Abstract
The testis specific protein Y encoded (TSPY) gene is a tandemly repeated gene on the mammalian Y chromosome. It encodes several slightly variant proteins that harbor a conserved domain of ~170 amino acids, termed TSPY/SET/NAP1 domain, capable of binding to cyclin B. The human TSPY is preferentially expressed in spermatogonia and to lesser extent the spermatids. Although rat harbors a single functional Tspy gene on its Y-chromosome, the human and rat genes differ in their expression patterns, suggesting that they might serve different or variant functions in the testis. Transcripts of rTspy were first detected in the testis of 28 days old rats, at which time the first wave of meiotic division was occurring. The rTspy protein was initially detected in stage-9 elongating spermatids and peaked at stage-13 spermatids in adult testis, but not in spermatogonia, unlike the expression pattern of the human TSPY gene. Using a GST pull-down assay, we demonstrated that rTspy could bind to the core histones H2A, H2B, H3 and H4. Rat Tspy co-localized with the histones in the cytoplasm of selected elongated spermatids. Our results suggest that the rTspy may play critical roles as a histone chaperone during maturation of the elongating spermatids in the rat testis.
Keywords: TSPY, spermatid, histone H2A, H2B, NAP/SET, Y-chromosome
Introduction
The testis specific protein Y-encoded (TSPY) is an evolutionarily conserved gene on the Y chromosome of mammals, including humans, primates, bovines and rodents [1–6]. The human TSPY (hTSPY) and bovine TSPY (bTSPY) genes are expressed primarily in the spermatogonia of adult testis. TSPY has been postulated to serve a vital function in spermatogonial proliferation and early meiotic division [1, 7, 8]. Significantly, the human TSPY is tandemly repeated 23–64 times and is principally located in the critical region of a cancer predisposition locus, gonadoblastoma on the Y chromosome (GBY) that is associated with gonadoblastoma development in the dysgenetic gonads of XY females and intersex patients [7, 9–11]. Indeed TSPY is strongly expressed in gonadoblastoma tissues and is considered to be the putative gene for GBY [12–14].
In rodents, the mouse harbors an apparently nonfunctional Tspy gene on its Y chromosome with multiple in-frame nonsense mutations in its open reading frame [15, 16]. These nonsense mutations seem to be results of recent evolutionary events since numerous related species, such as Apodemus sylvaticus [6], do possess a functional Tspy gene on their Y chromosome that is capable of encoding sizable proteins with the conserved TSPY/SET/NAP1 domain. The apparent inactivation of the mouse Tspy gene suggests that the autosomal Tspy-like (Tspyl) or other Y-located gene(s) would have assumed its function(s) in germ cell biology. Indeed, there are some examples in the mouse in which autosomal gene has substituted for its homologous human Y-chromosomal gene. One such example is the human heat shock transcription factor on Y-chromosome (HSFY) gene whose mouse homologue, the HSFY-like gene, is located on an autosome, and both genes seem to have an evolutionarily conserved function(s) in spermatogenesis [17]. The rat harbors a functional Tspy (rTspy) gene that encodes a 334 amino acid protein with the TSPY/SET/NAP1 domain [5, 6]. To gain some insights in the function(s) of the rodent Tspy in testis, we had characterized the postnatal expression of the rat Tspy gene in the maturing testis using reverse transcription polymerase chain reaction (RT-PCR) and immunohistochemistry analyses. Our results showed that the expression pattern for the rat Tspy is distinctly different from that of the human TSPY [7, 18]. The rTspy is specifically expressed in elongating spermatids at late stages of spermatogenesis while the hTSPY is expressed preferentially in spermatogonial cells. Cytologically, the rTspy protein was primarily localized in the cytoplasm while the hTSPY shows both cytoplasmic and nuclear locations [18]. Since the expression of the rTspy coincides with the chromatin remodeling stages in which the nuclear histones are being replaced by protamines and exported to the cytoplasm and several members of the SET/NAP proteins are capable of binding to histones in the nucleus involved in gene regulation, we explored the possibility that rTspy might interact with the displaced histones in the cytoplasm. To evaluate this possibility, we had performed additional GST pull-down assays, and demonstrated that the rTspy could interact with the core histones H2A, H2B, H3 and H4, and was co-localized with histone H2B in the cytoplasm of selected elongating spermatids, suggesting that the rTspy may serve a role in spermiogenesis in the rat.
Materials and Methods
Animals
Wistar rats and CD1 mice were purchased from the Charles River Laboratories, Inc. (Wilmington, MA). The Institutional Animal Care and Use Committee of the Veterans Affairs Medical Center, San Francisco approved all experimental procedures in accordance with the NIH Guide for Care and Use of Laboratory Animals.
