Abstract
The groESL operon of Bartonella bacilliformis, a facultative intracellular, Gram-negative bacterium and etiologic agent of Oroya Fever, was characterized. Sequence analysis revealed an operon containing two genes of 294 (groES) and 1632 nucleotides (groEL) separated by a 55-nt intergenic spacer. The operon is preceded by a 72-nt ORF (ORF1) that encodes a hypothetical protein with homology to a portion of the HrcA repressor for groESL. A divergent fumarate hydratase C (fumC) gene lies further upstream. Deduced amino acid sequences for B. bacilliformis GroEL and GroES revealed a high degree of identity with homologues from other Bartonella and α-Protebacteria. A single transcriptional start site (TSS) was mapped 79 nucleotides upstream of the groES start codon, regardless of incubation temperature. The TSS was located immediately 5′ to a potential controlling inverted repeat of chaperonin expression (CIRCE) element and is preceded by a σ70-like promoter. The operon is followed by a predicted rho-independent transcriptional terminator. Northern blot analysis indicated that groES and groEL are co-transcribed as a single mRNA of ∼2.4 kb. A 6-h time course analysis by qRT-PCR showed that groEL expression increases 1.3-fold within 30 min of a temperature upshift from 30 to 37 °C, with maximum transcription reached after 60 min (∼4.3-fold), followed by a steady decrease to background (30 °C) transcription levels by 6 h. Western blot analysis revealed a 1.4- and 1.5-fold increase in GroEL synthesis following a temperature upshift or by inhibiting DNA supercoiling with coumermycin A1, respectively. Functional expression and complementation of temperature-sensitive Escherichia coli groES or groEL mutants with the cloned operon allowed them to grow at otherwise restrictive temperatures.
Keywords: GroESL operon, Bartonella, Heat shock protein, Chaperonin, GroEL, GroES
1. Introduction
Bartonella bacilliformis is a facultative intracellular bacterial parasite of human erythrocytes and endothelial cells. The pathogen is transmitted between humans by the bite of phlebotamine sand flies and is responsible for Oroya fever and verruga peruana; life-threatening diseases endemic to Colombia, Ecuador, Chile and Peru (Brenner et al., 1991). Bartonellosis is a biphasic disease with remarkably disparate syndromes. During the acute/primary phase, the bacteria attach to and invade nearly all circulating erythrocytes (Benson et al., 1986). Because approximately 80% of the infected red cells are culled during acute infection, a life-threatening hemolytic anemia can result. During the secondary/chronic phase, the bacterium associates with endothelial cells of the vasculature (Benson et al., 1986) and this stage of the disease is accompanied by angiomatous skin eruptions, termed verruga peruana. The verrugae tend to cluster on the head and limbs of the patient and can last for months to years. B. bacilliformis produces a protein that triggers proliferation of human endothelial cells and it is believed to enhance formation of the vascular lesions (Garcia et al., 1990; Minnick et al., 2003b).
B. bacilliformis' parasitic strategy relies on an insect vector and a subsequent intracellular location in the human host, conditions that are accompanied by a variety of environmental stresses including reactive oxygen species, pH or temperature changes and hemin fluctuations (Coleman and Minnick, 2003). One counter-measure used by all organisms against environmental stress is the heat shock response, whose essential components include GroES (Hsp10) and GroEL (Hsp60) (Lindquist and Craig, 1988; Mayhew and Hartl, 1996). Although referred to as heat shock components, groES and groEL genes are essential for growth at all temperatures and are constitutively expressed. GroES and GroEL are co-chaperonins involved in the correct folding of polypeptides. GroEL is composed of two rings consisting of seven subunits of approximately 57 kDa each, while GroES is a heptamer comprised of 10-kDa subunits (Sigler et al., 1998). Recent studies have shown that the apical domain of GroEL binds polypeptides after their release from the ribosome and hydrophobic forces and ATP help drive this interaction (Weissman et al., 1996; Sigler et al., 1998). When GroEL contacts GroES through a flexible, hydrophobic loop, the volume of GroEL's central channel doubles as the apical domains open upward and outward (Weissman et al., 1996). ATP and GroES bind to the GroEL ring containing the polypeptide to create a stable GroEL–GroES cis assembly (Sigler et al., 1998) and, once bound, the polypeptide is released into the central cavity to begin folding (Chaudhry et al., 2003).
Our laboratory recently showed that the B. bacilliformis GroEL protein contributes to the enhanced proliferation observed when cultured human vascular endothelial cells (HUVECs) are treated with the soluble fraction of a B. bacilliformis cell lysate (Minnick et al., 2003b). Data showed that HUVEC mitogenicity of the bacterial lysate: a) was augmented by upshifting the growth temperature of the bacterial source culture, b) could be inhibited by antibodies specific to GroEL, and c) correlated with levels of GroEL in the fraction supplement. In addition, GroEL is actively secreted by B. bacilliformis and is found in both soluble and insoluble fractions of the bacterium (Minnick et al., 2003b). To continue exploring GroEL's involvement in human endothelial cell proliferation and its suspected role in pathogenesis, we characterized a Bartonella groESL operon and analyzed its expression in response to thermal stress; a biologically relevant environmental cue for this bacterial pathogen.
2. Materials and methods
2.1. Bacterial strains and plasmids
Bacterial strains used in this study are listed in Table 1. B. bacilliformis strains KC583 and LSS001 were cultivated on heart infusion agar containing 5% sheep erythrocytes (HIAB) at 30 °C with 100% relative humidity and were harvested at 4 days post-inoculation. E. coli TOP10F' (Invitrogen) was used as a host for plasmid propagation and was grown in Luria Bertani (LB) medium for 16 h at 37 °C. Temperature-sensitive mutants of E. coli (strains CG2244 and NRK117) were used in groES and groEL complementation studies. Antibiotics including kanamycin (25 µg ml−1), ampicillin (100 µg ml−1), tetracycline (10 µg ml−1) or coumermycin A1 (0.05 µg ml−1) were added to the growth medium, as required.
Table 1.
