Abstract
The Drosophila brain is tracheated by the cerebral trachea, a branch of the first segmental trachea of the embryo. During larval stages the cerebral trachea splits into several main (primary) branches that grow around the neuropile, forming a perineuropilar tracheal plexus (PNP) at the neuropile surface. Five primary tracheal branches whose spatial relationship to brain compartments is relatively invariant can be distinguished, although the exact trajectories and branching pattern of the brain tracheae is surprisingly variable. Immuno-histochemical and electron microscopic demonstrate that all brain tracheae grow in direct contact with the glial cell processes that surround the neuropile. To investigate the effect of glia on tracheal development, embryos and larvae lacking glial cells as a result of a genetic mutation or a directed ablation were analyzed. In these animals, the tracheal branching pattern was highly abnormal. In particular, the number of secondary branches entering the central neuropile was increased. Wild type larvae possess only two central tracheae, typically associated with the mushroom body and the antenno-cerebral tract. In larvae lacking glial cells, six to ten tracheal branches penetrate the neuropile in a variable pattern. This finding indicates that glia-derived signals constrained tracheal growth in the Drosophila brain and restrict the number of branches entering the neuropile.
Keywords: Trachea, glia, brain, Drosophila, larva, pattern
Introduction
To meet its high oxygen and energy demand, the vertebrate nervous system is permeated by a dense network of capillaries and small blood vessels. Vascularization of the nervous system begins in the early embryo shortly after neurulation (Strong, 1964; Rovainen and Kakarala, 1989; Gerhardt et al., 1999; 2004). Angioblasts derived from the lateral plate mesoderm migrate dorsally and coalesce into a plexus of blood vessels that surrounds the somites and neural tube. From this perineural vascular plexus capillary sprouts penetrate into the neural tube. Interacting closely with the radial glial cells (the forerunners of astrocytes), capillaries initially follow a radial course. Before reaching the ependymal layer, capillaries branch and form a deep vascular plexus within the neural primordium, the subependymal vascular plexus.
Drosophila has an open vascular system in which the vasculature is reduced to a contractile dorsal vessel. Gas exchange is mediated by a branched network of air-filled tubes called tracheae, which are independent of the dorsal vessel. The insect tracheal system is not homologous to the mesodermally derived vascular system of vertebrates, because tracheae develop as invaginations from the epidermis. The tracheal system evolved in terrestrial arthropods, analogous to the way in which terrestrial vertebrates acquired a lung as an outgrowth from the foregut. However, molecular mechanisms underlying patterning and differentiation of the Drosophila tracheal system appear to be similar in many respects to the mechanisms controlling blood vessel (and lung) development in vertebrates (Metzger and Krasnow, 1999; Affolter et al., 2003). This can be probably understood in view of the fact that cells of all branched tubular organs, irrespective of their later function in the mature organism, have to go through a similar sequence of steps during morphogenesis. One of the central mechanisms controlling tracheal morphogenesis in Drosophila, shared with vertebrate lung and vascular development, is FGF signaling. The invaginating ectodermal placodes that give rise to the tracheal tree express the FGF receptor breathless (Klambt et al., 1992), through which they interact with groups of epidermal and intestinal cells that present the FGF signal, branchless (Sutherland et al., 1996). The pattern of tracheal branches is therefore specified by a ‘pre-pattern’ engraved in the environment of the tracheal primordium. FGF signaling is also responsible for later tracheal growth and branching, which is in part dependent on the oxygen demand of the tissue permeated by the tracheae (Metzger and Krasnow, 1999).
Tracheation of the Drosophila nervous system has recently been studied in a series of papers that focused on the ganglionic tracheal branches growing towards the ventral nerve cord (VNC) in the late embryo (Englund et al., 1999). These tracheae follow the peripheral nerves into the VNC primordium towards the ventral midline, guided by the FGF signaling pathway as well as the Slit-Robo pathway (Englund et al., 2002). FGF and Robo/Slit dependent transcription factors and signal transducers, among them the adrift (Englund et al., 1999) and the Rho-GAP vilse (Lundstrom et al., 2004), were identified as factors that connect membrane bound receptors with the molecular apparatus of cell movement.
In this paper we have reconstructed the development of the tracheal system of the Drosophila brain during the embryonic and larval stages and have analyzed the relationship between brain glia and tracheae. We conclude that, very similar to the above mentioned relationship between vascular precursors and glial precursors in vertebrates, tracheae grow continuously along glial processes. A single trachea, the cerebral trachea, enters the embryonic brain and extends along the glia-covered neuropile surface. Branching during the embryonic period is minimal and occurs mostly at the tip of the cerebral trachea. During early larval stages several stem branches appear close to the point of contact of the cerebral trachea with the brain neuropile. The stem branches subsequently grow around the neuropile and develop secondary and higher order branches that form a dense tracheal plexus (perieuropilar plexus, PNP) at the neuropile surface in the late larval stage. Two secondary tracheae penetrate the center of the brain neuropile. All brain tracheae grow in direct contact with glial cells, which form a sheath around the neuropile and individual neuropile compartments. To investigate the effect of glia on tracheal development we analyzed embryos and larvae lacking glial cells, using embryos mutant for the glial cells-missing (gcm; Jones et al., 1995) gene and larvae in which an apoptosis-inducing UAS-hid;rpr construct (Wing et al., 1998) was expressed by a glial-specific Gal4 construct. Despite the total lack of glia the cerebral trachea enter the brain and form a perineuropilar plexus. However, the branching pattern is abnormal and the overall density of branches entering into the neuropile is increased. We conclude that glia-derived signals restrict and guide tracheal growth in the Drosophila brain.
Material and Methods
Markers and Stocks
The following structures were labeled with monoclonal antibodies acquired from Developmental Studies Hybridoma Bank: embryonic/larval neuropile using anti-DN-cadherin (DN-Ex#8), glial cells using anti-repo (8D12), secondary axon tracts using anti-neurotactin (BP106) and embryonic trachea using anti-crumbs (Cq4). Glia were labeled with Nrv2-Gal4,UAS-GFP (Sun et al., 1999) and genetically ablated using the gcm null fly line gcmrA87Δ1/CyO (Bloomington Stock Center 5445). The UAS-hid,UAS-rpr;;UAS-lacZ (Zhou et al., 1997) line was used to remove glial cells during larval development. Trachea were labeled with btl-Gal4, UAS-GFP (Bloomington Stock Center, BSC, 8807). Trachea were visualized in the larva using a 30MW diode laser emitting at 405nm. Excitation using this wavelength caused unlabeled trachea to fluoresce, as confirmed using the tracheal marker btl-Gal4>UAS-GFP.
Immunohistochemistry and histology
The antibody against DN-cadherin (rat) was diluted 1:20. Anti-repo (mouse) was diluted 1:10. Anti-neurotactin (mouse) was diluted 1:10. Anti-crumbs (mouse) was diluted 1:10. Secondary antibodies were Alexa546-conjugated anti-rat used at a 1:50 dilution (Invitrogen A11081), Alexa546-conjugated anti-mouse used at a 1:500 dilution (Invitrogen A11030), fluorescein-conjugated anti-mouse used at a 1:200 dilution (Jackson Immuno Research 115-095-166) and Cy5-conjugated anti-mouse used at a 1:100 dilution (Jackson Immuno Research 115-175-166). Drosophila embryos were staged (Campos-Ortega and Hartenstein, 1997) and larval brains were dissected. Specimens were viewed as whole-mounts in a confocal microscope. Figures 1-5 were done on a Bio-Rad MRC 1024ES microscope with Radiance 2000 using Bio-Rad Lasersharp 2000 version 5.2 build 824 software. The images in Figure 6 were recorded with a Zeiss LSM510 confocal microscope on a Zeiss AxioImager Z1 microscope using the LSM510 release version 4.0 software. Complete series of optical sections were taken using a 40x oil lens at 2-μm intervals for at least five specimens per stage.
Fig.1.