RT-PCR
Total RNAs were isolated from dissected tissues using the TRIZOL reagent (Invitrogen, Carlsbad, CA) and purified by RQ1-DNase treatment (Promega, Madison, WI) to remove any contaminant DNA. Total RNA (0.36 μg) was reverse-transcribed in a volume of 21 μl using the SuperScript II reverse transcriptase kit (Invitrogen, Carlsbad, CA). One μl of reverse transcribed product was subjected to PCR using a touchdown procedure [18] and specific primer sets for respective genes (Table 1). The PCR products were analyzed by 1.2% agarose gel electrophoresis and visualized with ethidium bromide staining.
Table 1.
Sequences of primer sets used in RT-PCR analysis and construction of cDNA.
| Name | Gene | Direction | Sequence (5′ −> 3′) |
|---|---|---|---|
| rTSPY1F | rTspy (exon 1) | forward | ATGGAGAATTCTGAGGAGGAGAGTGTGG |
| rTSPY1R | rTspy (exon 1) | reverse | GCATTCACAAAGCTGAGCTCCAGTTG |
| rTSPY4F | rTspy (1–334) | forward | GGATCCATGGAGACTCTAGAGGAGGAGAGTGT |
| rTSPY4R | rTspy (1–334) | reverse | CGGTGGATCCCGTGAGTGGTCTTCCTTAGGGTAGTAGT |
| rTSPY5R | rTspy (1–96) | reverse | GCCTCGAGTTAATTCTTGACATCTACCACCTCTGT |
| rTSPY6R | rTspy (1–162) | reverse | GCCTCGAGTTACACAAAGCTGAGCTCCAGTTG |
| rTSPY7R | rTspy (1–184) | reverse | GCCTCGAGTTAAAAGTGAGGTCTGCGCATTTT |
| rTSPY8F | rTspy (187–206) | forward | GATCCCGCAGAAAGACCATAATCCAGGGCATTCCAGGC–TTCTGGGCTAAAGCTATGATGAACCATC |
| rTSPY8R | rTspy (187–206) | reverse | TCGAGATGGTTCATCATAGCTTTAGCCCAGAAGCCTGG-AATGCCCTGGATTATGGTCTTTCTGCGG |
| rTSPY9F | rTspy (213–232) | forward | GATCCATCAGCAACCAAGATGAAGACTTACTGAGCTAC-ATGTTGAGCTTGGAGGTGGAGGAGTACC |
| rTSPY9R | rTspy (213–232) | reverse | TCGAGGTACTCCTCCACCTCCAAGCTCAACATGTAGCTC-AGTAAGTCTTCATCTTGGTTGCTGATG |
| Ssty1F | Ssty1 | forward | AAAGGCAAGCCAGCTCCTGAACTCCA |
| Ssty1R | Ssty1 | reverse | TTCCATTGGGTGACAGGCTCATTACC |
| Eif2s3yF | Eif2s3y | forward | AGCCGCATCTTTCTCGTCAGGATCTT |
| Eif2s3yR | Eif2s3y | reverse | TTCAATGGCAGCCAGGTGTTCAGAAG |
| TH2BF | testis H2B | forward | GCGAATTCTATGCCGGAGGTGTCGGCAAAGGGG |
| TH2BR | testis H2B | reverse | GCGGATCCTCACTTGGAGCTGGTGTACTTGGT |
| β–actin5 | β–actin | forward | TCACCCACACTGTGCCCATCTACGA |
| β–actin2R | β–actin | reverse | CCACGTCACACTTCATGATGGA |
Generation of rTspy antiserum
The full-length rat Tspy cDNA was initially isolated from adult testis by RT-PCR with primers rTSPY4F and rTSPY4R, and cloned into the pGEM-T Easy Vector System (Promega, Madison, WI). After verification of its sequence, the rTspy cDNA was inserted into the BamHI site of pGEX-4T-3 vector (Amersham Biosciences Corp., Piscataway, NJ). GST-rTspy fusion protein was expressed in bacterial host, BL21DE3, and purified with affinity chromatography using glutathione-Sepharose 4B (Amersham Biosciences Corp, Piscataway, NJ). One mg of GST-rTspy protein per animal was used to immunize two New Zealand white rabbits at multiple time points to generate polyclonal antibodies using the service of a commercial facility (Sigma-Genosys, Woodlands, TX). The anti-rTspy and pre-immune antisera were further purified by absorbing with GST-Sepharose resins and total mouse testis extracts as described by Conlon and Rossant [19]. The resulting supernatants were used as primary antibodies for immunohistochemistry studies. The respective antisera were confirmed by Western-blot with recombinant rTspy protein, as described below.
