 100% 200 proof ethanol (Sigma) and was resuspended in 3 ml of 0.1mg/ml polyvinylpyrolidone in 100% ethanol. The gold was then loaded into the tubing using the tubing prep station (Bio-Rad), and the gold loaded tubing was cut into 0.5-inch pieces to load into the cartridges. The bullet containing cartridges were loaded into the gene gun and delivered into the mouse epidermis at a helium pressure of 450 psi.
2.8 Immunization Schemes
Immune monitoring of the E7IR Vaccine
Mice were anesthetized by i.p. injection of 80/kg of ketamine (Abbott Laboratories, Chicago, IL) and 10 mg/kg of xylazine (Sigma) mixed in 80 μl of PBS. The abdominal area was shaved, and the DNA was delivered into the epidermis. Vaccination was administered at day 0 and 30. Ten days following the last vaccination, mice were sacrificed and spleens were harvested in order to conduct in vivo CTL assays.
Tumor protection experiments
Vaccination was administered at day 0 and 14. At day 21 mice were challenged with 6×104 TC-1 tumor cells in 100 μl of HBSS (Sigma). Individual mice were monitored two or three times a week for 57 days for tumor growth and survival. TC-1 cells were derived from a large batch of TC-1 tumor cells of the same passage, tested for tumor formation, and frozen in liquid nitrogen. For every tumor injection a vial from this batch was thawed and grown according to a standard procedure for 10 days to obtain the required number of tumor cells. On the day of injection, 80% confluent cell cultures were trypsinized (Invitrogen) and washed three times with HBSS (Sigma). Cells were concentrated at 6×105 cells/ml, and 100 μl was injected s.c. in the left flank of the mice.
Tumor therapy
Mice in therapy experiments received a tumor s.c. injection with 2×105 cells/ml of TC-1 cells in 100 μl of HBSS (Sigma) 10 days before DNA introduction. By day 10, all mice had palpable tumors defined as the presence of a small mass bearing an approximate volume of 0.125mm3; DNA administration was initiated on day 10 and repeated 7 days later. During this time the tumor sizes in the mice were recorded two or three times a week.
2.9 Statistical Analysis
Graphs were generated and one tailed T tests were performed using GraphPad Prism version 4.00 for Windows, (GraphPad Software, San Diego California USA, www.graphpad.com). Composite one-tailed P values for tumor challenge studies were generated from two independent experiments with equal variance by combining z values generated from the mean and standard deviation in a Fischer’s method meta-analysis using the program Quick Calcs Graphpad Prism Software (http://www.graphpad.com/quickcalcs).