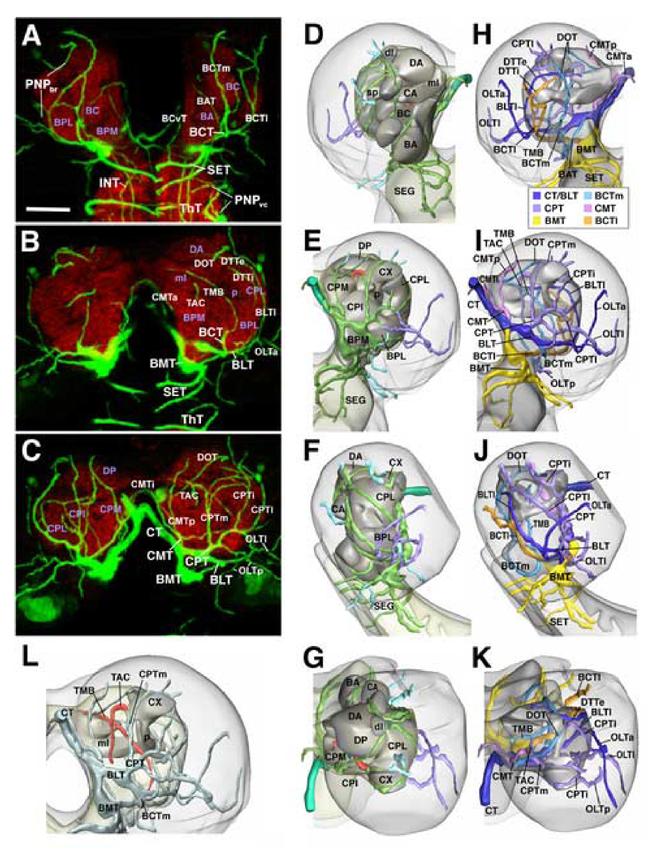
Pattern of brain tracheae at mid larval stage (early third instar; 72h af). A-C: Z-projections of stacks of confocal cross sections of 72h larval brain in which tracheae are visualized by the expression of btl-Gal4 driving UAS-GFP (green); the neuropile is labeled with anti-DN-cadherin (red). Each Z-projection is generated from 8 consecutive sections at 2μm interval. Panel A shows the anterior part of the brain (20-35μm behind anterior tip); B represents the central part of the brain (35-50μm behind anterior tip), C the posterior brain (50-70μm behind anterior tip). Tracheae are identified by white lettering, neuropile compartments by purple lettering. For description of tracheal pattern see text. D-G and H-K: Digital 3D models of right brain hemisphere showing neuropile compartments in grey and major tracheae in different colors. Models of first row (D, H) represent anterior view (dorsal up, lateral to the left); second row (E, I) shows posterior view (dorsal up, lateral to the right); third row (F, J) represents lateral view (dorsal up, anterior to the left), and bottom row (G, K) dorsal view (anterior up, lateral to the right). In models of central column (D-G), coloring indicates depth of tracheae. Tracheae forming the perineuropilar plexus (surrounding the neuropile surface) are depicted in green; secondary branches turning externally into the cortex of the central brain are shown in light blue; optic lobe tracheae in purple. Two secondary branches turning centrally into the neuropile are shown in red. Panel L provides a clearer view of the central tracheae (red color). In this model, presenting postero-dorsal view of right brain hemisphere, neuropile compartments are rendered semi-transparent, and all tracheae (except central ones) are shaded light blue. In models of right column (H-K) neuropile is also semi-transparent, and each primary brain trachea together with its belonging secondary branches is depicted in its own color (see color key at bottom of panel H), which allows one to follow the trajectories of tracheae. All panels except L are presented at the same scale. Scale bar in A corresponds to 30μm. Model in panel L is shown approximately 25% larger than models in D-K.
Description of the brain tracheal pattern: The basomedial trachea (BMT) extends straight anteriorly, contacting the ventral surface of the BMP compartment, which it supplies with several thin branches. The BMT then continues towards the ventral nerve cord, forming several ring-shaped tracheae (SET) around the neuropile of the subesophageal ganglion (B,D-F,H-J;3H,K,N,Q). A secondary branch of the BMT extending dorsally, in between esophagus and inner surface of the basocervical compartment (BCvT), branches off the BMT or the SET (A). The subesophageal ganglion, formed by the three gnathal neuromeres that form the anterior tip of the ventral nerve cord, does not possess segmental tracheae like those present in the thoracic and abdominal neuromeres. Thus, the first (most anterior) segmental trachea is formed at the T1/T2 boundary. The cerebral trachea, branching off the T2 trachea, supplies the entire brain, including the supraesophageal ganglion and the subesophageal ganglion.
The basolateral trachea (BLT) follows an antero-lateral course (B, C, I, J). Traversing the ventral surface of the BPM and BPL compartment, it reaches the lateral neuropile. Here, it turns dorsally and medially and extends over the surface of the BPL compartment as the external dorsal transverse trachea (DTTe).
The basocentral trachea (BCT) grows anteriorly along the boundary between BPL and BPM compartment until reaching the basocentral (BC) compartment. After giving off a branch to the BA compartment (BAT) the BCT encircles the BC compartment and curves dorsally at its anterior surface, branching into two ascending branches, the medial and lateral basocentral trachea (BCTm, BCTl; A-I). The BCTm branch continues as the dorsal oblique cerebral trachea (DOT) in the dorsal brain neuropile (C, J, K; see below). Another branch of the BCTm enters the center of the neuropile and forms the trachea of the mushroom body (TMB in I, L; see below). The BCTl extends dorsally and then postero-medially, often penetrating into the CPL neuropile forming the internal dorsal transverse trachea (DTTi; B,H). Frequently the medial and lateral branch of the BCT are separate from the beginning. In the brain sample shown in Figure 1, the BCTm branch originates from the BLT, whereas the BCTl branches off the BMT.
The centromedial trachea (CMT) branches off the cerebral trachea or the basolateral trunk derived from it and forms three ascending branches (CMTa, CMTi, CMTp, respectively; A-C; H, I). The CMTa and CMTi tracheae project dorsally, at the inner surface of the CM compartment, and anastomose with their contralateral counterparts underneath the cerebral commissure. CMTp continues upward at the posterior surface of the CM compartment and enters the DP compartment from posteriorly.
The centroposterior trachea (CPT) represents the fifth main branch of the cerebral trachea. CPT branches cover the posterior surface of the neuropile (C, E, I). Typically, a medial and an intermediate branch curve upward over the CPI compartment to reach the calyx of the mushroom body. Encircling the calyx medially and laterally, the two branches anastomose with each other dorsal of the calyx. The CPl branch grows in a antero-dorsal direction, curving upward over the lateral surface of the CPLd compartment.
Abbreviations: BA baso-anterior (antennal) neuropile compartment; BC baso-central neuropile compartment; BAT baso-anterior trachea; BCT baso-central trachea; BCTl lateral baso-central trachea; BCTm medial baso-central trachea; BCvT baso-cervical trachea; BLT baso-lateral trachea; BLTl lateral branch of baso-lateral trachea; BPL baso-posterior lateral neuropile compartment; BPM baso-posterior medial neuropile compartment; CA centro-anterior neuropile compartment; CMT centro-medial trachea; CMTa anterior branch of centro-medial trachea; CMTi intermediate branch of centro-medial trachea; CMTp posterior branch of centro-medial trachea; CPI centro-posterior intermediate neuropile compartment; CPL centro-posterior lateral neuropile compartment; CPM centro-posterior medial compartment; CPT centro-posterior trachea; CPTm medial branch of centroposterior trachea; CPTi intermediate branch of centro-posterior trachea; CPTl lateral branch of centro-posterior trachea; CT cerebral trachea; CX calyx of mushroom body; DA dorso-anterior neuropile compartment; dl dorsal lobe of mushroom body; DP dorso-posterior compartment; DOT dorsal oblique trachea; ml medial lobe of mushroom body; DTTe external dorsal transverse trachea; DTTi internal dorsal transverse trachea; INT intraneuropilat tracheae of ventral nerve cord; OLTa anterior optic lobe trachea; OLTl lateral optic lobe trachea; OLTp posterior optic lobe trachea; p peduncle of mushroom body; PNPbr perineuropilar plexus of brain; PNPvc perineuropilear plexus of ventral nerve cord; SEG subesophageal neuropile; SET subesophageal tracheae; sp spur of mushroom body; TAC trachea of the antenno-cerebral tract; ThT thoracic trachea; TMB trachea of the mushroom body;
Fig.5.