Western blot analysis
The rTspy cDNA was inserted into the BamHI site of mammalian expression vector, pCS2plus [20, 21], and transfected to COS7 cells using FuGENE6 (Roche, Indianapolis, IN), according to the manufacturer’s instructions. The cells were harvested 48 h after the transfection, washed with PBS and lysed with 100 μl of TNBT buffer containing 50 mM Tris-HCl (pH 7.0), 150 mM NaCl, 1% NP-40, and 0.25% deoxycholic acid. Five μl of cell lysates or 50 μg of rat testis extracts were subjected to SDS-PAGE on 12% (w/v) polyacrylamide gel and processed for Western blot using a semi-dry transfer method [22] and the purified rTspy antiserum. The binding of the primary antibody was detected with a peroxidase-conjugated anti-rabbit IgG antibody and visualized by the ECL-plus chemiluminescence system (Amersham Biosciences Corp, Piscataway, NJ) or diaminobenzine (DAB) (Vector Laboratories, Burlingame, CA).
Histology and immunohistochemistry
Rat tissues were fixed for 12 h in Bouin’s fluid (Sigma-Aldrich, St. Louis, MO) or paraformaldehyde-PB solution, 4% (w/v) paraformaldehyde, 100 mM phosphate buffer (pH 7.4) at 4 °C, dehydrated in ethanol series and embedded in paraffin wax. Five μm sections were dewaxed and rehydrated through alcohol series and finally in distilled water. Antigen retrieval was performed in 100 mM Tris-HCl (pH 10) at 95 °C for 30 min. The sections were incubated in 3% hydrogen peroxide, washed in PBS, and nonspecific protein binding was blocked using 3% BSA–PBS solution. Sections were incubated overnight in a humid chamber at 4 °C with anti-rTspy (1:400) or anti-histone H2B (1:200, Upstate, Charlottesville, VA) antibody. The immunoreactive sites were detected with the SuperPicTure Polymer Detection kit (ZYMED/Invitrogen, Carlsbad, CA) and sections were counterstained with hematoxylin (Fisher Scientific, Hampton, NH). For identification of rTspy-positive germ cells, the Tyramide Signal Amplification Kit (with biotin-XX; Molecular Probes/Invitrogen, Carlsbad, CA) was used to amplify the immunoreacting signals following the incubation with the SuperPicTure Polymer Detection kit.
Immunofluorescence was performed with paraformaldehyde-fixed sections and processed similarly, as described above. The signals were visualized by Alexa488 conjugated anti-rabbit IgG (Molecular Probes/Invitrogen). For double immunofluorescence, the anti-rTspy and anti-H2B antibodies were directly labeled in red and green respectively by Zenon Rabbit IgG Labeling Kit (Molecular Probes/Invitrogen, Carlsbad, CA) according to the manufacturer’s instructions. DNA was stained with DAPI (Roche, Indianapolis, IN).
GST pull-down assay
Various rat Tspy cDNAs harboring different lengths of its coding sequences were initially generated by PCR with the full-length rTspy cDNA and primers (Table 1), and cloned into the pGEM-T Easy Vector System (Promega, Madison, WI). After verification of their sequences, the cDNAs were individually inserted between BamHI and XhoI sites of the expression vector, pGEX-4T-3. The hTSPY cDNA [3] was inserted in the plasmid pGEX-4T-3 and processed similarly for fusion protein production in bacterial host. Approximately 250 μg of GST, GST-rTspy or GST-hTSPY proteins were bound separately to glutathione-Sepharose 4B (Amersham Biosciences Corp, Piscataway, NJ) and reacted independently with 5 μg of histone H2A or H2B (Upstate Biochemicals, Waltham, MA) in 100 μl of buffer A (PBS, 20% glycerol, 0.1% NP40, 5 mM DTT, proteinase inhibitors with EDTA [Roche]) overnight at 4 °C. The GST pull-down assays with histones H3 and H4 (Upstate, Waltham, MA) were performed similarly with modified buffer A, containing additional 1% NP40 and 250 mM NaCl to reduce non-specific binding of GST. Protein complexes bound to the Sepharose beads were precipitated by centrifugation and washed with 500 μl of buffer A for 10 min. Proteins were eluted with 40 μl of SDS sample buffer, analyzed by SDS-PAGE on 10% to 20 % (w/v) polyacrylamide gradient gels (Bio-Rad Laboratories, Hercules, CA), and visualized by Coomassie blue staining or Western-blot with appropriate antibodies (Upstate, Waltham, MA). The relative amounts of bound and unbound histones were estimated by the NIH Imager Program (http://rsb.info.nih.gov/nih-image/index.html).
Immunocytochemical analysis
The C-terminal truncated rat Tspy cDNAs, rTspy(1–96) and rTspy(1–162), were individually inserted between BamHI and XhoI sites of the mammalian expression vector, pCS2plus. The testis type histone H2B (TH2B) cDNA was initially isolated from adult testis by RT-PCR with primers TH2BF and TH2BR (Table 1), and cloned into the pGEM-T Easy Vector System. After sequence verification, the cDNA was inserted between EcoRI and BamHI sites of the green fluorescence protein expression vector pEGFP-C1 (Clonthech Laboratories, Mountain View, CA) to express EGFP-TH2B fusion protein. The resultant pCS2-rTspy variants and/or pEGFP-TH2B plasmid were transiently transfected to COS7 cell using FuGENE6. Forty eight hours after the transfection, the cells were fixed by 10% formalin-PBS solution for 5 min, and processed for immunofluorescence with the anti-rTspy antiserum and anti-EGFP mouse IgG (Clontech Laboratories, Mountain View, CA). The immunoreactive signals were detected by Alexa594 conjugated anti-rabbit IgG and Alexa488 conjugated anti-mouse IgG (Molecular Probes/Invitrogen, Carlsbad, CA) and analyzed by fluorescence microscopy.