3.Results
3.1 Analysis, Protein Expression and Transformation Capacity of the E7IR vaccine construct
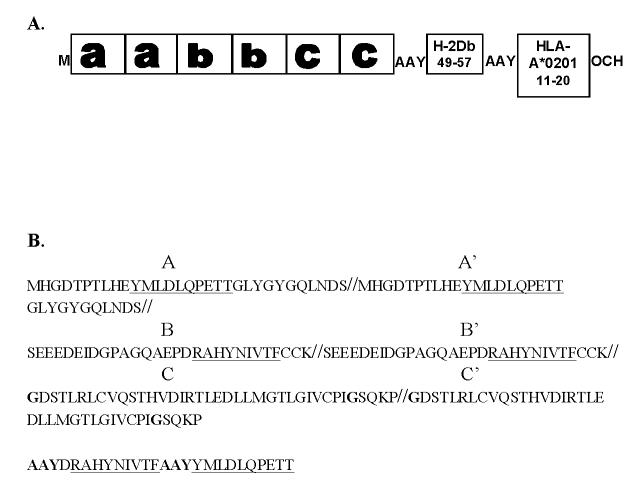
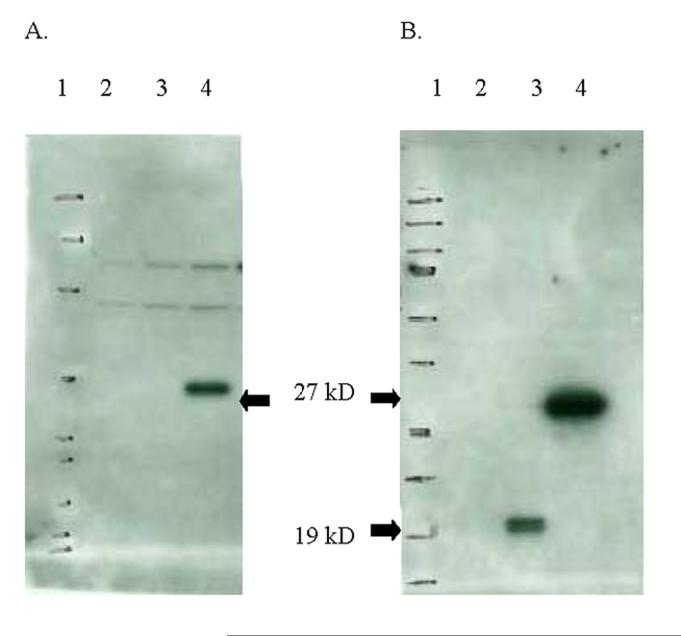
The Construction of the E7IR DNA construct utilized for vaccination studies is represented in figure 1. Protein expression of the E7IR construct in the APL023 mammalian expression vector was confirmed and compared to expression of the HPV-16 E7 WT by western blot analysis using anti-HA antibodies to show the E7IR construct with an HA tag (figure 2 A) and the E7WT and E7IR protein with an HPV-16 E7 antibody (figure 2 B). Retroviral infected NIH 3T3 cells confirmed the background level transformation efficiency of the E7IR construct compared to the wild type in a soft agar transformation assay (figure 3).
Figure 1.
E7IR Vaccine Construction. A: Design of the E7IR vaccine construct. The native E7 protein was split into three arbitrary regions designated as a, b, or c. In the a region amino acids 21, 24, and 26 were modified from DLYCYEQ to GLYGYGQ to reduce transformation activity. In the b region a split was introduced between two cystine residues at position 31 and 32 to reduce transformation activity. In the c region two zinc finger binding motifs were disrupted by a split between amino acids 60 and 61 and modifications of C61 to G61 and C94 to G94 (both in bold). All three regions, a, b, and c were replicated to enhance expression and H-2Db and HLA-A*0201 epitopes were added downstream of the aabbcc construct with AAY as a spacer. Start (M) and stop (OCH) condons were included on the 5’ and 3’ end respectively. Figure 1B shows the amino acid sequence of the vaccine used for vaccination with the human HLA-A*0201 and the H2Db epitopes underlined and with the AAY spacers in bold.
Figure 2.
Protein Expression of the E7IR Vaccine. Cellular protein extracts from transfected NIH 3T3 cells were separated by SDS-PAGE and blotted onto nitrocellulose. Lanes are labeled as follows: lane 1 protein marker, lane 2 empty vector, lane 3 E7WT, and lane 4 E7IR vaccine construct. Panel A shows the 27kD E7IR protein detected with an HA-specific polyclonal antibody. Panel B shows detection of the E7WT protein, lane 3, 19kD and the E7IR protein, lane 4, 27kD with an HPV-16 E7 specific monoclonal antibody.
Figure 3.