Bacterial strains and plasmids used in this study
| Strain/plasmid | Relevant genotype/description | Source/reference |
|---|---|---|
| Bartonella | ||
| B. bacilliformis | KC583 (neotype strain) | ATCC/Brenner et al., 1991 |
| LSS001 (control strain for groESL construct) | Minnick et al., 2003b | |
| E. coli | ||
| TOP10F′ | Host for plasmid propagation | Invitrogen |
| CG2244 | B178, groES619, TcR | Landry et al., 1993 |
| NRK117 | groEL44 | Kusukawa et al., 1989 |
| Plasmids | ||
| pBK-CMV | Expression vector | Stratagene |
| pCR2.1-TOPO | High copy-number TA cloning vector | Invitrogen |
| pACYC177 | Low copy-number cloning vector | New England Biolabs |
| pGROESL-TOPO | B. bacilliformis groESL operon cloned into pCR2.1-TOPO | Minnick et al., 2003b |
| pGROESL177 | B. bacilliformis groESL operon cloned into pACYC177 | This study |
| pGROESL-CMV | B. bacilliformis groESL operon cloned into pBK-CMV | Minnick et al., 2003b |
2.2. Nucleic acid preparation and manipulation
Plasmids used in this study are described in Table 1. The plasmid, pGROESL-CMV, contains a functional groESL operon derived from a λ ZAP expression library of B. bacilliformis KC583 and was previously described (Minnick et al., 2003b). pGROESL-CMV served as a template to PCR amplify the operon using primers: BbGroESL_Operon-For-TTAAATCGACGTATAAAGAAATTTG and BbGroESL_Operon-Rev-CTTTTACAATGCAGCTCTTTCAATG (Sigma Genosys). The resulting amplicon was recloned into pCR2.1-TOPO (Invitrogen) to produce pGROESL-TOPO, a plasmid with minimal flanking sequences and with the operon in opposite orientation to the vector's lacZ' promoter. Plasmids were routinely purified and screened using a Midi Prep Kit (Qiagen). DNA sequencing templates were purified with a Wizard Plasmid Prep kit (Promega). Total RNA was isolated using a RiboPure Bacteria kit as instructed by the manufacturer (Ambion). The mRNA was subsequently purified from total RNA using a MICROBexpress kit (Ambion). Sequencing template and RNA concentrations were determined at 260 nm using a Spectronic Genesys 2 spectrophotometer (Milton Roy). DNA fragments and amplicons were purified from ethidium bromide-stained gels using a GeneClean II kit per the manufacturer’s instructions (QBIOgene). Chemical transformation of E. coli was done by standard methods (Ausubel et al., 1995).
2.3. Automated DNA sequencing and analysis
Approximately 100 ng template DNA was sequenced in both strands using oligodeoxynucleotide primers (Sigma Genosys), a BigDye Terminator Cycle Sequencing Ready Reaction kit (ABI/Roche) and an automated DNA sequencer (ABI; model 377). Resulting chromatograms were analyzed using Chromas software (Technelysium). Sequence data were analyzed using a variety of software including BLAST (http://www.ncbi.nlm.nih.gov/BLAST/), Mfold 3.0 (http://mfold.burnet.edu.au/), the Sequence Manipulation Suite (http://www.ualberta.ca/∼stothard/javascript/index.html) and the BCM Search Launcher (http://searchlauncher.bcm.tmc.edu/).
2.4. Transcriptional start site determination
The transcriptional start site for the B. bacilliformis groESL operon was determined using a 5′ RACE System, Version 2.0, per the manufacturer's instructions (Invitrogen). RACE was performed on RNA samples isolated from temperature-upshifted cultures (30 to 37 °C for 30 min) or control samples (30 °C) as above. [Note: a temperature upshift from 30 °C (normal in vitro growth temperature) to 37 °C for 30 min induces several novel 35S-labeled gene products when organisms are pulse-labeled with 35Smethionine and total proteins are analyzed by two-dimensional SDS-PAGE (Minnick; unpublished results)]. RACE was done using GROESLREV2 primer (which anneals 920 nucleotides downstream of the groES start codon): GATCAAACTGCATTCCTTC, to generate cDNA from RNA in a single primer reaction. A second primer, BBGROESREV1: TCCCACCAGCTGTCTTG was used in combination with the 5′ RACE Abridged Anchor Primer-(AAP) GGCCACGCGTCGACTAGTACGGGIIGGGIIGGGIIG to convert TdT C-tailed cDNA into double-stranded DNA. Dilution of the resulting DNA product and a second PCR reaction with BBGROESREV1and AAP resulted in single 200-bp PCR products, on ethidium bromide-stained agarose gels, that were subsequently purified and cloned into pCR2.1TOPO. Three clones per temperature regimen were sequenced as above using M13R and T7 primers.
2.5. Southern blots, Northern blots and qRT-PCR
Southern blots were done at high stringency (∼7% DNA mismatch) using a 32P-labeled groEL-specific probe produced by PCR with the primers BbGroelFor-ATGGCTGCTAAAGAAGTAAAATTTG and BbGroelRev-TTAGAAATCCATTCCGCCCATTC. Southern blotting, probe labeling by random primer extension and washing were done by standard protocols (Ausubel et al., 1995).
B. bacilliformis cultures were temperature-upshifted by transferring culture plates from a 30 to 37 °C incubator and incubating for 0.5–6 h. Plates were then harvested into 350 µl of cold, heart infusion broth and centrifuged for 5 min at 5000 ×g/4 °C. Pellets were resuspended in 350 µl cold RNAwiz (Ambion) to inhibit RNases. Cultures maintained at 30 °C were harvested in parallel, as a control.