Results
Developmental expression of rTspy in postnatal testes
The tissue-specific and developmental expression of the rTspy was analyzed by RT-PCR. The open reading frame of the rTspy transcript encodes a protein with 334 amino acids spanning from exon 1 to exon 5 (Genbank accession no. AF074880) [5]. In adult animals, rTspy transcripts were detected abundantly in testis, but not in other tissues examined (Fig. 1A), consistent with the results of Dechend and coworker [5]. To determine the overall time course of rTspy expression in developing testis, similar studies were conducted with total RNAs derived from 2 male rats each at ages 4, 12, 20, 28, 36, and 44 days. Our results showed that rTspy expression was initially detected in testes of rats at 28 days of age, when the first wave of meiotic division occurred. The level of rTspy expression increased with age of the animals (Fig. 1B, rTspy). As references, expression of the Y-located Ssty1 and Eif2s3y genes was analyzed similarly. In the mouse, Ssty1 is expressed specifically in spermatids [23, 24] while Eif2s3y is primarily expressed in spermatogonial cells and germ cells through the meiotic prophase [25]. Similar to rTspy, transcript or the rat Ssty1 was detected only at or after 28 days of age while Eif2s3y transcript was more ubiquitous among testes of different ages (Fig. 1B, Ssty1 and Eif2s3y), reflecting their spermatid and spermatogonia/early germ cell expression patterns, respectively. The authenticity of the amplified products was confirmed to be those of Ssty1 and Eif2s3y respectively by DNA sequencing analysis of the RT-PCR products (data not shown). Our results suggested that rTspy mRNA was expressed specifically in haploid germ cells.
Figure 1.
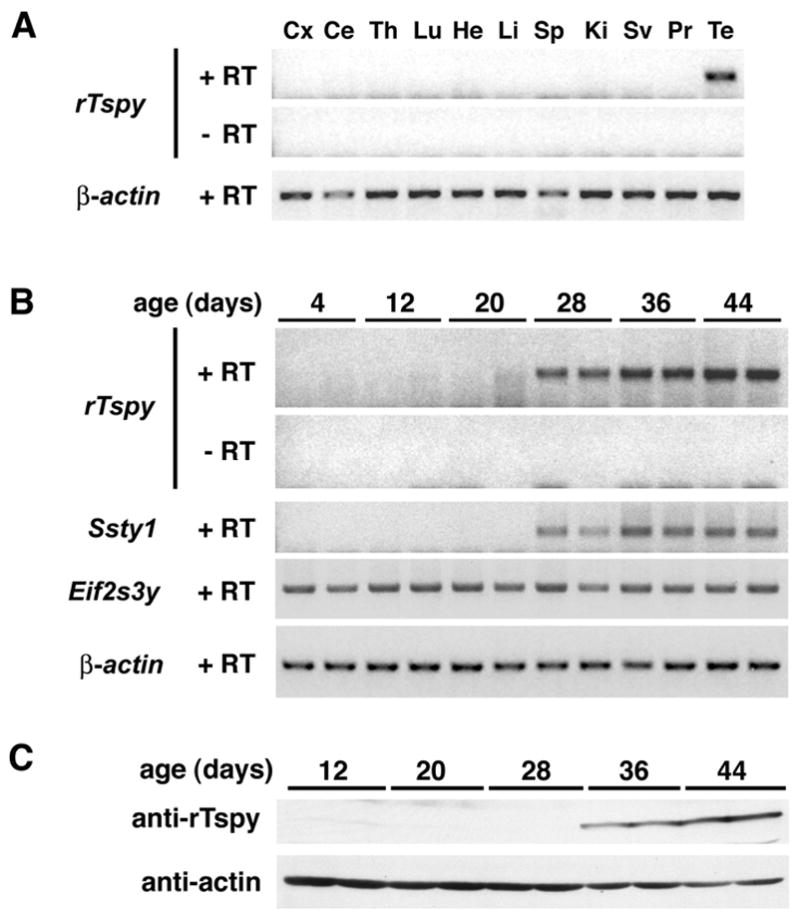
The tissue specific expression in rat Tspy in developmental and adult testes. (A) Tissue specificity was analyzed by RT-PCR. Tspy was detected only in adult rat testes. (B) Tspy mRNA expression in rat postnatal testes. Total RNAs from testes of 2 rats at 4, 12, 20, 28, 36, and 44 days of age were individually analyzed by RT-PCR with a primer set derived from exon 1 of the rat Tspy gene. Significant signals were detected in the testes of 28 days and older rats. The expression of rat Ssty1 and Eif2s3y were analyzed in parallel with rTspy. In the case of the mouse, Ssty1 is expressed in elongated spermatids while Eif2s3y is expressed in spermatogonia and prophase spermatocytes. The similar patterns between the rTspy and Ssty1 suggest that they are expressed in similar manner in the postnatal rat testes. (C) Tspy protein expression in postnatal rat testes. Total proteins from testes of 2 rats at 12, 20, 28, 36, and 44 days of age were analyzed by Western-blot by using anti-rTspy and anti-actin antibodies, respectively. Significant signals were detected in the testes of 36 days and older rats. Abbreviations; Cx, cerebral cortex; Ce, cerebellum; Th, thymus; Lu, lung; He, heart; Li, liver; Sp, spleen; Ki, kidney; Sv, seminal vesicle; Pr, prostate; Te, testis; +RT, with reverse transcription;-RT, without reverse transcription. β-actin was used as a positive control for all samples.