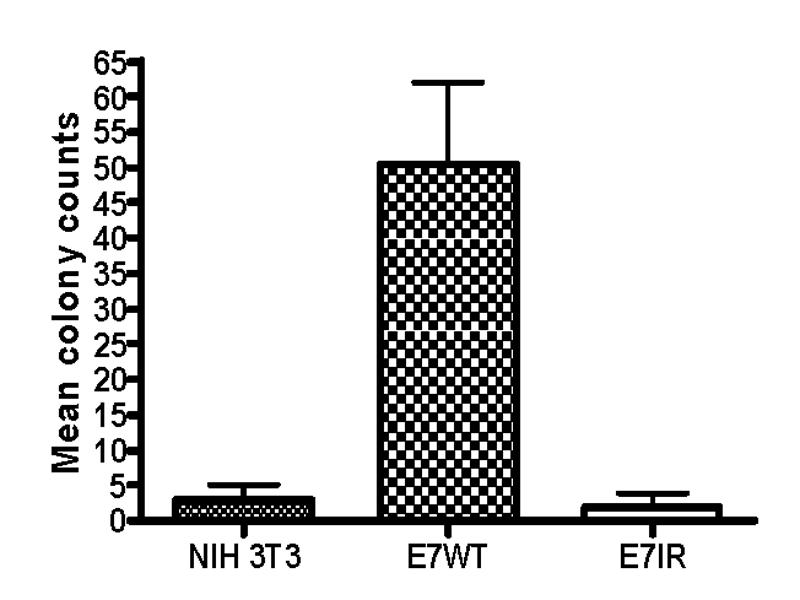
Soft Agar Transformation Assay. Anchorage-independent growth was determined by the colony formation efficiency of cells suspended in soft agar. E7WT and E7IR transformed NIH 3T3 cells were plated in triplicate dilutions (1×104, 2×104, and 4×104) and colonies were stained and counted after a two week incubation. Mean colony counts were determined as a function of normalized triplicates in each dilution series. The E7IR construct is showing a background level of efficiency of transformation compared to the E7WT construct.
3.2 Anti-tumor efficacy of the E7IR vaccine in a prophylactic setting
Groups of five C57BL/6 mice were vaccinated with 4 μg of DNA constructs containing the E7IR gene, the E7WT gene, or the APL023 vector alone twice at two week intervals and challenged with 6x104 TC-1 tumor cells seven days later. The mice were then monitored and measured for 57 days for tumor growth and survival. By day 33 all naïve and vector vaccinated mice were tumor bearing, 3/5 mice vaccinated with the WT construct were tumor bearing and none of the animals vaccinated with the E7IR construct were tumor bearing. As a result, vaccination with the E7IR construct promoted 100% survival in these mice to 57 days (figure 4).
Figure 4.
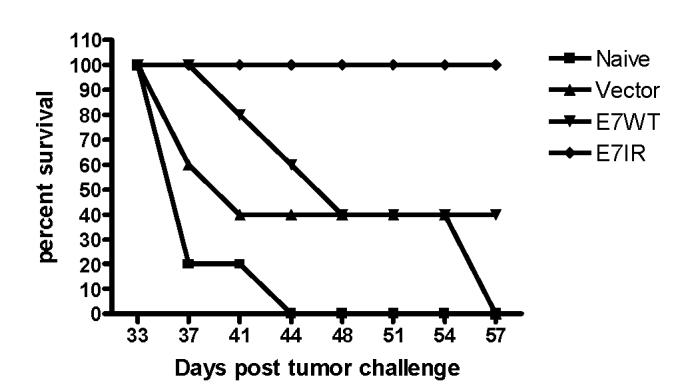
Percent Survival in Prophylactically Vaccinated Mice. Survival was monitored for 57 days in mice prophylactically vaccinated with the E7IR construct. Groups of five mice were vaccinated twice at two week intervals and challenged with TC-1 cells seven days later. At 57 days post TC-1 challenge, 100% of E7IR mice survived compared to 40% of mice vaccinated with the E7WT construct or 0% of naïve or vector alone treated animals. Data is representative of two experiments.
3.3 Efficacy of the E7IR vaccine to treat established tumors
Given the promising results of the prophylactic tumor studies, a therapeutic study was performed to test the ability of the E7IR vaccine to treat existing tumors. Groups of five C57BL/6 mice received 2×104TC-1 tumor cells. After 10 days, all mice had palpable tumors defined as the presence of a small mass bearing an approximate volume of 0.125mm3, and were immunized with either E7WT, vector alone or the E7IR DNA vaccine. Therapeutic treatment was initiated at day 10 and repeated 7 days later. Figure 5 demonstrates that vaccination with the E7IR construct resulted in a decreased tumor growth compared to control groups on day 34, the last time point before mice had to be sacrificed due to overt tumor induced pathology. A repeat of this experiment, combined with the data shown in figure 5 produced a composite p value (p= 0.01) that indicates that vaccination with the E7IR construct showed a significant decrease in tumor volume compared to vaccination with the E7WT construct. Figure 6 indicates that although therapeutic vaccination did not always eradicate the tumor completely, it greatly enhanced survival in these animals past 100 days.