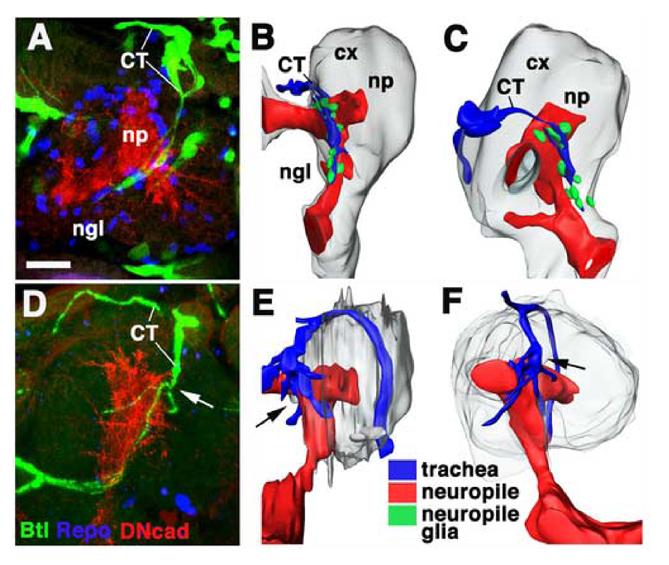
Tracheal patterning in glial cells missing (gcm) mutant embryo. A, D: Z-projections of confocal stacks prepared from brains of late embryonic preparations (A: wild type; D: gcm mutant) in which tracheae are visualized by the expression of btl-Gal4 driving UAS-GFP (green); the neuropile is labeled with anti-DN-cadherin (red), glial cells are labeled by anti-Repo (blue). B, C, E, F: Digital 3D models of wild-type (B, C) and gcm mutant (E, F) embryo. Neuropile (np) is shown in red, brain cortex (cx) in gray, cerebral trachea (CT) in blue, and neuropile glia (ngl) in green. B and E are posterior views (midline left, dorsal up), C and F are lateral views (anterior to the left, dorsal up). Note absence of neuropile glia and increased tracheal branching (arrow) in mutant (D, E, F). Scale bar (for A-F): 10μm
Fig.6.
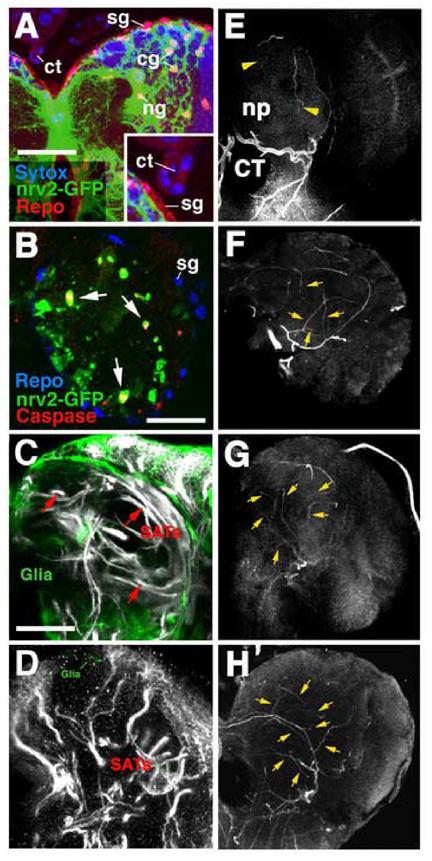
Tracheal patterning in late larval brain lacking glial cells. A: Confocal section of late larval brain showing expression of the driver line nrv2-Gal4 (green) in cortex glia (cg) and neuropile glia (ng). Red label indicates expression of the Repo antigen in all glial nuclei; blue shows sytox labeling of nuclei. Note absence of nrv2-gal4 expression in surface glia (sg) and cerebral trachea (ct; see enlarged view in inset). B: Confocal section of early second instar brain following activation of UAS-hid;rpr by nrv2-Gal4 (embryos raised at 18°C and shifted to 25°C right after hatching). Green label shows residual glial cells; most of these cells are in the process of apoptosis, as indicated by their expression of caspase-3, a marker for cell death (red). Repo (blue) is virtually absent in cortex and neuropile glia, but is expressed normally in surface glia. C: Labeling of glia (green) by nrv2-Gal4 driven GFP. Normal trajectories of secondary axon tracts (SATs; white). D: Labeling of glia (green) and secondary axon tracts (white) in brain of larva in which a UAS-hid;rpr construct was driven by nrv2-Gal4. Note virtual absence of glial cells and abnormal, short/branched trajectories of axon tracts. E-H: Z-projections of confocal sections of wild-type brain (E; anterior view; lateral to the right, dorsal up) and similarly oriented brains in which glia was ablated using nrv2-Gal4>UAS;hid;rpr (F-H). Tracheae were visualized using a 30MW diode laser emitting at 405nm which causes unlabeled tracheae to fluoresce. In wild type (E) only two secondary tracheal branches (yellow arrowheads; TAC trachea of antenno-cerebral tract; TMB trachea of mushroom body) penetrate into the neuropile (np). In brains lacking glia one observes a greatly increased number of intra-neuropilar tracheal branches (yellow arrowheads).
Other abbreviations: BCT baso-central trachea; CT cerebral trachea. Scale bar (A): 35μml (B): 25μm; (C-H): 50μm.
For histology and electron microscopy, larval brains were dissected and fixed in 2% glutaraldehyde in PBS for 20 minutes, followed by a post-fixation for 30 minutes in a mixture of 1% osmium tetroxide and 2% glutaraldehyde in 0.15 M cacodylate buffer (on ice). Specimens were washed several times in PBS and dehydrated in graded ethanol and acetone (all steps on ice). Preparations were left overnight in a 1:1 mixture of Epon and acetone and then for 5-10 hours in unpolymerized Epon. They were transferred to molds, oriented, and placed at 60°C for 24 hours to permit polymerization of the Epon. Blocks were sectioned (0.1μm). Sections were mounted on net grids (Ted Pella) and treated with uranyl acetate and lead citrate.
Generation of Three-Dimensional Models
Surface-rendered digital models of the brain surface, neuropile compartments, glia and trachea were generated using the Image Segmentation module of Amira 3.0 (Mercury Computer Systems). The model of the brain surface in Figure 1 was generated using a plugin for ImageJ (‘A 3D editing plugin’, http://www.pensament.net/java/) and brought into registration with the other components of the model within Amira.
Results
Basic pattern of tracheae in the larval nervous system
The tracheae of the ventral nerve cord at mid-larval stages (72h AEL) are the ganglionic branches (GB) of the segmental tracheal stems. GBs are arranged as metamerically reiterated rings surrounding the neuropile (ThT in Fig.1A). Numerous secondary branches that form anastomoses between adjacent GB rings give the tracheal system of the ventral nerve cord the appearance of a plexus (perineuropilar plexus, PNP; Fig.1A, B). Secondary branches entering within the neuropile (INT in Fig.1A) become aligned longitudinally alongside discrete axonal fascicles (V.H., unpubl. obs.). The formation of the PNP begins during stage 16 of embryonic development when GBs arise as ventral branches of the segmental tracheae and invade the ventral nerve cord (Englund et al., 1999; Fig.2B, arrowhead ‘1’). Advancing medially, GBs pass underneath the neuropile of the ventral nerve cord (Fig.2B, arrowhead ‘2’). Shortly before reaching the midline, GBs turn dorsally (arrowhead ‘3’) and then laterally (arrowhead ‘4’), always growing along the cortex-neuropile boundary. During larval stages, the advancing tips of the GBs have formed a circle and fuse with a more proximal part of the same or adjacent GBs. A similar pattern of ring-shaped tracheae is generated in the brain. Here, a branch of the first segmental trachea, called the cerebral trachea, reaches the medial surface of the brain neuropile in the embryo (Manning and Krasnow, 1993; Hartenstein, 1993; Fig.2B, arrowhead ‘5’). From here, multiple branches (called the primary tracheae of the brain in the following) grow laterally and medially around the neuropile surface (see below). Eventually, medial and lateral branches will meet at some point over the dorsal brain neuropile. Formation of complete tracheal rings is not completed before the early to mid third larval instar.
Fig.2.