For Northern blots, a 255-bp groEL-specific probe was generated by PCR using the primers GroESLFor2/M13F3-CTATTTCTGCTAATGGCG and T7GroELR-TAATACGACTCACTATAGGGAGACAAAGATTGC. The T7GroELR primer has a 23-base T7 promoter sequence added to the 5′ end for in vitro synthesis of a transcript. The resulting RNA probe was labeled with [α-32P] CTP (Perkin Elmer) using a MAXIscript in vitro transcription kit as instructed (Ambion). Unincorporated nucleotides were removed using a NucAway kit (Ambion). The isolated mRNA and the labeled probe were used in a Northern blot following the NorthernMax-Gly kit protocol (Ambion).
qRT-PCR was essentially done as previously described (Minnick et al., 2003a) using a 96-well format and a TaqMan 7700 system (ABI). RNA was prepared as above from control (30 °C) and temperature-upshifted cultures (37 °C for 0.5–6 h). The primers for groEL consisted of BbGroEL Forward: CTTAAGCGCGGAATTGATGC, BbGroEL Reverse: CTGAGGTTTGGATCTTCTTCGC (ABI) and were used with a fluorescent probe: CTGTTGAAGCGGTTGTTGCAGATCTTTTC, covalently linked at its 5′ end with 5-Carboxyfluorescein and at its 3′ end with N,N′,N′-tetramethyl-6-Carboxyrhodamine (Sigma). The control primer/probe set for the 16S rRNA was previously described (Minnick et al., 2003a). Raw data were analyzed using SDS software version 1.7 (ABI).
2.6. SDS-PAGE and Western blots
B. bacilliformis cells were harvested at 6 h following transfer of the culture plates from 30 to 37 °C. Cultures maintained at 30 °C were harvested in parallel as a control. B. bacilliformis cultures grown on HIAB plates supplemented with coumermycin A1 (0.05 µg ml−1) were also prepared to determine whether gyrase inhibition would enhance GroEL synthesis as previously observed in E. coli (Mizushima et al., 1993). Protein concentrations were determined with a BCA assay (Pierce). SDS-PAGE and Coomassie blue staining were done using standard methods (Ausubel et al., 1995), 20 µg protein per lane and 12.5% (ω / ν) or 7–15% (ω / ν) continuous gradient acrylamide gels. For Western blots, unfixed gels were electrophoretically transferred to nitrocellulose filters (0.45 µm pore size) by standard protocol (Ausubel et al., 1995). Resulting blots were probed 2 h at 25 °C with rabbit anti-B. bacilliformis GroEL polyclonal antiserum (1 :1000 dilution), as previously described (Minnick et al., 2003b), and reactive bands were visualized as before using goat anti-rabbit IgG-horse-radish peroxidase-conjugated antibody (Sigma) (1 :1000 dilution), a hydrogen peroxide substrate and 4-chloronapthol as the chromogen. Densitometry of immunoblots was performed using a UMAX Astra 1200S scanner and OneDscan software (Scanalytics).
2.7. Functional expression and complementation analysis
E. coli groES and groEL mutants (CG2244 and NRK117) were complemented in trans with the cloned B. bacilliformis groESL operon by transforming with pGROESL-TOPO or pGROESL177, (high- and low-copy number plasmids, respectively). Transformants were screened for plasmid content as above and subsequently tested on selective LB plates at both permissive and restrictive temperatures (determined to be 30 and 47 °C, respectively). E. coli NRK117 was complemented in trans in a similar fashion, at a restrictive temperature determined to be 44 °C.
3. Results
3.1. Sequence analysis of the B. bacilliformis groESL operon
The entire groESL operon of B. bacilliformis was sequenced in both strands and its linkage map is shown in Fig. 1 (GenBank accession no. AY664491). The operon is preceded by an unusual 72-bp open reading frame (ORF1) encoding a hypothetical 24-residue protein with a 71% identity to a 14-amino acid stretch (residues 100–113) of the 392-amino acid heat shock gene repressor and negative regulator of a controlling inverted repeat of chaperonin expression (CIRCE; Zuber and Schumann, 1994) element (HrcA) from Chlamydia muridarum (GenBank accession no. U52216). A divergent gene, fumC, is located 282 bp upstream of groES and, based on amino acid sequence homology, encodes fumarate hydratase C. The groES gene is 294 nucleotides long and encodes a protein of 98 amino acids with a predicted molecular mass of 10.76 kDa. A diagram showing the unusual region upstream of the operon including ORF1 and groES gene sequences is shown in Fig. 2A. To our knowledge, this is the first sequence analysis of a B. bacilliformis groES gene and its flanking sequences. The B. henselae and B. quintana groES genes are of the same length as the B. bacilliformis gene. The B. bacilliformis GroES protein is most closely related to B. henselae and B. quintana homologues with 89% and 90% identity, respectively, but shares only 51.0% identity to E. coli GroES (Fig. 3A).
Fig. 1.
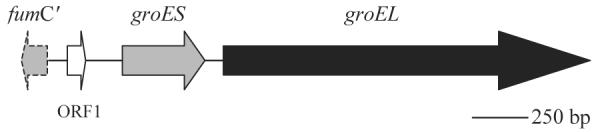
Linkage map of the groESL operon of B. bacilliformis. Closed, continuous arrows indicate positions of the open reading frames. The dashed arrow indicates a predicted, partial and divergent fumarate hydratase c-encoding gene (fumC′) upstream.
Fig. 2.
Nucleotide sequence of the B. bacilliformis groESL operon. A) Sequence of the region 5′ to the operon showing fumC′ and ORF1, followed by groES and the 5′ end of groEL. B) Sequence of the 3′ end of the operon and region immediately downstream with 12-base inverted repeats (arrows) comprising a predicted rho factor-independent terminator. (The complete groEL sequence is listed as GenBank accession no. AY664491.) Corresponding, deduced amino acids are shown below their respective codons. Putative-35 and -10 regions of the σ70-like promoter are indicated. The potential Shine – Dalgarno site (SD) for each gene is shown. The TSS determined by RACE analysis is shown by a down arrowhead. Diamond arrows designate the potential CIRCE element.
Fig. 3.
Comparison of the deduced amino acid sequences of: (A) GroES and (B) GroEL proteins from B. bacilliformis (Bb; this study; GenBank accession no. AY664491) with the closest homologues in the database, i.e. B. henselae (Bh; GenBank accession no. AJ749669 and NC005956) and B. quintana (Bq; GenBank accession no. U78515 and NC005955), plus E. coli (Ec; GenBank accession no.U14003) for comparison.