Figure 5.
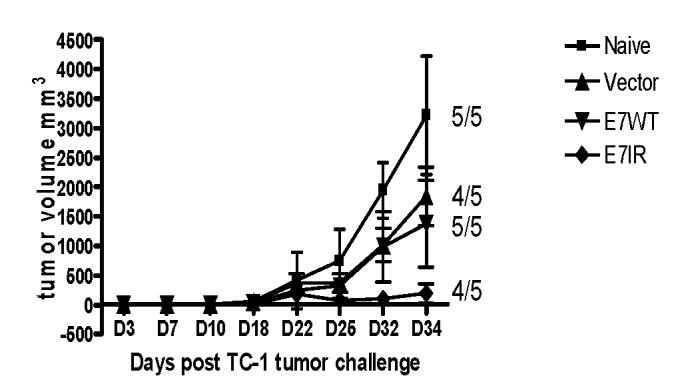
Mean Tumor Volume in Therapeutic E7IR Vaccine Treatment. In therapeutic experiments with the E7IR vaccine, 2×104 TC-1 cells per mouse were injected into 5 mice per group on day 0. After all mice in all groups had a palpable tumor (day 10), the first gene gun vaccination was given, followed by a second vaccination 7 days later. Numbers equal the number of tumor bearing mice in each group at day 34, the last day that all mice were living. Data is representative of two experiments. A composite p value generated from these two experiments indicates that vaccination with the E7IR construct showed a significant decrease in tumor volume compared to vaccination with the E7WT construct (p = 0.01).
Figure 6.
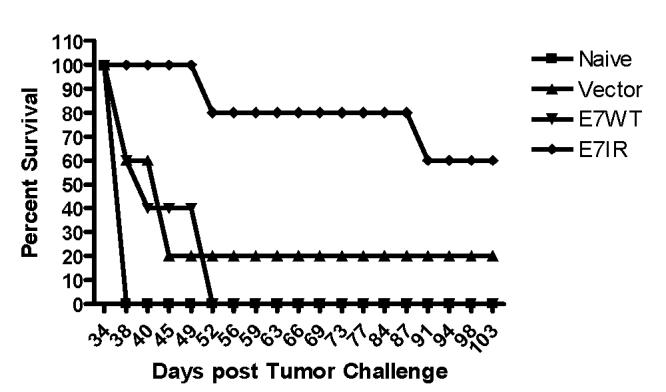
Percent Survival in Therapeutically Treated Mice. Survival was monitored for 103 days in groups of five mice therapeutically treated with the E7IR construct. At 103 days post TC-1 challenge, 60% of E7IR mice survived compared to 0% of naïve or E7WT vaccinated mice. Data is representative of two experiments.
3.4 Cellular immune responses induced by vaccination with E7IR DNA Construct
Immune responses generated by vaccination with the E7IR construct were evaluated in a cellular immune assay. Mice were vaccinated twice with the E7WT, vector alone or E7IR DNA vaccine. Splenocytes from vaccinated and naive animals were then tested for MHC class I-restricted T-cell responses to the RAHYNIVTF H-2Db peptide by in vivo CTL. The in vivo CTL assay measured the specific CTL-mediated killing of naïve splenocyte target cells loaded with the RAHYNIVTF peptide and a PSA5 irrelevant control peptide. Figure 7 demonstrates the percent lysis of loaded targets in the different vaccinated groups based on the remaining ratio of relevant to irrelevant peptide. Mice that were immunized with the E7IR construct showed significantly increased lysis as compared to WT vaccinated mice (27% versus 15%, p=0.0001).
Figure 7.