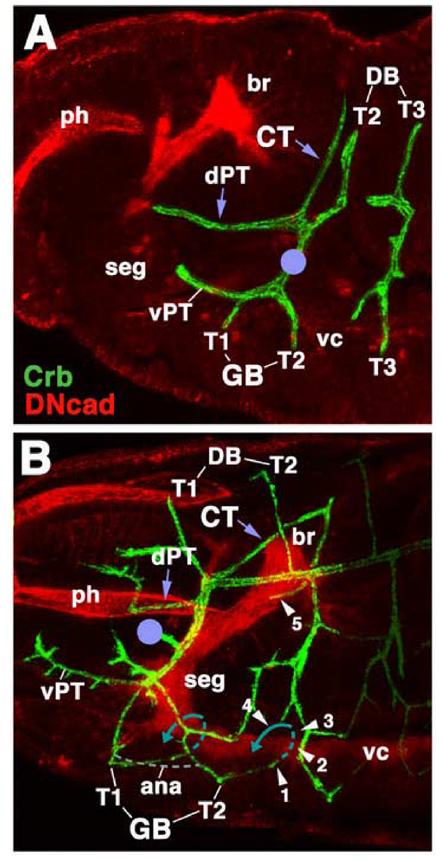
Embryonic origin of the cerebral trachea and ganglionic tracheal branches. Both panels show Z-projections of confocal sections of embryos (lateral view, anteriior to the left) labeled with anti-Crb (green) to visualize tracheae. Anti-DN-cadherin (red) labels neuropile and other embryonic structures. A: Stage 14. Cerebral trachea (CT) and dorsal pharyngeal trachea (dPT) form a Y-shaped, anteriorly directed branch of the first segmental trachea (I) that grow around the posterior surface of the brain (br). Other branches of the first segmental trachea are the dorsal branches (DB) of segments T2 and T1 (formed later than stage 14), the ventral ganglionic branches (GB) of segments T1 and T2, and the ventral pharyngeal trachea. The location of the anterior spiracle is indicated by violet circle. B: Stage 15 late. Segmental tracheae have fused, primary branches have increased in length, and some secondary branches have been initiated. Note position of the cerebral trachea (CT) and dorsal pharyngeal trachea (dPT). The cerebral trachea has reached the medial surface of the brain neuropile (arrowhead ‘5’). Ventral ganglionic branches contact the ventral surface of the neuropile of the ventral nerve cord (vc; arrowhead ‘1). During later stages ganglionic branches will extend underneath the neuropile, turn dorsally (hatched blue line; arrowheads ‘2’ and ‘3’) and then laterally (solid blue line, arrowhead ‘4’). Anastomoses (ana; gray hatched line) will interconnect ganglionic branches of neighboring segments.
Other abbreviations: ph pharynx; seg subesophageal ganglion
Tracheae of the larval brain
The cerebral trachea (CT) reaches the larval brain at a dorsomedial position, enters the cortex and extends ventrally along the posterior-medial neuropile surface. At the level of the BPM compartment (see Younossi-Hartenstein et al., 2003 for larval neuropile anatomy), located at the base of the brain neuropile, the CT branches into five primary brain tracheae which will be called the basomedial cerebral trachea (BMT), basolateral cerebral trachea (BLT), basocentral cerebral trachea (BCT), centromedial cerebral trachea (CMT), and centroposterior cerebral trachea (CPT). The pattern of these brain tracheae is illustrated and described in Fig.2. Note that the exact pattern in which even the primary brain trachea (not to mention higher order branches) relate to each other is quite variable. For example, in the brain sample shown in Fig.2, the CT of the right hemisphere initially branches dichotomously into a medial and a lateral trunk. The medial trunk gives rise to the BMT and BCT, the lateral trunk to the BLT, CPT, and CMT. In the left hemisphere, the CT branches into three main trunks, one that forms the BMT, one giving rise to the BCT and BLT, and the third one to the CPT and CMT.
As shown and described in detail in Fig.1, the primary and most higher order tracheae of the brain all grow along the cortex-neuropile interface. Only two or three secondary tracheae enter the center of the neuropile (Fig.1L). These are the trachea of the mushroom body (TMB), the trachea of the antennocerebral tract (AC), and the internal dorsal transverse trachea (DT; not always found). The mushroom body trachea represents a branch of the BCT trachea. Traveling upward in the septum between BC and BPL compartment, it reaches the spur region of the mushroom body and then curves medially, alongside the posterior surface of the medial lobe of the mushroom body. The trachea of the antennocerebral tract (TAC) typically constitutes a branch of the CPTm trachea, given off near the point where this trachea reaches the medial surface of the calyx. The TAC follows the antennocerebral tract antero-ventrally towards the BC-BPM boundary.
In addition to the TAC and TMB tracheae which are directed inward, into the center of the neuropile, a number of secondary tracheal branches project outward into the cortex and the optic lobe (Fig.1D-G). Noteworthy are the three tracheae that extend towards the optic lobe. Throughout the first half of the larval period, the optic lobe consists of two horseshoe-shaped epithelia, the inner and outer optic anlagen (IOA, OOA), which are attached to the basolateral brain surface. During the third larval instar the epithelia transform into neuroblasts that produce lineages of neurons, occupying the space between the IOA and OOA (Meinertzhagen and Hanson, 1993). Two tracheae, the posterior and lateral optic lobe trachea (OLTp, OLTl), branch off the CPT and grow towards the posterior and the dorsal edge of the optic lobe, respectively (Fig. 1C,H-K). The anterior optic lobe tracheae (OLTa) is formed by the BLT (Fig.1B,H-K).
Development of the tracheal tree from embryo to late larva
In the late embryo the cerebral trachea is visible as a thick, posteriorly directed branch of the first segmental trachea that belongs to the second thoracic segment (Fig.2A,B;3A-C). The CT arches over the brain surface; reaching the dorso-medial apex of the brain, posterior to the supraesophageal commissure (SEC), the CT turns ventrally, following the medial surface of the brain. Here, the neuropile (that at all other positions is surrounded by a multilayer of neuronal cell bodies) lies close to the surface, merely covered by a thin layer of surface glia (Fig.4A,B). The cerebral trachea, embedded in this glial layer, follows the neuropile surface ventrally. Reaching the point where the incipient BPL and BPM neuropile compartments depart from each other, the CT bifurcates into a thick main branch that continues ventrally, along the BPM, and a thinner lateral branch that projects dorso-laterally along the surface of the BPL and CPL compartments, reaching a point where the calyx of the mushroom body forms (Fig.3A-C). This lateral embryonic tracheal branch foreshadows the BLT/CPT tracheae of the larval brain, which, as described in the previous section, ramify over the dorso-posterior surface of the neuropile (see Fig.1A,B,H-J;3). The ventrally directed tracheal branch of the late embryo probably pioneers the BMT, since, like the larval BMT, it follows the medial edge of the BPM compartment. A short branch projecting laterally foreshadows the BCT (Fig.3B). The BMT continues ventrally into the subesophageal ganglion where it splits into two or three thin terminal branches (the SET pioneers) associated with the subesophageal neuropile.
Fig.3.
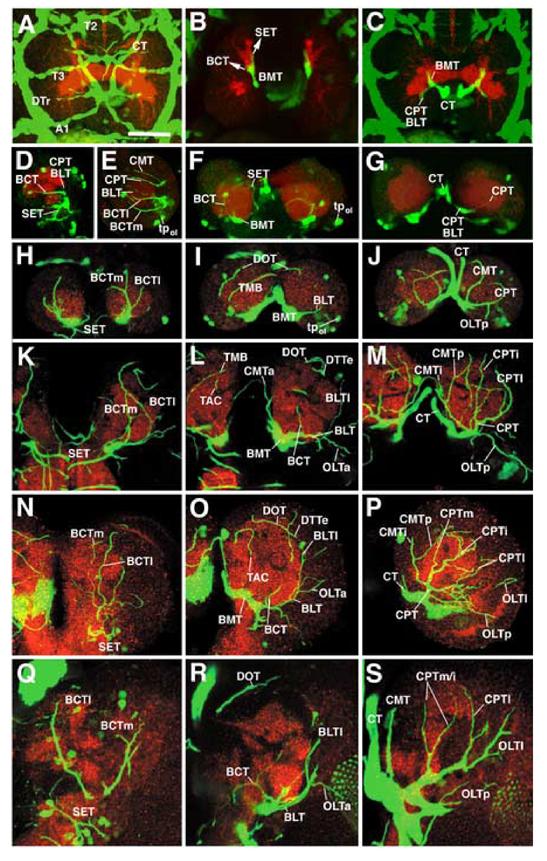
Development of the brain tracheae. All panels show Z-projections of confocal stacks prepared from brains of embryonic (A-C) and larval (D-S) preparations in which tracheae are visualized by the expression of btl-Gal4 driving UAS-GFP (green); the neuropile is labeled with anti-DN-cadherin (red). A-C: Stage 16 embryo, dorsal view, anterior to the top. Z-projection in A (15 consecutive sections at 2μm interval, starting at the dorsal surface) shows the anterior segmental tracheae (T2, T3, A1), the dorsal longitudinal trunk (DTr) and the cerebral trachea (CT), wich passes diagonally over the brain surface (arrows) before turning ventrally right behind the supraesophageal commissure (SEC). Z-projections in B (basal brain; 10 consecutive sections at 2μm intervals, 40-60μm ventral of the dorsal surface) and C (dorsal brain; 10 consecutive sections, 10-30μm below dorsal surface) show the trajectory of the cerebral trachea (CT). Reaching the central neuropile from posteriorly, the CT bifurcates into a short lateral branch that pioneers the CPT and BLT tracheae and a branch that continues ventrally, representing the BMT. The BMT bifurcates again further ventrally (B) into the forerunner of the BCT and the terminal branches that reach the subesophageal neuropile (SET). D, F, G: Early first instar larva (24h af). Panel D (12 sections starting at lateral brain surface) shows tracheae of lateral brain in side view (anterior to the left, dorsal to the top). F and G represent dorsal views (F: 25-40 μm below dorsal surface; G: 10-25μm below dorsal surface) of both brain hemispheres. E shows lateral view of early second instar brain (Z-projection of 12 sections starting at lateral surface). Note isolated clusters of btl-Gal4-positive cells which may constitute tracheal progenitors in the region around the optic lobe primordium (tpol). Remainder of panels (H-S) is organized in such a way that each row corresponds to one stage (H-J: early second instar, 48h af; K-M: early third instar, 72h af; N-P: mid third instar, 120h af; Q-S: late third instar, 144h af) and each column to a particular level along the anterior-posterior axis (left column: anterior brain, 8-12 sections, approx. 20-40μm behind anterior tip of brain; middle column: center of the brain, 8-12 sections, approx. 40-60μm behind anterior tip; right column: posterior brain; 10-15 sections, approx. 60-90μm behind anterior tip). For abbreviations see legend of Fig.1. Figures of all panels are shown at the same scale. All brains are shown at the same magnification. Scale bar (A): 30μm
Fig.4.