The B. bacilliformis groEL gene is 1632 nucleotides long and encodes a protein of 544 amino acid residues, with a predicted molecular mass of 57.5 kDa. The B. bacilliformis GroEL is most closely related to homologues from B. henselae and B. quintana (Haake et al., 1997; Alsmark et al., 2004) and shares about 92% identity with both proteins. B. bacilliformis GroEL shares roughly 86% identity to homologues from related Brucella species but only 66% identity with E. coli GroEL (Fig. 3B). The B. bacilliformis GroEL protein from this study is identical to the immunoreactive Bb63 protein of B. bacilliformis (unpublished; GenBank accession no. M98257). A GMR motif (Gly-Gly-Met repeats) occurs in the C-terminus of nearly all GroEL proteins (McLennan et al., 1993) and this motif is conserved in the Bartonella homologues as well (see Fig. 3B).
3.2. Regulatory elements of the operon
The transcriptional start site of the B. bacilliformis operon was determined by RACE analysis and located 79 bases upstream of the start codon of groES, a position one base 5′ to the potential CIRCE regulatory region (Fig. 2A). The TSS was the same regardless of ambient temperature (30 or 37 °C). A potential promoter that closely resembles an E. coli (σ70) consensus promoter is located just upstream of the TSS (TTGACA-N19-TATAAC). The putative CIRCE element (TTGGCGCTC-N9-GAGTGCTAA) is located 78 bp upstream of the groES initiation codon (Fig. 2A) and is nearly identical to CIRCE elements from other α-Proteo-bacteria (Babst et al., 1996). Examination of genomic sequence data for other Bartonella also reveals potential CIRCE elements [TTAGCACTC-N9-GAG(C/T)GCTAA] located 76 and 78 nucleotides upstream of B. henselae and B. quintana groESL operons, respectively, with a C-to-T substitution in the B. quintana CIRCE as indicated.
A 12-base, imperfect, inverted repeat likely constituting a rho-independent transcriptional termination signal is found 23 nucleotides downstream of the B. bacilliformis groEL stop codon (Fig. 2B). The secondary structure of the predicted RNA stem loop structure formed from this inverted repeat has a ΔG of −17.0 kkal/mol by Mfold and the stem loop is immediately followed by 12 Us and 2 As in a length of 17 nucleotides. Potential rho-independent transcriptional termination signals can be found 3′ to the B. henselae and B. quintana groESL operons as well (data not shown).
3.3. B. bacilliformis contains a single groEL gene expressed on a polycistronic transcript
Previous studies have shown that bacteria that are closely related to Bartonella may contain more than one copy of groEL (Babst et al., 1996). Further, there are discrepancies between two unpublished B. bacilliformis groEL sequences previously deposited in GenBank (accession nos. Z15160 and M98257), which may reflect sequence variation in different copies of the gene. To examine this possibility, we performed DNA hybridizations at high stringency. Southern blotting of B. bacilliformis chromosomal DNA restriction endonuclease digested with KpnI or XbaI revealed single hybridization signal bands (data not shown). The groEL gene was also PCR-amplified from B. bacilliformis chromosomal DNA and the resulting amplicons cloned into pCR2.1-TOPO (Invitrogen). Sequence analysis of inserts from twenty randomly selected TOPO clones revealed perfect identity to the groEL sequence obtained in this study and to the B. bacilliformis immunoreactive Bb63 (GenBank accession no. M98257) (data not shown). Taken as a whole, these data suggest that B. bacilliformis contains a single copy of the groEL gene and this conclusion is in agreement with very recent genome sequence data for two other Bartonella species (Alsmark et al., 2004). Northern blot analysis of B. bacilliformis mRNA revealed a full-length and therefore polycistronic transcript of approximately 2.4 kb, together with a smaller 1.9-kb RNA fragment (Fig. 4). The size of the full-length transcript estimated from Northern blots is similar to that predicted from analysis of the corresponding sequence from the TSS to the end of the potential rho-factor independent terminator (i.e., 2116 nucleotides).
Fig. 4.
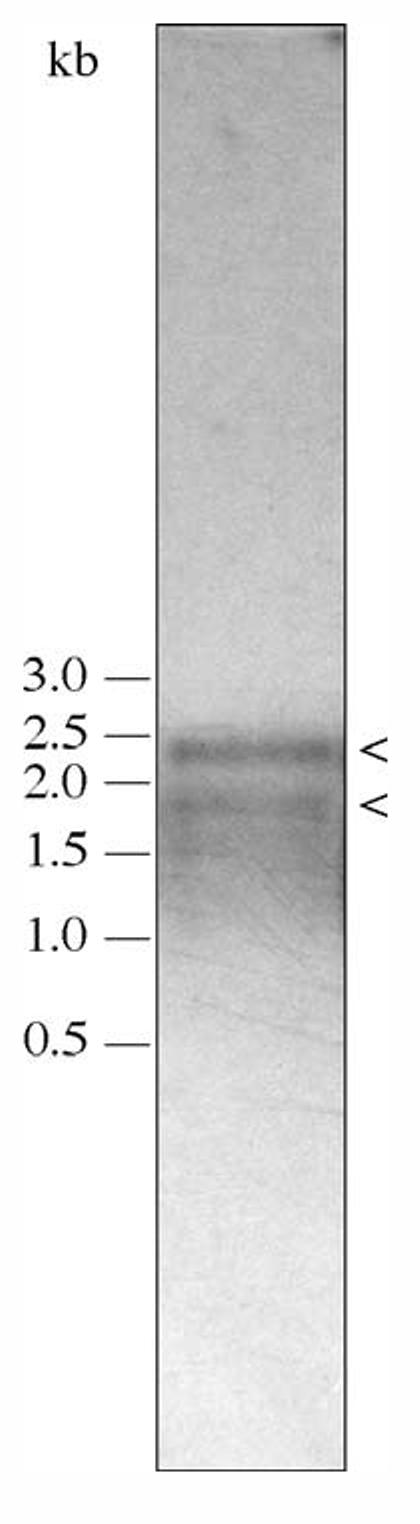
Size of the B. bacilliformis groESL transcript as determined by Northern blot analysis. The polycistronic, full-length transcript is ∼2.4 kb in size (arrowed). A ∼1.9-kb cleavage product is also indicated (arrowed). RNA on the blot was prepared from a culture grown at 30 °C. A 1.1-µg total RNA sample is shown.