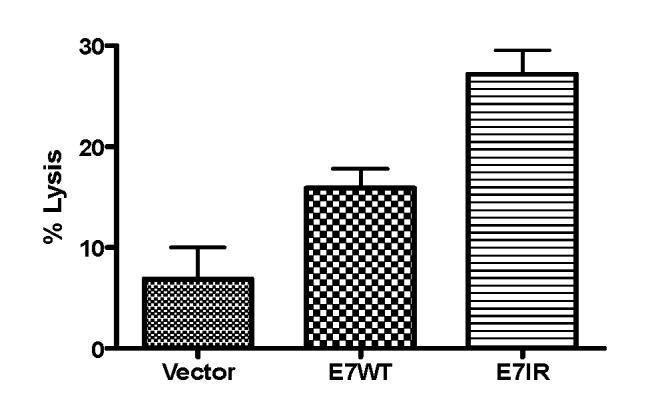
In Vivo CTL. Five mice per group were vaccinated with the E7IR vaccine twice in a thirty day interval, CFSE-labeled target cells loaded with E7 (49-57) peptide or control peptide were injected into vaccinated mice. After 18 hours, splenocytes from vaccinated mice were harvested and flow cytometry was utilized to determine percentages of CFSElow and CFSEhigh target populations and were used to perform an in vivo cytotoxicity assay. Data were normalized compared to background lysis seen in naïve, non-tumor challenged mice. Administration of the E7IR vaccine caused significantly more lysis of target cells than administration of the wild type E7 gene (p=0.0001).
4.Discussion
High risk infections with HPV are highly prevalent but generally transient and are believed to be controlled by the cell mediated immune response since a majority of HPV-induced lesions spontaneously regress [21]. The essential factors necessary for HPV-induced malignant progression are largely unknown but may be related to suboptimal or inappropriate T cell responses to HPV antigens during a natural infection [21]. The human HPV E6 and E7 proteins are selectively expressed in cervical cancer cells, making them excellent targets for T cell mediated immunotherapy [22]. Therefore, the goal of immunization against HPV-induced carcinomas is to activate cellular components of the immune system to recognize and attack cells infected with HPV. Numerous vaccination strategies against HPV-induced tumors have yielded encouraging results in mice. Among them, DNA vaccination has emerged as an attractive strategy because of low production costs, overall stability and ease of storage [14]. Additionally, naked DNA vaccines lack unwanted immune responses directed against other components of the vaccine that is commonly observed in the case of familiar vectors in vector based prime-boost regimens [14, 23]. Lastly, it has been shown that delivery of gold coated DNA particles into the skin will activate dendritic cells and induce migration to the draining lymph nodes to initiate an immune response [14].
The E7IR DNA vaccine candidate presented in the current study is based on the E7 oncogene of HPV-16. HPV-16 is the most common type among cervical cancer cases worldwide with about 54% of cervical carcinomas attributed to the HPV-16 genotype. [24]. As the HPV-16 E7 oncoprotein is constitutively expressed in cervical cancers, it is considered a good target for immune therapy of existing lesions and several E7-specific therapeutic vaccines are currently under development in preclinical models [25]. Additionally, the E7 oncoprotein contains several identified MHC I CTL epitopes [26]. However, although DNA vaccination appears to be a safe means to deliver HPV antigens, DNA vaccination with oncogenes still harbors the risk of transformation for the cells that receive and express the oncogenes [27, 28]. Thus E7 based vaccine strategies seek to simultaneously eliminate or reduce the transforming ability and enhance the immunogenic potential of the E7 oncoprotein.