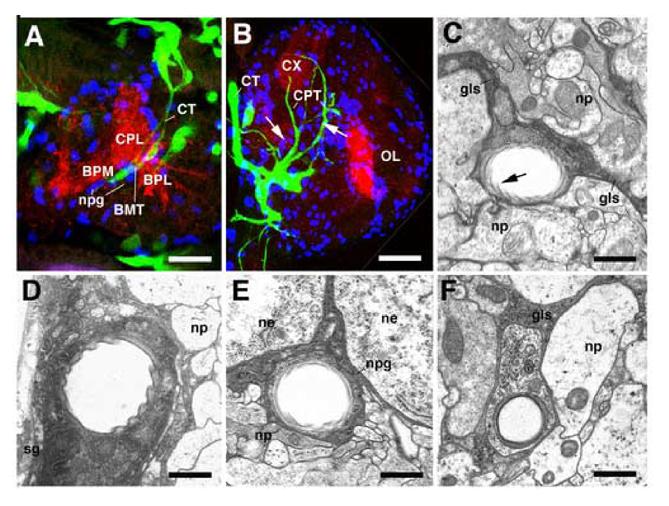
Tracheae grow alongside glial cells. A, B: Z-projections of confocal stacks prepared from brains of late embryonic (A) and late larval (B) preparation in which tracheae are visualized by the expression of btl-Gal4 driving UAS-GFP (green); the neuropile is labeled with anti-DN-cadherin (red), glial cells are labeled by anti-Repo (blue). In the embryo (A), the cerebral trachea (CT) projects along the dense population of neuropile glial cells (npg) that covers the medial neuropile surface. The close relationship between tracheae and glia is maintained in the larva (B). Note clusters of newly formed glial cells (recognizable by their small size; examples shown by arrows) that always occur alongside tracheae. C-F: transmission electron micrographs of cross sections of larval brain (early third instar, 72h af). Tracheae are recognizable by their rifled internal cuticular lining (arrow in C); glial cells/processes stand out by their high electron density and characteristic leaf-like shape. Representative tracheae at different depths within the brain are shown: C: secondary branch in center of neuropile (np), accompanied by inter-compartmental glial septum (gls); D: main cerebral trachea at medial brain surface, covered by surface glia (sg); E: trachea of perineuropilar plexus, located between cortex (ne neuronal cell body) and neuropile (np), surrounded by neuropile glia (npg); F: tracheole in neuropile, attached to thin glial process (gls). Other abbreviations: see legend of Fig.1. Scale bars: 10μm (A); 30μm (B); 0.5 μm (C, F); 3μm (D); 2μm (E).
During the first larval instar, all of the primary brain tracheae have become established. Shortly after hatching (24h post fertilization, af) the CT of the brain sample shown in Fig.3 (panels D, F, G) splits into a laterally and a ventrally directed trunk. The lateral trunk gives rise to the CMT, CPT and BLT, each of them representing, thin, short and unbranched tracheae. The ventral trunk bifurcates into BMT and BCT. By 48h af (early second instar), each of these tracheae has grown in length and diameter. Two BCTs (BCTl and BCTm) are present which curve anteriorly and then dorsally; one of them reaches the dorsal neuropile, pioneering the DOT (Fig.3H, I). Still, few secondary branches are present. Exhibiting a striking left-right asymmetry in the 72h af brain sample shown in Fig.3E,H-J, the secondary trachea branching off the BCTm towards the mushroom body (TMB) is present in the left, but completely absent in the right hemisphere (Fig.3I). A single short secondary branch is given off by the CPT towards the optic lobe.
During the second half of the larval period, following the stage (72h af) described in detail in the previous section, the pattern of primary and secondary tracheae does not appear to change significantly. Thus, despite a large increase in neuropile volume (largely due to the outgrowth of the axons of secondary lineages, as well as the proliferation of the optic lobe; Pereanu et al., 2006), no new major tracheal branches are formed (Fig.3N-S) in the central brain. Only the tracheae reaching the optic lobe become longer and more elaborate, a process we did not follow in detail. It should be noted that the pattern of tracheae described here only includes the major branches which exceed 1-2μm in diameter, and which can be followed by light microscope. Aside from these major trachea, the neuropile is permeated by a tree of thin higher order branches (tracheoles) with a diameter in the range of 0.4-0.6μm, which appear as a filigrane network on confocal sections and cannot be reconstructed by following the approach utilized in this study (serial transmission electron microscopy would be the only reliable way of reconstructing structures with such small dimensions). It stands to reason that the tracheoles increase in length and number during larval growth to adapt to the increase in neuropile volume, similar to the oxygen demand-related growth of tracheoles in the epidermis (Jarecki et al., 1999; Arquier et al., 2006).
Tracheal growth as described so far occurs through elongation and branching of preexisting tracheal tubes. In addition, and somewhat unexpectedly, btl-Gal4 expression occurs in isolated cell clusters which are not connected to the already established tracheal tree. Thus, in the first and early second instar brain, several groups of btl-Gal4-positive cells appear at more or less invariant positions, in particular around the optic lobe primordium (Fig.3E,F,I). In some instances, fine processes that could be interpreted as nascent tracheal tubes emanate from these isolated cell clusters. Around the transition of second and third instar (72h af) no more isolated btl-positive clusters are observed, suggesting that they have become incorporated into the tracheal network. However, it should be noted that the identity of the isolated btl-Gal4 expressing cells as tracheal precursors should be considered with caution. In our (larval) material, btl-Gal4 was also expressed in subsets of retinal cells, and the cells we here tentatively identify as isolated, brain-associated tracheal precursors may actually represent a different cell type.
The role of glia in tracheal development
Most, if not all, major tracheae of the brain grow along glial cells. In the embryo, the cerebral trachea on its ventrally directed path follows the population of neuropile glial cells (Fig.4A) that cover the neuropile surface. Likewise, during larval stages, the primary and secondary tracheae forming the perineuropilar plexus are enclosed within the layer of neuropile glia (Fig.4E); tracheae invading the neuropile are juxtaposed to the glial septa in between neuropile compartments (Fig.4C), and even tracheolae are accompanied by glial processes (Fig.4F). The close spatial relationship between glia and tracheae prompts the question of mutual inductive interactions. We therefore analyzed tracheal development in embryos and larvae that lacked glial cells, using the glial cells missing (gcm) gene and a UAS-hid;rpr construct expressed in glial cells by the nrv2-Gal4 driver.
gcm is expressed and required for glial cells and hemocytes (Jones et al., 1995; Jones, 2001). Loss of gcm results in embryonic lethality accompanied (and probably caused) by the almost complete absence of glial cells and reduction in macrophages (Bernardoni et al., 1997; Fig.5). Glial loss can also be achieved by expressing a UAS-hid;rpr construct under the control of the nrv2-Gal4 driver line. This driver line is expressed in all cortex and neuropile glia from late embryonic stages onward (Younossi-Hartenstein et al., 2003; 2006; Fig.6A). No expression of the nrv2 driver can be observed at any stage (Fig.6A); the same is true for gcm in the embryo or larva (Jones et al., 1995; Hosoya et al., 1995). By raising nrv-2-Gal4>UAS-hid;rpr embryos at 18°C and switching to a high temperature (25°C) shortly after hatching, most animals develop to late larval stages. In these larvae, glial cell death (visualized with an antibody against caspase-3) sets in around hatching and is more or less complete by the early 2nd instar (after 24h; Fig.6B, D). Larvae lacking cortex and neuropile glia (but retaining surface glia in which the driver line is not expressed; Fig.6A, B) show severe behavioral deficits, characterized by reduced peristaltic movement and feeding, and a constant ‘tremor’ of the pharyngeal and body wall musculature (W.P. and S.F., unbpublished observation).