The developmental expression of rTspy protein was analyzed by Western-blot with testicular protein extracts from 12, 20, 28, 36, and 44 days old rats. The result showed that rTspy protein was initially detected in the testis of 36-day old rats (Fig. 1C, anti-rTspy), while the rTspy mRNA was detected in those at 28 days and later (Fig. 1B, rTspy). The most differentiated type of germ cells was spermatocytes, round spermatids and elongating spermatids of 20, 28, and 36 days old rats respectively. Our results, taken together, suggested that rTspy is initially transcribed in round spermatids and translated in elongated spermatids.
The rTspy is specifically located in the elongated spermatids in adult testis
To confirm the results obtained from Western-blot of developmental testes, we had performed an immunochemical staining study on adult rat testis using a polyclonal antiserum against the rTspy protein. The specificity of this antiserum was initially demonstrated by Western-blot of total protein lysates derived from COS7 cells transfected with rTspy expression vector (Fig. 2I, lane 2, arrowhead). The respective rTspy band could be abolished when the antiserum was pre-absorbed with excess GST-rTspy recombinant protein (Fig. 2I, lane 4). Although the conserved TSPY/SET/NAP1 domain is also present in the products of other members of the TSPY/TSPY-like/SET/NAP-1 (TTSN) superfamily (e.g. Tspy-like protein 1 [26] and DENTT/Tspy-like 2 protein [27]), no cross reactivity with hTSPY on Western-blot was observed (data not shown), suggesting that our anti-rTspy antiserum was specific for the rTspy protein.
Figure 2.
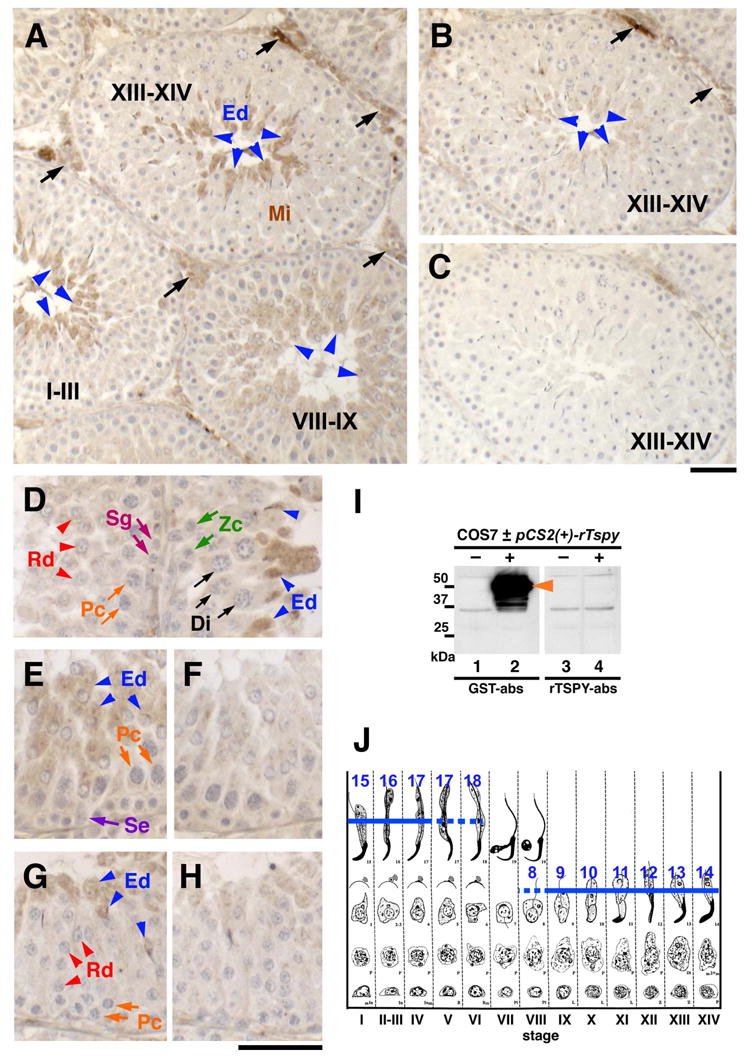
Detection of spermatid-specific expression of rat Tspy using immunohistochemistry. (A) Only elongating spermatids at various spermatogenic stages were stained positive for the anti-rTspy antiserum (arrowheads). Roman numbers indicate spermatogenic stages as described in J. (B) Image of an adjacent section immunostained similarly by a control antiserum preabsorbed with GST-rTspy. The staining of elongating spermatids was significantly reduced (arrowheads). (C) Image of an adjacent section immunostained by a preimmune serum as a negative control. (D) Elongating spermatids at stage XIII are positively stained (blue arrowheads), but spermatids of stage II–III were negative (red arrowheads). No other cells, including somatic cells, were positive for the antiserum (Sg, Zc, Pc and Di). (E–H) Sections of seminiferous tubules at spermatogenic stage VIII (E and F) and II–IV (G and H) immunostained by the anti-rTspy antiserum (E and G), and control antiserum preabsorbed with GST-rTspy (F and H). Only elongating seprmatids are faintly positive (blue arrowheads). Abbreviations; Di, diplotene spermatocytes; Ed, elongating spermatids; Mi, meiotic dividing