The E7IR vaccine design attempts to resolve these challenges first by utilizing well characterized mutations that interfere with binding to the host cell Rb protein and by creating a gene coding for an artificial E7 protein that is unlikely to keep its original function through inserted and replicated sequences thus reducing the transformation capacity of the construct [17]. Another commonly utilized method to reduce the transforming capacity of the E7 oncogene has been to create epitope based vaccines typically containing a few common human epitopes [12]. Comparatively, an advantage offered by the E7IR vaccine strategy is that the entire coding region is intact and thus there are no HLA restrictions that require consideration in wide-spread vaccine application. In addition, duplication of the gene sequence should cause duplication of all inherent CTL epitopes and the E7IR construct was designed with codon modifications that favor translation in mice, a strategy that has proved effective in enhancing translation of the E7 gene [10, 29]. Mechanistically, enhanced expression of CTL epitopes is probably accomplished by increased protein production as the western blot in figure 2b would suggest. In the experiments presented here, the immunological effects of the vaccine are likely due to the H-2Db RAHYNIVTF (49-57) epitope, the only E7 CTL epitope described for the C57BL/6 mouse. The in vivo CTL assay shown in figure 7, in which target cells are coated with the RAHYNIVTF peptide, is showing roughly two times higher lysis in the mice vaccinated with the E7IR than those vaccinated with the wild type. Additionally, H-2Db and HLA-A*0201 epitopes were added downstream of the construct using AAY spacers, which has been shown to increase epitope processing resulting in higher CTL precursors [7]. One limitation to this approach in which it was necessary to attach unique sequences onto the 3’ end for the purposes of cloning, is that in it’s current form we can not definitively decipher the contribution that additional H-2Db and HLA-A*0201 epitopes are making to the immune response. However, we felt that in contrast to attaching an irrelevant epitope, attachment of these two well characterized CTL epitopes would serve to maximize vaccine potency. The results indicate that indeed this construct did enhance the immune response to E7. Therefore the E7IR may provide a good and safe backbone representation of mutated E7, especially when combined with other well characterized modifications such as targeting antigens to endosomal/lysosomal compartments [30]. One last advantage of the E7IR vaccine strategy is that it could easily be incorporated into vector based approaches, such as Venezuelan Equine Encephalitis virus (VEE) replicons, for which there is no pre-existing immunity in a majority of the human population and which specifically target dendritic cells in order to initiate an immune response [25]. Other possibilities include combining sophisticated and elegant methods of targeting antigens to the MHC I and II pathways [31] with the E7IR gene as a backbone. Ultimately the pairing of some of these adaptations to the modified E7IR gene, which on its own has demonstrated protection and immunity, could be synergistic and result in a superior DNA vaccine which would be useful in prime boost regimes to treat women with pre-cancerous lesions.
In the present study, vaccination with the modified E7IR construct was able to significantly reduce tumor volume and enhance survival in both prophylactic and therapeutic experiments in mice compared to the WT E7 gene. Additionally, vaccination with E7IR lead to a significant increase in CTL-mediated lysis of peptide loaded target cells compared to vaccination with the WT construct. Taken together these results indicate that this design may represent an innovative way to reduce the transforming capacity and simultaneously enhance the immunogenicity of the HPV-16 E7 gene for use in therapeutic vaccination regimens to treat cervical cancer.
Acknowledgements