The total loss of glia in gcm mutant embryos causes neuronal pathfinding defects in stage 16 embryos (Takizawa and Hotta, 2001). The cerebral trachea grows towards the dorso-medial surface of the neuropile normally. However, upon reaching the neuropile surface, it gives off numerous thick, long branches that are never observed in wild type embryos (Fig.5D-F). A similar abnormality manifests itself in late larvae in which glia was ablated postembryonically (Fig.6). Here, six or more secondary branches split off the perineuropilar plexus at variable positions and penetrate the neuropile (Fig.6F-H, yellow arrowheads). In wild-type brains, secondary branches in the neuropile typically amount to two, the TAC and TMB, and were never observed to exceed three (Fig.6E). A similar, if less pronounced neuropile tracheal overgrowth was observed in the ventral nerve cord of glia-ablated animals (data not shown). These findings support the idea that glial cells have an inhibitory effect on tracheal branching and/or elongation. Specifically, the glial layer at the cortex-neuropile appears to restrict entry of tracheae into the neuropile.
Discussion
Tracheal patterning in the Drosophila nervous system
Outgrowth and branching morphogenesis of the tracheal system has been investigated in great detail for the tracheal network underlying the epidermis (Shilo et al., 1997; Metzger and Krasnow, 1999; Rosin and Shilo, 2002; Affolter at al., 2003; Lubarsky and Krasnow, 2003). Three phases, all of them dependent on the FGF signaling pathway, have been distinguished. In the early embryo, shortly after gastrulation, the epidermal ectoderm of segments T2-A8 gives rise to metameric pairs of tracheal placodes. Shortly after invagination, placodes resemble simple, round cups. Soon primary tracheal branches grow out at stereotypic positions from each cup. These branches grow in length and form secondary and tertiary branches. Growth of the primary (and some of the secondary) tracheal branches is induced by the FGF homolog Bnl which is expressed in strategically positioned clusters of epidermal cells. The pattern of the Bnl expressing epidermal cells foreshadows the later pattern of primary branches; both patterns are highly invariant.
The second phase of tracheal branching morphogenesis includes the formation of secondary branches which occur mainly at the growing tips of primary branches. High levels of the Bnl signal induce the expression of intrinsic activators of branching behavior, such as the ETS transcription factor Pointed (Metzger and Krasnow, 1999), as well as inhibitory factors (e.g., sprouty; Hacohen et al., 1998) that restrict the formation of secondary branches to positions that are somewhat remote from the primary branch tip. The resulting pattern of secondary and higher order branches is considerably more variable, given that no ‘hard-wired’ prepattern exists. The third and terminal phase of tracheal branching describes the formation of a filigrane network of tracheoles that sprout from the tips of secondary tracheal branches. This process occurs largely after hatching of the embryo and depends on extrinsic factors, among them local oxygen levels (Guiellemin et al., 1996). It is therefore a plastic, demand-based process. Again, the FGF signaling cascade plays a central role in terminal branching.
The development of the ganglionic tracheal branches that supply oxygen to the ventral nerve cord is initiated as a typical ‘phase 1’ process where a ventrally located cluster of epidermal cells guides the formation of a secondary branch towards the edge of the neural primordium (Englund et al., 1999). Subsequently, the tip of the GB follows the roots of the peripheral nerve towards the neuropile. It curves around the ventral edge of the neuropile and then turns dorsally. The formation of short tertiary tracheal branches that interconnect the corresponding ganglionic branches of the left and right side, as well as neighboring ganglionic branches of the same side, constitutes a ‘phase 2’ event and, accordingly, leads to a relatively variable plexus of tertiary tracheae. Aside from FGF signaling, the Slit/Robo pathway forms part of the molecular network that controls the phase 2 branching morphogenesis of the GB. Slit is expressed by midline glial cells, a subset of neuropile glia derived from the mesectoderm. GB cells express the Slit receptors Robo and Robo 2. The latter is required to attract the GB towards midline; the former inhibits crossing (Englund et al., 2002).
Tracheation of the brain appears to be fundamentally similar to that of the epidermis and ventral nerve cord, and it is reasonable to apply to brain tracheation the three-step model proposed by Krasnow and other workers (Metzger and Krasnow, 1999). Complicating this analysis is the fact that the entire brain, including the gnathal part of the ventral nerve cord that later becomes the subesophageal ganglion, is tracheated by a single trachea, the cerebral trachea, which splits into five relatively invariant main branches (called ‘primary’ branches in this paper) that form the perineuropilar plexus around the brain neuropile. Strictly speaking, the cerebral trachea represents a primary branch of the first tracheal primordium, and the main tracheae (BMT, BLT, BCT, CMT, CPT) would therefore constitute secondary branches. In line with such interpretation, the trajectories and branching patterns of the brain tracheae is much more variable than that of typical primary branches in the trunk (see Fig.1,2). On the other hand, with respect to their diameter and expansion, brain tracheae resemble primary tracheal branches of the trunk. More analysis is required to evaluate brain tracheation in comparison to the segmentally organized tracheation of the ventral nerve cord. Of particular importance will be comparative studies that reconstruct tracheation patterns in more primitive insects. Thus, it is likely that the lack of segmental tracheal primordia in the gnathal segments and, possibly, even in the preoral head segments of Drosophila, is derived from a primitive condition in which such segmental primordia were present. The main (‘primary’) brain tracheae which in Drosophila arise as secondary branches of a single tracheal primordium could in the primitive condition have been formed as primary branches of one or more segmental tracheal primordia.
From the existing descriptions of tracheation of the ventral nerve cord (Englund et al., 1999) it seems likely that throughout development, the GBs and their branches are in contact with glial cell precursors that surround the peripheral nerve roots and the neuropile. A direct contact between tracheae and glia does certainly occur for all tracheae of the brain, as shown in this paper. Furthermore, experimental ablation of glia reveals inhibitory interactions between glial and tracheal cells. The molecular nature of these interactions remains to be established. It is tempting to invoke FGF signaling as part of the mechanism, given its pervasive involvement in other aspects of tracheal development, and the fact that the FGF receptor heartless (htl) is expressed and required for glial development (Shishido et al., 1992). Loss of Htl prevents the formation of glial processes enwrapping the neuropile, and application of FGF soaked beads in grasshopper embryos provokes the outgrowth of such processes (Condron, 1999). We thus have a scenario where two different FGF receptors, Htl and Btl, are expressed by adjacent tissues, namely glia and trachea, respectively. Competition for a common source of the FGF signal (from brain neurons?) or other, more complex interactions could guide the formation of glia and tracheae, and form the basis of the inhibitory action of the former on the latter.
Vascularization/tracheation of the brain in vertebrates and Drosophila
The comparison of vascular patterning in the vertebrate brain and tracheal patterning in Drosophila reveals a number of similarities. In both systems, epithelial vessels penetrate the neural primordium from the outside, follow radially organized glial elements, and branch out to form a vascular/tracheal plexus, the subependymal plexus in vertebrates and the perineuropilar plexus in Drosophila. Furthermore, vascular growth in vertebrates and tracheal growth in flies is guided by specialized tip cells (Manning and Kransnow, 1993; Gerhardt et al., 2003; 2004). Similar to the tips of extending axons, vascular/tracheal tip cells have a growth cone whose filopodia explore cues of the microenvironment, which primarily consists of glia and the extracellular matrix produced by these cells.
Numerous signaling mechanisms guiding vascular/tracheal tip cells are shared between vertebrates and Drosophila, among them FGF signaling which plays a preeminent role. In addition to FGF signaling, other receptor tyrosine kinases and their ligands were identified as mediators of vascular patterning in the vertebrate brain. During their radial growth into the neural primordium, vertebrate capillary tip cells, expressing VEGF and PDGF receptors, follow a gradient of VEGF (Gerhardt et al., 2003). A corresponding role of the Drosophila VEGFR/PDGFR homolog, PVR, cannot be ascertained, at least during the embryonic and larval phase. Drosophila PVR is expressed and required for hemocyte differentiation and migration, but has not been found in the tracheal system (Cho et al., 2002; Bruckner et al., 2004).