3.4. Thermal stress and DNA topology—effect on groEL expression and GroEL synthesis
To determine if expression of the B. bacilliformis operon is induced by thermal stress, in vivo transcription was assayed using qRT-PCR. Total RNA was isolated from bartonellae subjected to 30 or 37 °C temperatures. qRT-PCR analysis showed a ∼1.3-fold increase in groEL transcript as compared to 30 °C controls within 30 min of incubation. Transcription increased to a maximum of 4.25-fold at 60 min and subsequently fell to background (30 °C) levels by 6 h (Table 2). On Western blots, the B. bacilliformis GroEL has an estimated mass of 61 kDa and temperature-upshifted cultures showed a 1.4-fold increase in GroEL protein levels after 6 h of incubation, as compared to controls (Fig. 5).
Table 2.
qRT-PCRa of B. bacilliformis groEL and 16S rDNA during a temperature upshift timecourse
| Target gene (time at 37 °C) | bAvg. 37 °C CT ± SEM | ΔCTqc | ΔΔCTd | Fold increase at 37 °C relative to 30 °C (2−ΔΔCT) |
|---|---|---|---|---|
| 16s rDNA (0.5 h) | 15.15±0.12 | 0.27 | 0 | 1.00 |
| groEL (0.5 h) | 20.63±0.05 | −0.06 | −0.33 | 1.26 |
| 16s rDNA (1.0 h) | 15.91±0.17 | 1.03 | 0 | 1.00 |
| groEL (1.0 h) | 19.63±0.14 | −1.06 | −2.09 | 4.25 |
| 16s rDNA (1.5 h) | 15.45±0.18 | 0.57 | 0 | 1.00 |
| groEL (1.5 h) | 19.68±0.04 | −1.01 | −1.58 | 2.99 |
| 16s rDNA (3.0 h) | 14.34±0.18 | −0.54 | 0 | 1.00 |
| groEL (3.0 h) | 19.72±0.18 | −0.97 | −0.43 | 1.34 |
| 16s rDNA (6.0 h) | 15.25±0.11 | 0.37 | 0 | 1.00 |
| groEL (6.0 h) | 21.05±0 | 0.36 | −0.01 | 1.00 |
Reactions were performed in triplicate and analyzed as previously described (Minnick et al., 2003a). A representative data set from six independent determinations is shown.
0.7 ng total RNA per 25 μl reaction.
Avg. 37 °C CT −Avg. 30 °C CT (where Avg. 30 °C CT for 16s rDNA= 14.88±0.24 and for groEL = 20.69±0.28).
Avg. ΔCTq −Avg. ΔCT16s.
Fig. 5.
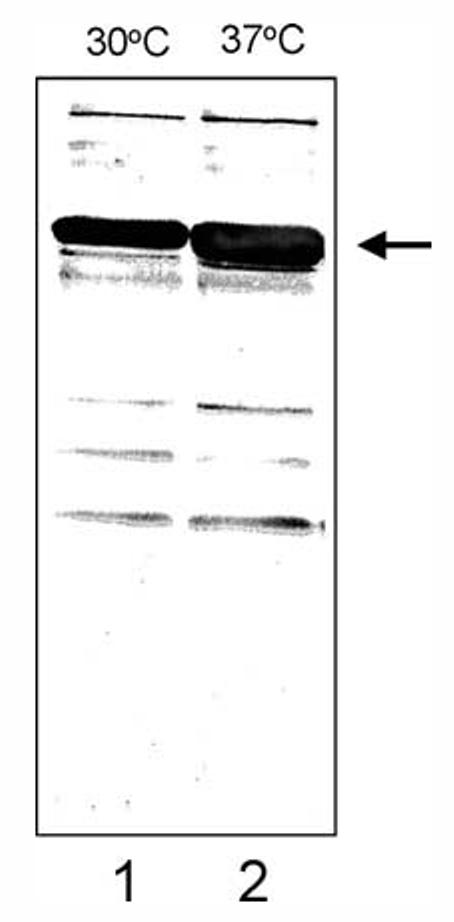
Western blot showing increased GroEL synthesis in B. bacilliformis (LSS001) in response to thermal stress. A blot reacted with rabbit anti-B. bacilliformis GroEL antiserum is shown. Densitometry of the GroEL protein on the immunoblot (arrowed) revealed a ∼1.4-fold increase in GroEL protein during a 6-h temperature upshift from 30 to 37 °C.
Immunoblots were also used to examine GroEL protein levels in response to coumermycin A1, a drug that inhibits DNA supercoiling reactions catalyzed by DNA gyrase. Earlier work with E. coli showed that gyrA mutation or inhibition of gyrase by nalidixic acid or novobiocin correlated with an increased synthesis of GroEL and DnaK (Mizushima et al., 1993). Similarly, B. bacilliformis cultures grown in the presence of coumermycin A1 showed a ∼1.5-fold increase in GroEL levels compared to controls when analyzed by Western blots (data not shown), a value that is commensurate to what is observed following temperature upshift (Fig. 5). Up-regulation of the Bartonella groESL locus is presumably due to inhibition of gyrase activity by coumermycin, resulting in decreased DNA supercoiling and enhanced expression of groESL.