Parts of the studies mentioned in this article were supported through grant NCI RO1 CA74397. WM Kast holds the Walter A. Richter Cancer Research Chair. Joeli A. Brinkman holds the John H. Richardson/Margaret Kersten Ponty Endowed Fellowship award of the Achievement Rewards for College Scientists Foundation (ARCS).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference List
- [1].Human Papillomaviruses. World Health Organization; 2004. http://www.who.int/vaccine_research/documents/new_vaccines/en/index8.html [Google Scholar]
- [2].Walboomers JM, Jacobs MV, Manos MM, et al. Human papillomavirus is a necessary cause of invasive cervical cancer worldwide. J.Pathol. 1999;189(1):12–9. doi: 10.1002/(SICI)1096-9896(199909)189:1<12::AID-PATH431>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- [3].Koutsky LA. Epidemiology of genital human papillomavirus infection. American Journal of Medicine. 1997;102(5A):3–8. doi: 10.1016/s0002-9343(97)00177-0. [DOI] [PubMed] [Google Scholar]
- [4].Eiben GL, Da Silva DM, Fausch SC, Le Poole IC, Nishimura MI, Kast WM. Cervical cancer vaccines: recent advances in HPV research. Viral Immunol. 2003;16(2):111–21. doi: 10.1089/088282403322017866. [DOI] [PubMed] [Google Scholar]
- [5].Ji H, Wang TL, Chen CH, et al. Targeting human papillomavirus type 16 E7 to the endosomal/lysosomal compartment enhances the antitumor immunity of DNA vaccines against murine human papillomavirus type 16 E7-expressing tumors. Hum.Gene Ther. 1999;10(17):2727–40. doi: 10.1089/10430349950016474. [DOI] [PubMed] [Google Scholar]
- [6].Liu WJ, Zhao KN, Gao FG, Leggatt GR, Fernando GJ, Frazer IH. Polynucleotide viral vaccines: codon optimisation and ubiquitin conjugation enhances prophylactic and therapeutic efficacy. Vaccine. 2001;20(56):862–9. doi: 10.1016/s0264-410x(01)00406-6. [DOI] [PubMed] [Google Scholar]
- [7].Velders MP, Weijzen S, Eiben GL, et al. Defined flanking spacers and enhanced proteolysis is essential for eradication of established tumors by an epitope string DNA vaccine. J.Immunol. 2001;166(9):5366–73. doi: 10.4049/jimmunol.166.9.5366. [DOI] [PubMed] [Google Scholar]
- [8].Hung CF, Cheng WF, He L, et al. Enhancing major histocompatibility complex class I antigen presentation by targeting antigen to centrosomes. Cancer Res. 2003;63(10):2393–8. [PubMed] [Google Scholar]
- [9].Peng S, Ji H, Trimble C, et al. Development of a DNA vaccine targeting human papillomavirus type 16 oncoprotein E6. J.Virol. 2004;78(16):8468–76. doi: 10.1128/JVI.78.16.8468-8476.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Cid-Arregui A, Juarez V, zur Hausen H. A synthetic E7 gene of human papillomavirus type 16 that yields enhanced expression of the protein in mammalian cells and is useful for DNA immunization studies. J.Virol. 2003;77(8):4928–37. doi: 10.1128/JVI.77.8.4928-4937.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Liu WJ, Gao F, Zhao KN, et al. Codon modified human papillomavirus type 16 E7 DNA vaccine enhances cytotoxic T-lymphocyte induction and anti-tumour activity. Virology. 2002;301(1):43–52. doi: 10.1006/viro.2002.1584. [DOI] [PubMed] [Google Scholar]
- [12].Doan T, Herd K, Ramshaw I, Thomson S, Tindle RW. A polytope DNA vaccine elicits multiple effector and memory CTL responses and protects against human papillomavirus 16 E7-expressing tumour. Cancer Immunol.Immunother. 2005;54(2):71. doi: 10.1007/s00262-004-0544-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Osen W, Peiler T, Ohlschlager P, et al. A DNA vaccine based on a shuffled E7 oncogene of the human papillomavirus type 16 (HPV 16) induces E7-specific cytotoxic T cells but lacks transforming activity. Vaccine. 2001;19(30):4276–86. doi: 10.1016/s0264-410x(01)00154-2. [DOI] [PubMed] [Google Scholar]
- [14].Ohlschlager P, Pes M, Osen W, et al. An improved rearranged Human Papillomavirus Type 16 E7 DNA vaccine candidate (HPV-16 E7SH) induces an E7 wildtype-specific T cell response. Vaccine. 2006;24(15):2880–93. doi: 10.1016/j.vaccine.2005.12.061. [DOI] [PubMed] [Google Scholar]
- [15].Hung CF, He L, Juang J, Lin TJ, Ling M, Wu TC. Improving DNA vaccine potency by linking Marek′s disease virus type 1 VP22 to an antigen. J.Virol. 2002;76(6):2676–82. doi: 10.1128/JVI.76.6.2676-2682.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Lin KY, Guarnieri FG, Staveley-O′Carroll KF, et al. Treatment of established tumors with a novel vaccine that enhances major histocompatibility class II presentation of tumor antigen. Cancer Res. 1996;56(1):21–6. [PubMed] [Google Scholar]