Other signal-receptor systems, among them members of the semaphorin and neuropilin families of proteins, as well as adhesion molecules such as N-cadherin and integrin, modify the response of developing capillaries to FGF and VEGF (Gerhardt et al., 1999). Many of these signaling mechanisms require the close interaction between capillaries and radial glia/astrocytes. Astrocyte-derived cues are also responsible for the structural changes in capillary endothelia that underlie the formation of the blood-brain barrier (BBB), which consists of specialized tight junctions, as well as a number of enzymatic transport systems which regulate molecular movements across the endothelial cell membrane (Risau and Wolburg, 1990; Esser et al., 1998). In addition, factors derived from endothelial cells, including leukemia inhibitory factor (LIF), modulate astrocytic differentiation, demonstrating that endothelium and astrocytes are engaged in a two-way inductive process (Abbott, 2006). In Drosophila, cadherins (e.g. E-cadherin) are essential for tracheal morphogenesis (Tanaka-Matakatsu et al., 1996; Tepass et al., 1996; Uemura et al., 1996). A more specific role of these and other adhesion systems in the tracheation of the nervous system awaits to be studied.
In conclusion, as in so many other instances where comparisons between organogenetic processes in Drosophila and vertebrates were conducted, one is confronted with a surprising degree of similarity, both in structural morphogenetic mechanisms and their molecular control. Given that there is little support for true homologies [tracheae are highly derived structures that only appear in insects, and even glia as a tissue might not have been present in the bilaterian ancestor (Radojcic and Pentreath, 1979; Pereanu et al., 2005). it is reasonable to assume that the constraints that acted during the independent evolution of a tracheated insect nervous system and a vascularized vertebrate nervous system were similar. In both scenarios, epithelial tubes, branching in a dichotomous way, penetrate the neural primordium at an early stage. In branching out and permeating the volume of the growing nervous system, tracheae/blood vessels had to adopt to the special microenvironment presented by the neural primordium, a microenvironment that is composed of conserved elements like axonal growth cones, a specific extracellular matrix, signaling systems controlling midline crossing, and others. Some of these elements are most likely homologous, i.e., existed in the bilaterian ancestor; one only need to look at the conserved role of the Slit/Robo system, which regulates midline crossing of axons in animals ranging from flies to humans (Nguyen-Ba-Charvet and Chedotal, 2002). Novel elements that, later in evolution and possibly multiple times, were added to this conserved microenvironment presented by the neural primordium, made use of the elements already present for their own development, and thereby convergently evolved a large number of similar characteristics.
Acknowledgments
This work was supported by National Institutes of Health Grant NS29367 to V.H., the United States Public Health Service National Research Service Award GM07185 to W.P.and to Shana Fawcett. The monoclonal antibodies developed by T. Uemura, C. Goodman and E. Knust were obtained from the Developmental Studies Hybridoma Bank developed under the auspices of the NICHD and maintained by The University of Iowa, Department of Biological Sciences, Iowa City, IA 52242.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abbott NJ, Ronnback L, Hansson E. Astrocyte-endothelial interactions at the blood-brain barrier. Nat. Rev. Neurosci. 2006;7(1):41–53. doi: 10.1038/nrn1824. [DOI] [PubMed] [Google Scholar]
- Affolter M, Bellusci S, Itoh N, Shilo B, Theiry JP, Werb Z. Tube or not tube: remodeling epithelial tissues by branching morphogenesis. Dev. Cell. 2003;4(1):11–18. doi: 10.1016/s1534-5807(02)00410-0. [DOI] [PubMed] [Google Scholar]
- Arquier N, Vigne P, Duplan E, Hsu T, Therond PP, Frelin C, D’Angelo G. Analysis of the hypoxia-sensing pathway in Drosophila melanogaster. Biochem. J. 2006;393(2):471–480. doi: 10.1042/BJ20050675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M, editor. Drosophila. A Laboratory Manual. Cold Spring Harbor Laboratory Press; New York: 1989. [Google Scholar]
- Bernardoni R, Vivancos V, Giangrande A. glide/gcm is expressed and required in the scavenger cell lineage. Dev. Biol. 1997;191(1):118–130. doi: 10.1006/dbio.1997.8702. [DOI] [PubMed] [Google Scholar]
- Bruckner K, Kockel L, Duchek P, Luque CM, Rorth P, Perrimon N. The PDGF/VEGF receptor controls blood cell survival in Drosophila. Dev. Cell. 2004;7(1):73–84. doi: 10.1016/j.devcel.2004.06.007. [DOI] [PubMed] [Google Scholar]
- Campos-Ortega JA, Hartenstein V, editors. The Embryonic Development of Drosophila melanogaster. 2nd edition Springer Press; New York: 1997. [Google Scholar]
- Cho NK, Keyes L, Johnson E, Heller J, Ryner L, Karim F, Krasnow MA. Developmental control of blood cell migration by the Drosophila VEGF pathway. Cell. 2002;108(6):865–876. doi: 10.1016/s0092-8674(02)00676-1. [DOI] [PubMed] [Google Scholar]
- Condron B. Spatially discrete FGF-mediated signaling directs glial morphogenesis. Development. 1999;126(20):4635–4641. doi: 10.1242/dev.126.20.4635. [DOI] [PubMed] [Google Scholar]
- Englund C, Uv AE, Cantera R, Mathies LD, Krasnow MA, Samakovlis C. adrift, a novel bnl-induced Drosophila gene, required for tracheal pathfinding into the CNS. Development. 1999;126(7):1505–1514. doi: 10.1242/dev.126.7.1505. [DOI] [PubMed] [Google Scholar]
- Englund C, Steneberg P, Falileeva L, Xylourgidis N, Samakovlis C. Attractive and repulsive functions of Slit are mediated by different receptors in the Drosophila trachea. Development. 2002;129(21):4941–4951. doi: 10.1242/dev.129.21.4941. [DOI] [PubMed] [Google Scholar]
- Esser S, Wolburg K, Wolburg H, Breier G, Kurzchalia T, Risau W. Vascular endothelial growth factors induces endothelial fenestration in vitro. J. Cell Biol. 1998;140(4):947–959. doi: 10.1083/jcb.140.4.947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerhardt H, Golding M, Fruttiger M, Ruhrberg C, Lundkvist A, Abramsson A, Jeltsch M, Mitchell C, Alitalo K, Shima D, Betsholtz C. VEGF guides angiogenic sprouting utilizing endothelial tip cell filopodia. J. Cell Biol. 2003;161(6):1163–1177. doi: 10.1083/jcb.200302047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerhardt H, Ruhrberg C, Abramsson A, Fujisawa H, Shima D, Betsholtz C. Neuropilin-1 is required for endothelial tip cell guidance in the developing central nervous system. Dev. Dyn. 2004;231(3):503–509. doi: 10.1002/dvdy.20148. [DOI] [PubMed] [Google Scholar]
- Gerhardt H, Liebner S, Redies C, Wolburg H. N-cadherin expression in endothelial cells during early angiogenesis in the eye and brain of the chicken: relation to blood-retina and blood-brain barrier development. Eur. J. Neurosci. 1999;11(4):1191–1201. doi: 10.1046/j.1460-9568.1999.00526.x. [DOI] [PubMed] [Google Scholar]
- Guillemin K, Groppe J, Ducker K, Treisman R, Hafen E, Affolter M, Krasnow MA. The pruned gene encodes the Drosophila serum response factor and regulates cytoplasmic outgrowth during terminal branching of the tracheal system. Development. 1996;122(5):1353–1362. doi: 10.1242/dev.122.5.1353. [DOI] [PubMed] [Google Scholar]
- Hacohen N, Kramer S, Sutherland D, Hiromi Y, Krasnow MA. sprouty encodes a novel antagonist of FGF signaling that patterns apical branching of the Drosophila airways. Cell. 1998;92(2):253–263. doi: 10.1016/s0092-8674(00)80919-8. [DOI] [PubMed] [Google Scholar]