3.5. Functional expression of B. bacilliformis groESL and complementation of E. coli mutants
To determine if the B. bacilliformis operon and the encoded chaperonins can be functionally expressed in E. coli, a complementation analysis of temperature-sensitive mutants for groES and groEL was undertaken (Table 3). The pGROESL-TOPO construct was initially transformed into E. coli NRK117 (groEL) and the plasmid allowed the strain to grow at a restrictive temperature of 44 °C, whereas NRK117 transformed with the respective pCR2.1-TOPO cloning vector could not grow. However, similar complementation attempts using pGROESL-TOPO and E. coli CG2244 (groES) were not successful. To examine whether the high copy number of the construct was responsible for negative results with the groES mutant, the groESL operon was re-cloned into the low-copy number vector, pACYC177, to produce pGROESL177. Subsequent transformation of E. coli CG2244 with pGROESL177 enabled the strains to grow overnight at a restrictive temperature of 47 °C, whereas transformation with the respective vector, pACYC177, did not.
Table 3.
Complementation of E. coli groES and groEL mutations using the cloned operon from B. bacilliformis
| E. coli strain (relevant mutation) | Growth at restrictive temperaturea when transformed with the following plasmids: |
|||
|---|---|---|---|---|
| pCR2.1-TOPO | pGROESL-TOPO | pACYC177 | pGROESL177 | |
| NRK117 (groEL) | − | + | − | + |
| CG2244 (groES) | − | − | − | + |
Restrictive temperature of 44 (NRK117) and 47 °C(CG2244). All strains grew at a permissive temperature of 30 °C.
4. Discussion
Transmission of B. bacilliformis between sandflies and the human host is undoubtedly accompanied by several types of cellular stress, including ambient temperature upshift, oxidative stress, pH change or hemin limitation (Coleman and Minnick, 2003), and the pathogen must be able to rapidly adapt if it is to survive. One counter-measure used by all bacteria against such stress is the heat shock response, which induces a group of highly conserved genes encoding so-called Hsps. Two essential components of the heat shock response in bacteria are GroES and GroEL (Mayhew and Hartl, 1996). GroEL proteins are major antigens for several pathogenic bacteria, including Bartonella (Knobloch and Schreiber, 1990; Chenoweth et al., 2004). Our laboratory recently discovered that B. bacilliformis GroEL is also involved in the pathogen's mitogenicity for human endothelial cells in vitro (Minnick et al., 2003b). In this study, we describe a molecular and functional analysis of the B. bacilliformis groESL operon.
Analysis of sequence data revealed a gene linkage of groES–groEL, in keeping with the majority of eubacteria (Segal and Ron, 1996b). Surprisingly, a small ORF (ORF1) was discovered upstream of the groES gene of B. bacilliformis (Fig. 2A). However, ORF1 is apparently not present in B. henselae or B. quintana (Alsmark et al., 2004; GenBank accession nos. NC005956 and NC005955, respectively). ORF1 encodes a hypothetical protein that is similar to a short stretch of HrcA, a heat shock gene repressor that binds to the CIRCE element. Whether a gene product from ORF1 is synthesized and involved in negative regulation of the operon via CIRCE is unknown.
The GroES and GroEL from this study are most closely related to homologues from B. quintana and B. henselae (>89% sequence identity) (Fig. 3). This finding is not surprising, as GroES and GroEL are some of the most highly conserved proteins in nature and are essential for viability (Segal and Ron, 1996b). However, sequence identity to the E. coli GroEL homologue falls to about 66% and identity between B. bacilliformis and E. coli GroES proteins is lower still at 51% (Fig. 3). It is possible that certain of these different amino acids may contribute to the mitogenic activities attributed to the B. bacilliformis GroEL (Minnick et al., 2003b).
Previous studies have shown that B. bacilliformis GroEL is located in both soluble and insoluble fractions of the bacterial cell, including inner and outer membranes, and that it is actively secreted (Minnick et al., 2003b). Likewise, GroEL has also been found in the outer membrane of B. henselae (Chenoweth et al., 2004). However, analysis with SignalP 3.0 software (http://www.cbs.dtu.dk/services/SignalP/) does not reveal a predicted signal sequence at the N-terminus of any Bartonella GroEL. These data suggest that another secretory system is likely responsible for the translocation of these polypeptides. It is also interesting to note that all Bartonella GroELs characterized to date (see Fig. 3B) possess a C-terminal phenylalanine, a feature that is common to many secreted, integral membrane proteins (Struyve et al., 1991).
groESL operons from a variety of bacteria are often transcribed from two promoters, including a σ32-like promoter for transcription under stressful conditions and a σ70-like promoter for transcription in the absence of σ32. However, results of this study show a single TSS for the operon regardless of incubation temperature and the TSS is located immediately downstream of a predicted σ70 -like promoter (Fig. 2A). Another system for regulation of groESL expression in both Gram-positive and Gram-negative bacteria is a highly conserved inverted repeat called CIRCE, which acts as an operator element bound by HrcA (Segal and Ron, 1996a). A putative B. bacilliformis CIRCE immediately follows the groESL TSS (Fig. 2A). The B. henselae and B. quintana operons also contain similar CIRCE elements at essentially the same position relative to the groESL genes and both bacteria possess hrcA homologues in their genomes (GenBank accession nos. NC005956 and NC005955). We hypothesize that all Bartonella species possess hrcA genes that encode a repressor for the CIRCE elements in their genomes.
Finally, a potential rho-independent transcriptional termination signal for the B. bacilliformis operon is located 23 nucleotides downstream of the groEL stop codon and consists of a 12-base inverted repeat, whose RNA secondary structure has a ΔG of − 17.0 kkal/mol by Mfold, suggesting it would form spontaneously during transcription (Fig. 2B). This type of terminator is typical for groESL operons in eubacteria (Zeilstra-Ryalls et al., 1991).