cells; Pc, pachytene spermatocytes; Rd, round spermatids; Se, Sertoli cells; Sg, spermatogonia; Zc, zygotene spermatocytes. Scale bars = 50 μm in A–C and D–H, respectively. (I) Western-blot of COS7 cells transfected with a rat Tspy expression vector using the anti-rTspy antiserum pre-absorbed with GST alone (lanes 1 and 2; GST-abs) and with GST-rTspy fusion protein (lanes 3 and 4; rTspy-abs). Positive band for rat Tspy protein was detected in COS7 cells transfected with rat Tspy expression vector (lane 2), but not in non-transfected cells (lanes 1, 3) or those reacted with the same antiserum subtracted with GST-rTspy protein (lane 3, 4). Anti-rTspy serum detected recombinant rTspy (lane 2). (J) Spermatogenic stages of the rat testis. Blue lines indicate the immuno-positive cells in steps 8–18 of spermatid development. Romanic numbers indicate stages of spermatogenesis. Dotted lines represent sections showing faint staining of spermatids in these stages. Modified from review by Russell et al. [28].
Immunohistochemical analysis was performed on sections of rat testes fixed in Bouin’s fluid. To minimize any non-specific reactivity, we treated the antiserum with total mouse testis protein extract before using it in the immunostaining procedure. Although the mouse Tspy gene was transcribed, its transcripts were incapable of coding for the full-length or any protein of significant size. Hence, we did not anticipate any depletion of the rTspy antibodies in antiserum with this pre-absorption procedure. Using this processed rTspy antiserum, we observed specific immunostaining in the elongating spermatids at spermatogenic stages XIII to XIV (Fig. 2A, arrowheads in stage XIII–XIV, and Fig. 2J [28]). Again, the immunostaining was significantly decreased by pre-absorption with GST-rTspy (Fig. 2B, blue arrowheads). The Sertoli cells and germ cells from spermatogonia to stage 7 round spermatids did not react with this antiserum (Fig. 2D–G; Sg, Zc, Pc, Di, Rd, and Se; Fig. 2J). The immunostaining signals were detected faintly in stage 8 spermatids that were in the process of nuclear elongation (Fig. 2E, blue arrowheads; Fig. 2J). They reached maximal levels in stage 12 elongating spermatids (Fig. 2D, blue arrowheads), but rapidly decreased and became minimally detectable at stage 16–18 spermatids (Fig. 2G, blue arrowheads; Fig. 2J). The interstitial cells including Leydig cells showed some non-specific staining (Fig. 2A, black arrows) that did not diminish even after a pre-absorption with GST-rTspy (Fig. 2B, black arrows). Our results, taken together, suggested that the rTspy protein initially expressed in spermatids at approximately stage 8, peaked at in stage 12–14 elongating spermatids, and decreased during spermatogenic stage II to V elongated spermatids (Fig. 2J, blue lines; modified from Russell et al. [28]).