- Hartenstein V. Atlas of Drosophila Development. In: Bate M, Martinez-Arias A, editors. The Development of Drosophila. Cold Spring Harbor Laboratory Press; Cold Spring Harbor: 1993. [Google Scholar]
- Hosoya T, Takizawa K, Nitta K, Hotta Y. glial cells missing: a binary switch between neuronal and glial determination in Drosophila. Cell. 1995;82(6):1025–1036. doi: 10.1016/0092-8674(95)90281-3. [DOI] [PubMed] [Google Scholar]
- Ito K, Awano W, Suzuki K, Hiromi Y, Yamamoto D. The Drosophila mushroom body is a quadruple structure of clonal units each of which contains a virtually identical set of neurons and glial cells. Development. 1997;124(4):761–771. doi: 10.1242/dev.124.4.761. [DOI] [PubMed] [Google Scholar]
- Jarecki J, Johnson E, Krasnow MA. Oxygen regulation of airway branching in Drosophila is mediated by branchless FGF. Cell. 1999;99(2):211–220. doi: 10.1016/s0092-8674(00)81652-9. [DOI] [PubMed] [Google Scholar]
- Jones BW, Fetter RD, Tear G, Goodman CS. glial cells missing: a genetic switch that controls glial versus neuronal fate. Cell. 1995;82(6):1013–1023. doi: 10.1016/0092-8674(95)90280-5. [DOI] [PubMed] [Google Scholar]
- Jones BW. Glial cell development in the Drosophila embryo. Bioessays. 2001;23(10):877–887. doi: 10.1002/bies.1129. [DOI] [PubMed] [Google Scholar]
- Klambt C, Glazer L, Shilo BZ. breathless, a Drosophila FGF receptor homolog, is essential for migration of tracheal and specific midline glial cells. Genes Dev. 1992;6(9):1668–1678. doi: 10.1101/gad.6.9.1668. [DOI] [PubMed] [Google Scholar]
- Lubarsky B, Krasnow MA. Tube morphogenesis: making and shaping biological tubes. Cell. 2003;112(1):19–28. doi: 10.1016/s0092-8674(02)01283-7. [DOI] [PubMed] [Google Scholar]
- Lundstrom A, Gallio M, Englund C, Steneberg P, Hemphala J, Aspenstrom P, Keleman K, Falileeva L, Dickson BJ, Samakovlis C. Vilse, a conserved Rac/Cdc42 GAP mediating Robo repulsion in tracheal cells and axons. Genes Dev. 2004;18(17):2161–2171. doi: 10.1101/gad.310204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning G, Krasnow MA. Development of the Drosophila tracheal system. In: Bate M, Arias AM, editors. The Development of Drosophila melanogaster. Cold Spring Harbor Laboratory Press; Cold Spring Harbor: 1993. pp. 609–685. [Google Scholar]
- Manseau L, Baradaran A, Brower D, Budhu A, Elefant F, Phan H, Philp AV, Yang M, Glover D, Kaiser K, Palter K, Selleck S. GAL4 enhancer traps expressed in the embryo, larval brain, imaginal discs, and ovary of Drosophila. Dev. Dyn. 1997;209(3):310–322. doi: 10.1002/(SICI)1097-0177(199707)209:3<310::AID-AJA6>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- Meinertzhagen IA, Hanson TE. The development of the optic lobe. In: Bate M, Martinez-Arias A, editors. The Development of Drosophila. Cold Spring Habor Laboratory Press; Cold Spring Habor, NY: 1993. pp. 1363–1492. [Google Scholar]
- Metzger RJ, Krasnow MA. Genetic control of branching morphogenesis. Science. 1999;284(5420):1635–1639. doi: 10.1126/science.284.5420.1635. [DOI] [PubMed] [Google Scholar]
- Nguyen-Ba-Charvet KV, Chedotal A. Role of Slit proteins in the vertebrate brain. J. Physiol. Paris. 2002;96(12):91–98. doi: 10.1016/s0928-4257(01)00084-5. [DOI] [PubMed] [Google Scholar]
- Pereanu W, Hartenstein V. Digital three-dimensional models of Drosophila development. Curr. Opin. Gen. Dev. 2004;14(4):382–391. doi: 10.1016/j.gde.2004.06.010. [DOI] [PubMed] [Google Scholar]
- Pereanu W, Shy D, Hartenstein V. Morphogenesis and proliferation of the larval brain glia in Drosophila. Dev. Biol. 2005;283(1):191–203. doi: 10.1016/j.ydbio.2005.04.024. [DOI] [PubMed] [Google Scholar]
- Radojcic T, Pentreath VW. Invertebrate glia. Prog. Neurobiol. 1979;12(2):115–179. doi: 10.1016/0301-0082(79)90002-9. [DOI] [PubMed] [Google Scholar]
- Risau W, Wolburg H. Development of the blood-brain barrier. Trends Neurosci. 1990;13(5):174–178. doi: 10.1016/0166-2236(90)90043-a. [DOI] [PubMed] [Google Scholar]
- Rosin D, Shilo BZ. Branch-specific migration cues in the Drosophila tracheal system. Bioessays. 2002;24(2):110–113. doi: 10.1002/bies.10041. [DOI] [PubMed] [Google Scholar]
- Rovainen CM, Kakarala MH. Angiogenesis on the optic tectum of albino Xenopus laevis tadpoles. Brain Res. Dev. Brain Res. 1989;48(2):197–213. doi: 10.1016/0165-3806(89)90076-x. [DOI] [PubMed] [Google Scholar]
- Shilo BZ, Gabay L, Glazer L, Reichman-Fried M, Wappner P, Wilk R, Zelzer E. Branching morphogenesis in the Drosophila tracheal system. Cold Spring Harb. Symp. Quant. Biol. 1997;62:241–247. [PubMed] [Google Scholar]
- Shishido E, Ono N, Kojima T, Saigo K. Requirements of DFR1/Heartless, a mesoderm-specific Drosophila FGF-receptor, for the formation of heart, visceral and somatic muscles, and ensheathing of longitudinal axon tracts in CNS. Development. 1997;124(11):2119–2128. doi: 10.1242/dev.124.11.2119. [DOI] [PubMed] [Google Scholar]
- Strong LH. The early embryonic pattern of internal vascularization of the mammalian cerebral cortex. J. Comp. Neur. 1964;123:121–138. doi: 10.1002/cne.901230111. [DOI] [PubMed] [Google Scholar]
- Sun B, Xu P, Salvaterra PM. Dynamic visualization of nervous system in live Drosophila. Proc. Natl. Acad. Sci. U.S.A. 1999;96(18):10438–10443. doi: 10.1073/pnas.96.18.10438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutherland D, Samakovlis C, Krasnow MA. branchless encodes a Drosophila FGF homolog that controls tracheal cell migration and the pattern of branching. Cell. 1996;87(6):1091–1101. doi: 10.1016/s0092-8674(00)81803-6. [DOI] [PubMed] [Google Scholar]
- Takizawa K, Hotta Y. Pathfinding analysis in a glia-less gcm mutant in Drosophila. Dev. Genes Evol. 2001;211(1):30–36. doi: 10.1007/s004270000117. [DOI] [PubMed] [Google Scholar]
- Tanaka-Matakatsu M, Uemura T, Oda H, Takeichi M, Hayashi S. Cadherin-mediated cell adhesion and cell motility in Drosophila trachea regulated by the transcription factor escargot. Development. 1996;122(12):3697–3705. doi: 10.1242/dev.122.12.3697. [DOI] [PubMed] [Google Scholar]
- Tepass U, Fessler LI, Aziz A, Hartenstien V. Embryonic origin of hemocytes and their relationship to cell death in Drosophila. Development. 1994;120(7):1829–1837. doi: 10.1242/dev.120.7.1829. [DOI] [PubMed] [Google Scholar]
- Tepass U, Gruszynski-DeFeo E, Haag TA, Omatyar L, Torok T, Hartenstein V. shotgun encodes Drosophila E-cadherin and is preferentially required during cell rearrangement in the neuroectoderm and other morphogenetically active epithelia. Genes Dev. 1996;10(6):672–685. doi: 10.1101/gad.10.6.672. [DOI] [PubMed] [Google Scholar]
- Uemura T, Oda H, Kraut R, Hayashi S, Kotaoka Y, Takeichi M. Zygotic Drosophila E-cadherin expression is required for processes of dynamic epithelial cell rearrangement in the Drosophila embryo. Genes Dev. 1996;10(6):659–671. doi: 10.1101/gad.10.6.659. [DOI] [PubMed] [Google Scholar]
- Wing JP, Zhou L, Schwartz LM, Nambu JR. Distinct cell killing properties of the Drosophila reaper, head involution defective, and grim genes. Cell Death Differ. 1998;5(11):930–939. doi: 10.1038/sj.cdd.4400423. [DOI] [PubMed] [Google Scholar]
- Younossi-Hartenstein A, Salvaterra PM, Hartenstein V. Early development of the Drosophila brain: IV. Larval neuropile compartments defined by glial septa. J Comp Neurol. 2003;455(4):435–450. doi: 10.1002/cne.10483. [DOI] [PubMed] [Google Scholar]
- Zhou L, Schnitzler A, Agapite J, Schwartz LM, Steller H, Nambu JR. Cooperative functions of the reaper and head involution defective genes in the programmed cell death of Drosophila central nervous system midline cells. Proc. Natl. Acad. Sci. U.S.A. 1997;94(10):5131–5136. doi: 10.1073/pnas.94.10.5131. [DOI] [PMC free article] [PubMed] [Google Scholar]