To examine the effects of thermal stress on expression of the groESL operon of B. bacilliformis, we performed temperature-upshift experiments followed by qRT-PCR, Northern and Western blot analyses. Northern blots revealed a full-length polycistronic mRNA of ∼2.4 kb (Fig. 4), a transcript size that would easily accommodate groES and groEL plus their respective intergenic spacer regions. In addition to the full-size transcript, a ∼1.9-kb band was also detected on Northern blots. This observation is similar to previous studies that describe a full-length transcript plus cleavage products of comparable size in groESL mRNA of Agrobacterium tumefaciens, E. coli (Segal and Ron, 1995) and Rickettsia typhi (Radulovic et al., 2002). Quantification of groEL transcript by qRT-PCR showed a 1.3-fold increase in mRNA within 30 min of a temperature upshift, relative to controls. The groEL transcripts reached a maximal 4.25-fold increase at 60 min and then fell steadily to 30 °C levels by 6 h (Table 2).
Densitometry of Western blots revealed a ∼1.4-fold increase in GroEL protein in cultures subjected to a temperature upshift from 30 to 37 °C as compared to 30 °C controls (Fig. 5). These findings were expected because the stress response is characterized by the upregulated synthesis of highly conserved proteins in response to thermal stress (Lindquist and Craig, 1988). About 6 h were needed to detect the increase in GroEL on immunoblots, which is similar to the lag time required to detect changes in B. bacilliformis IalB synthesis in response to pH or temperature change (Coleman and Minnick, 2003). The lag time probably reflects the slow, 6–8 h, generation time for this bacterium (Benson et al., 1986).
Growth of B. bacilliformis on medium supplemented with the gyrase inhibitor, coumermycin A1, increased GroEL protein levels ∼1.5-fold over cultures grown on standard HIAB medium (data not shown). These findings resemble previous studies showing that relaxation of super-coiled DNA with nalidixic acid or novobiocin elevates GroEL protein quantities in E. coli (Mizushima et al., 1993) and suggest that DNA topology may assist HrcA/CIRCE in regulating the Bartonella operon.
The B. bacilliformis groESL genes were functionally analyzed by testing their ability to complement temperature-sensitive mutations in homologous genes of E. coli. Results show that both B. bacilliformis groE genes were able to complement E. coli mutants (Table 3), suggesting that they are transcribed and translated in this host. Cloned groESL genes expressed from a high-copy number construct, pGROESL-TOPO, although able to complement the E. coli groEL (NRK117) mutant, could not complement E. coli groES (CG2244) (Table 3). These data are similar to a previous report where successful groES complementation in E. coli using Vibrio harveyi groES depended on the copy number of the cloning vector, apparently high-copy number plasmid constructs are ineffective (Kuchanny-Ardigò and Lipińska, 2003). Fortunately, B. bacilliformis groES cloned and expressed from a low-copy number pACYC177 construct (i.e., pGROESL177) was able to complement E. coli CG2244 (Table 3).
Previous studies on the B. bacilliformis GroES-EL proteins have shown that GroEL is a major antigen (Knobloch and Schreiber, 1990) and a participant in the pathogen's mitogenicity for human vascular endothelial cells (Minnick et al., 2003b). However, little is known about the operon's regulation in any Bartonella species. Results of this study suggest that the B. bacilliformis operon utilizes both CIRCE and DNA topology for its regulation. This study also demonstrates that the groESL operon of B. bacilliformis is upregulated in response to thermal stress, as would occur during transmission of the pathogen from the sandfly vector to human host. Further analysis of this locus should yield clues on its involvement in virulence, endothelial cell mitogenicity and production of angiomatous lesions during bartonellosis.
Acknowledgements
We gratefully acknowledge the contribution of E. coli strain CG2244 by Barbara Lipinska and E. coli NRK117 by Christopher Coker. Special thanks to Richard Birtles and Amanda Read for sharing unpublished data from the B. henselae groESL locus. Automated sequence analyses were done at the Murdock Sequencing Facility at The University of Montana. This work was supported by American Heart Association Established Investigator Grant 9940002N to MFM, who was also supported by Public Health Service grants AI52101 and AI053111.
This work was done in partial fulfillment of the Master of Science degree of JAC, from The University of Montana.
Abbreviations
- CIRCE
controlling inverted repeat of chaperonin expression
- Hsp
heat shock protein
- qRT-PCR
quantitative real-time polymerase chain reaction
- RACE
rapid amplification of cDNA ends
- TSS
transcriptional start site
References
- Alsmark CM, et al. The louse-borne human pathogen Bartonella quintana is a genomic derivative of the zoonotic agent Bartonella henselae. Proc. Natl. Acad. Sci. U. S. A. 2004;101:9716–9721. doi: 10.1073/pnas.0305659101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausubel FM, et al. Current Protocols in Molecular Biology. John Wiley and Sons Inc.; New York, N.Y.: 1995. [Google Scholar]
- Babst M, Hennecke H, Fischer HM. Two different mechanisms are involved in the heat-shock regulation of chaperonin gene expression in Bradyrhizobium japonicum. Mol. Microbiol. 1996;19:827–839. doi: 10.1046/j.1365-2958.1996.438968.x. [DOI] [PubMed] [Google Scholar]