rTspy and hTSPY bind directly to core histones H2A, H2B, H3 and H4
The human TSPY and rat Tspy proteins harbor a conserved ~170 amino acids domain, known as the TSPY/SET/NAP1 domain, shared by numerous members of this protein superfamily, represented by TSPY, SET oncoprotein and nucleosome assembly protein 1 (NAP1) (NCBI Conserved Domain Search #pfam00956 [29]) [26, 30]. This conserved domain is located at residues #150–328 of the rTspy and residues #110–288 of the hTSPY (Fig. 3A). Numerous studies suggested that the proteins containing the TSPY/SET/NAP1 domain are capable of binding to either the mitotic cyclin B [31, 32] or core histones [33–35], and are involved in cell cycle regulation and the nucleosome assembly and/or chromatin remodeling respectively. In particular, SET/TAFI-β can serve as a subunit of the inhibitor of acetyltransferases (INHAT). It binds to the core histones H2B, H3 and H4 in the nucleus and either inhibits enzymatic activities of acetyltransferases or facilitates transcription of target genes [36–38]. Since transcriptional activities of the elongating spermatids are drastically reduced as their chromatin is undergoing compaction, it is uncertain if the rTspy is involved in any transcription modulation. However, chromatin compaction is usually accompanied by replacement of histones by protamines in the nucleus and exportation of the replaced histones to the cytoplasm of maturing spermatids. Although the cytoplasmic location of the rTspy protein would have excluded it as an active participant of any transcriptional processes, it raises the possibility that rTspy might interact with the displaced histones in the cytoplasm.
Figure 3.
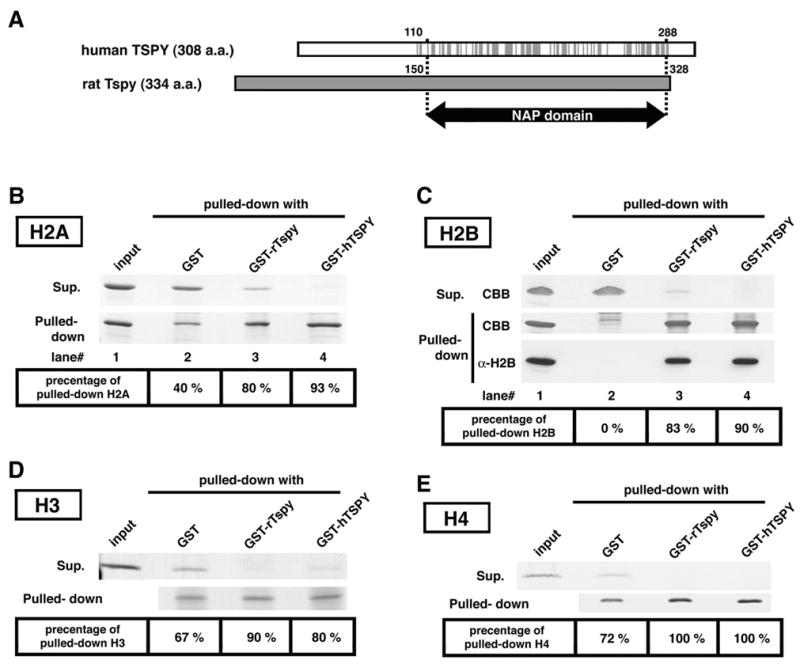
Interactions of rTspy and hTSPY with histones H2A, H2B, H3 and H4. (A) Alignment of hTSPY and rTspy proteins showing the homologous region encompassing the TSPY/SET/NAP1 domain. Gray lines on the hTSPY indicate positions with identical amino acid as the rTspy. The ~170 amino acid TSPY/SET/NAP1 domain is located between residue #110–288 of the hTSPY and residue #150–328 of the rTspy. The TSPY/SET/NAP1 domain is indicated by arrow (NAP domain). (B) GST pull-down assay of histone H2A using GST or GST-fusion proteins as indicated. Pulled-down proteins (pulled-down) and proteins of supernatant (supernatant) were subjected to SDS-PAGE, and visualized by Coomassie blue staining. Histone H2A was preferentially retained by Sepharose 4B resins conjugated with either GST-rTspy (lane 3) or GST-hTSPY (lane 4) but not by those conjugated with GST alone (lane 2). The numbers on bottom column indicate the approximate percentage of pulled-down H2A to total amount of H2A. Input represents equivalent material used for each GST pull-down assay. (C) GST pull-down assays of histone H2B using GST-fusion proteins. The experiment was performed similarly as described in B. Both GST-rTspy and GST-hTSPY fusion proteins (lane 3 and 4, respectively) showed significant affinity to histone H2B than GST alone (lane 2). The input represents the material used for GST pull-down assay. The authenticity of the pulled-down material as H2B was confirmed by Western-blot with anti-H2B antibody (bottom row, α-H2B). (D and E) GST pull-down assay of histones H3 (D) and H4 (E) performed similarly as described in B. Abbreviations; CBB, Coomassie blue stained band; α-H2B, Western-blot with anti-H2B antibody.
To evaluate this possibility, we performed a GST pull-down study between the rTspy/hTSPY and various purified core histones. This initial study demonstrated that core histones H2A, H2B, H3 and H4 was preferentially bound to GST-rTspy or -hTSPY fusion proteins, as compared to binding with GST alone (Fig. 3B–3E). Western blot analysis using anti-histone H2B antibody confirmed our observations that this histone was preferentially pulled down by both GST-rTspy and GST-hTSPY baits (Fig. 3C, α-H2B). These results suggest that both rTspy and hTSPY are capable of binding directly with the core histones H2A, H2B, H3 and H4.