- [17].Cassetti MC, McElhiney SP, Shahabi V, et al. Antitumor efficacy of Venezuelan equine encephalitis virus replicon particles encoding mutated HPV16 E6 and E7 genes. Vaccine. 2004;22(34):520–7. doi: 10.1016/j.vaccine.2003.07.003. [DOI] [PubMed] [Google Scholar]
- [18].Munger K, Basile JR, Duensing S, et al. Biological activities and molecular targets of the human papillomavirus E7 oncoprotein. Oncogene. 2001;20(54):7888–98. doi: 10.1038/sj.onc.1204860. [DOI] [PubMed] [Google Scholar]
- [19].Kozak M. Point mutations define a sequence flanking the AUG initiator codon that modulates translation by eukaryotic ribosomes. Cell. 1986;44(2):283–92. doi: 10.1016/0092-8674(86)90762-2. [DOI] [PubMed] [Google Scholar]
- [20].Feltkamp MC, Smits HL, Vierboom MP, et al. Vaccination with cytotoxic T lymphocyte epitope-containing peptide protects against a tumor induced by human papillomavirus type 16-transformed cells. Eur.J.Immunol. 1993;23(9):2242–9. doi: 10.1002/eji.1830230929. [DOI] [PubMed] [Google Scholar]
- [21].Eiben GL, Velders MP, Kast WM. The cell-mediated immune response to human papillomavirus-induced cervical cancer: implications for immunotherapy. Adv.Cancer Res. 2002;86:113–48. doi: 10.1016/s0065-230x(02)86004-3. [DOI] [PubMed] [Google Scholar]
- [22].Durst M, Glitz D, Schneider A, zur Hausen H. Human papillomavirus type 16 (HPV 16) gene expression and DNA replication in cervical neoplasia: analysis by in situ hybridization. Virology. 1992;189(1):132–40. doi: 10.1016/0042-6822(92)90688-l. [DOI] [PubMed] [Google Scholar]
- [23].Liu MA. Vaccine developments. Nat.Med. 1998;4(5 Suppl):515–9. doi: 10.1038/nm0598supp-515. [DOI] [PubMed] [Google Scholar]
- [24].Munoz N, Bosch FX, de Sanjose S, et al. Epidemiologic classification of human papillomavirus types associated with cervical cancer. N.Engl.J.Med. 2003;348(6):518–27. doi: 10.1056/NEJMoa021641. [DOI] [PubMed] [Google Scholar]
- [25].Brinkman JA, Caffrey AS, Muderspach LI, Roman LD, Kast WM. The impact of anti HPV vaccination on cervical cancer incidence and HPV induced cervical lesions: consequences for clinical management. Eur.J.Gynaecol.Oncol. 2005;26(2):42. [PubMed] [Google Scholar]
- [26].Kast WM, Brandt RM, Sidney J, et al. Role of HLA-A motifs in identification of potential CTL epitopes in human papillomavirus type 16 E6 and E7 proteins. J.Immunol. 1994;152(8):3904–12. [PubMed] [Google Scholar]
- [27].Munger K, Phelps WC, Bubb V, Howley PM, Schlegel R. The E6 and E7 genes of the human papillomavirus type 16 together are necessary and sufficient for transformation of primary human keratinocytes. J.Virol. 1989;63(10):4417–21. doi: 10.1128/jvi.63.10.4417-4421.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Phelps WC, Yee CL, Munger K, Howley PM. The human papillomavirus type 16 E7 gene encodes transactivation and transformation functions similar to those of adenovirus E1A. Cell. 1988;53(4):539–47. doi: 10.1016/0092-8674(88)90570-3. [DOI] [PubMed] [Google Scholar]
- [29].Steinberg T, Ohlschlager P, Sehr P, Osen W, Gissmann L. Modification of HPV 16 E7 genes: correlation between the level of protein expression and CTL response after immunization of C57BL/6 mice. Vaccine. 2005;23(9):1149–57. doi: 10.1016/j.vaccine.2004.08.027. [DOI] [PubMed] [Google Scholar]
- [30].Kang TH, Lee JH, Bae HC, et al. Enhancement of dendritic cell-based vaccine potency by targeting antigen to endosomal/lysosomal compartments. Immunol.Lett. 2006;106(2):126–34. doi: 10.1016/j.imlet.2006.05.004. [DOI] [PubMed] [Google Scholar]
- [31].Hung CF, Yang M, Wu TC. Modifying professional antigen-presenting cells to enhance DNA vaccine potency. Methods Mol.Med. 2006;127:199–220. doi: 10.1385/1-59745-168-1:199. [DOI] [PubMed] [Google Scholar]
- [32].Koh YT, Higgins SE, Weber JS, Kast WM. Comparison of immune responses induced by peptide based vaccines emulsified through three different techniques in incomplete Freund’s adjuvant. J. Transl. Med. 2006;4:42. doi: 10.1186/1479-5876-4-42. [DOI] [PMC free article] [PubMed] [Google Scholar]