- Benson LA, Kar S, McLaughlin G, Ihler GM. Entry of Bartonella bacilliformis into erythrocytes. Infect. Immun. 1986;54:347–353. doi: 10.1128/iai.54.2.347-353.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner DJ, O'Connor SP, Hollis DG, Weaver RE, Steigerwalt AG. Molecular characterization and proposal of a neotype strain for Bartonella bacilliformis. J. Clin. Microbiol. 1991;29:1299–1302. doi: 10.1128/jcm.29.7.1299-1302.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhry C, et al. Role of the γ-phosphate of ATP in triggering protein folding by GroEL–GroES: function, structure and energetics. EMBO J. 2003;22:4877–4887. doi: 10.1093/emboj/cdg477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chenoweth MR, Greene CE, Krause DC, Gherardini FC. Predominant outer membrane antigens of Bartonella henselae. Infect. Immun. 2004;72:3097–3105. doi: 10.1128/IAI.72.6.3097-3105.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman SA, Minnick MF. Differential expression of the invasion-associated locus B (ialB) gene of Bartonella bacilliformis in response to environmental cues. Microb. Pathog. 2003;34:179–186. doi: 10.1016/s0882-4010(03)00005-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia FU, Wojta J, Broadley KN, Davidson JM, Hoover RL. Bartonella bacilliformis stimulates endothelial cells in vitro and is angiogenic in vivo. Am. J. Pathol. 1990;136:1125–1135. [PMC free article] [PubMed] [Google Scholar]
- Haake DA, Summers TA, McCoy AM, Schwartzman W. Heat shock response and groEL sequence of Bartonella henselae and Bartonella quintana. Microbiology. 1997;143:2807–2815. doi: 10.1099/00221287-143-8-2807. [DOI] [PubMed] [Google Scholar]
- Knobloch J, Schreiber M. Bb65, a major immunoreactive protein of Bartonella bacilliformis. Am. J. Trop. Med. Hyg. 1990;43:373–379. doi: 10.4269/ajtmh.1990.43.373. [DOI] [PubMed] [Google Scholar]
- Kuchanny-Ardigò D, Lipińska B. Cloning and characterization of the groE heat-shock operon of the marine bacterium Vibrio harveyi. Microbiology. 2003;149:1483–1492. doi: 10.1099/mic.0.26273-0. [DOI] [PubMed] [Google Scholar]
- Kusukawa N, Yura T, Ueguchi C, Akiyama Y, Ito K. Effects of mutations in heat-shock genes groES and groEL on protein export in Escherichia coli. EMBO J. 1989;8:3517–3521. doi: 10.1002/j.1460-2075.1989.tb08517.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landry SJ, Zeilstra-Ryalls J, Fayet O, Georgopoulos C, Gierasch LM. Characterization of a functionally important mobile domain of GroES. Nature. 1993;364:255–258. doi: 10.1038/364255a0. [DOI] [PubMed] [Google Scholar]
- Lindquist S, Craig EA. The heat-shock proteins. Annu. Rev. Genet. 1988;22:631–677. doi: 10.1146/annurev.ge.22.120188.003215. [DOI] [PubMed] [Google Scholar]
- Mayhew M, Hartl F-U. Molecular chaperone proteins. Escherichia coli and Salmonella. In: Neidhardt FC, Curtiss R III, Ingraham JL, Lin ECC, Low KB, Magasanik B, Reznikoff WS, Riley M, Schaechter M, Umbarger EH, editors. Cellular and Molecular Biology. Vol. 1. ASM Press; Washington, D.C.: 1996. pp. 922–931. [Google Scholar]
- McLennan NF, Girshovich AS, Lissin NM, Charters Y, Masters M. The strongly conserved carboxyl-terminus glycine–methionine motif of the Escherichia coli GroEL chaperonin is dispensable. Mol. Microbiol. 1993;7:49–58. doi: 10.1111/j.1365-2958.1993.tb01096.x. [DOI] [PubMed] [Google Scholar]
- Minnick MF, Sappington KN, Smitherman LS, Andersson SGE, Karlberg O, Carroll JA. Five-member gene family of Bartonella quintana. Infect. Immun. 2003a;71:814–821. doi: 10.1128/IAI.71.2.814-821.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minnick MF, Smitherman LS, Samuels SD. Mitogenic effect of Bartonella bacilliformis on human vascular endothelial cells and involvement of GroEL. Infect. Immun. 2003b;71:6933–6942. doi: 10.1128/IAI.71.12.6933-6942.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima T, Natori S, Sekimizu K. Relaxation of supercoiled DNA associated with induction of heat shock response proteins in Escherichia coli. Mol. Gen. Genet. 1993;238:1–5. doi: 10.1007/BF00279523. [DOI] [PubMed] [Google Scholar]
- Radulovic S, Rahman SM, Beier MS, Azad AF. Molecular and functional analysis of the Rickettsia typhi groESL operon. Gene. 2002;298:41–48. doi: 10.1016/s0378-1119(02)00922-8. [DOI] [PubMed] [Google Scholar]
- Segal G, Ron EZ. The groESL operon of Agrobacterium tumefaciens: evidence for heat-shock dependent mRNA cleavage. J. Bacteriol. 1995;177:750–757. doi: 10.1128/jb.177.3.750-757.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segal G, Ron EZ. Heat shock activation of the groESL operon of Agrobacterium tumefaciens and the regulatory roles of the inverted repeat. J. Bacteriol. 1996a;178:3634–3640. doi: 10.1128/jb.178.12.3634-3640.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segal G, Ron EZ. Regulation and organization of the groE and dnaK operons in eubacteria. FEMS Microbiol. Lett. 1996b;138:1–10. doi: 10.1111/j.1574-6968.1996.tb08126.x. [DOI] [PubMed] [Google Scholar]
- Sigler PB, Xu Z, Rye HS, Burston SG, Fenton WA, Horwich AL. Structure and function in GroEL-mediated protein folding. Annu. Rev. Biochem. 1998;67:581–608. doi: 10.1146/annurev.biochem.67.1.581. [DOI] [PubMed] [Google Scholar]
- Struyve M, Moons M, Tommassen J. Carboxy-terminal phenylalanine is essential for the correct assembly of a bacterial outer membrane protein. J. Mol. Biol. 1991;218:141–148. doi: 10.1016/0022-2836(91)90880-f. [DOI] [PubMed] [Google Scholar]
- Weissman JS, Rye HS, Fenton WA, Beechem JM, Horwich AL. Characterization of the active intermediate of a GroEL–ES-mediated protein folding reaction. Cell. 1996;84:481–490. doi: 10.1016/s0092-8674(00)81293-3. [DOI] [PubMed] [Google Scholar]
- Zeilstra-Ryalls J, Fayet O, Georgopoulos C. The universally conserved GroE (Hsp60) chaperonins. Annu. Rev. Microbiol. 1991;45:301–325. doi: 10.1146/annurev.mi.45.100191.001505. [DOI] [PubMed] [Google Scholar]
- Zuber U, Schumann W. CIRCE, a novel heat shock element involved in regulation of heat shock operon dnaK of Bacillus subtilus. J. Bacteriol. 1994;176:1359–1363. doi: 10.1128/jb.176.5.1359-1363.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]