It was reported that SET/TAF-Iβ interacts with histones through its C-terminal acidic tail [39, 40]. Although neither rTspy nor hTSPY has such a long acidic tail, as in SET/TAF-Iβ, rTspy as a whole is highly acidic (with a predicted pI of 4.39). Rat Tspy contains abundant aspartic acid and glutamic acid residues, within its N-terminal region (Fig. 4A). Hence, we tested the histone binding ability for the N-terminal region and the acidic region of NAP-domain by GST-pull down assay using histone H2B as a probe. The N-terminal region of rTspy bound strongly to histone H2B at the similar level as that for the full-length rTspy (Fig. 4B, lanes 3 and 7). The acidic region in the NAP-domain (#213–232) also bound histone H2B, but at a weaker level than that of the full-length rTspy (Fig. 4B, lane 6). Non-acidic region of NAP-domain (#187–206), however, did not bind histone H2B with similar assay (Fig. 4B, lane 5). Based on these results, we postulate that rTspy preferentially interacts with the core histones at the N-terminal portion of its protein.
Figure 4.
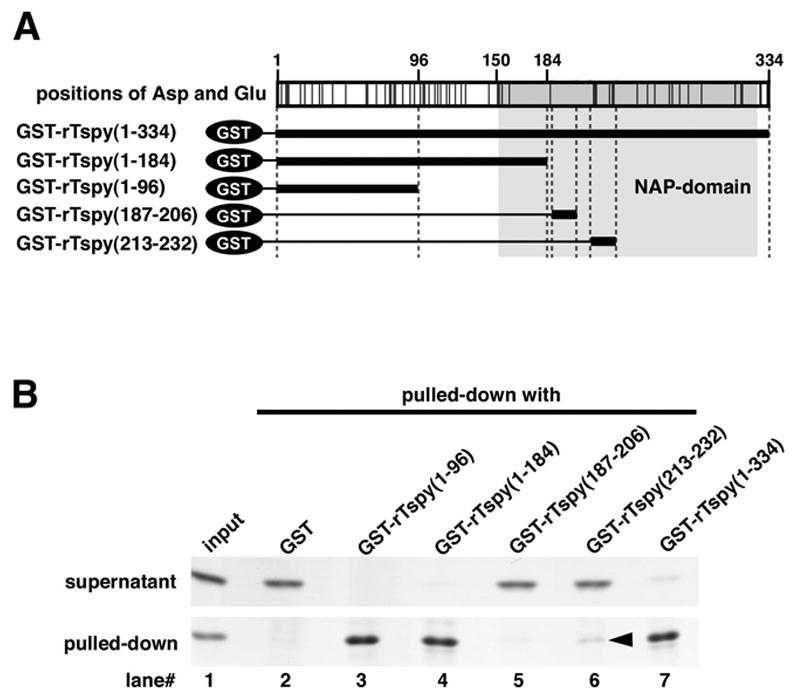
Analysis of the histone H2B binding region in rTspy. (A) Simplified structure of GST-fusion proteins used in GST pull-down assay. Top panel indicates the positions of aspartic acid and glutamic acid in rTspy. The positions of acidic amino acid are indicated by black lines. NAP-domain is shadowed by gray. In lower panel, bold lines indicate the GST-fused region analyzed in B. (B) GST pull-down assay of histone H2B using GST or GST-fusion proteins as indicated. Pulled-down proteins (pulled-down) and proteins of supernatant (supernatant) were subjected to SDS-PAGE, and visualized by Coomassie blue staining. The input represents the material used for GST pull-down assay.
Rat Tspy co-localizes with histone H2B in the cytoplasm of elongating spermatids
We further explored the possibility that both rTspy and displaced histones were co-localized in the cytoplasm of the spermatids using immunofluorescence and individually labeled primary antibodies. Our results showed that positive signals for H2B was detected in the nuclei of elongated spermatids at spermatogenic stage XI–XII (Fig. 5A and 5C, arrowheads), but not detectable in the nuclei of elongated spermatids at spermatogenic stage XIII–XIV (Fig. 5B and 5D, arrowheads). Under high magnification, histone H2B was exclusively localized in the cytoplasm and co-localized with rTspy in the elongated spermatids at spermatogenic stage XII–XIII (Fig. 5E–G, arrowheads). Immunohistochemical staining of adjacent sections of the rat testis independently with anti-rTspy and anti-H2B antibodies confirmed that H2B and rTspy were indeed co-localized in the cytoplasm of the elongating spermatids at stage XIII–XIV (Fig. 5H and 5I, arrowheads). The cytoplasmic localization of histone H2B was initially detected in stage 11 elongating spermatids, and reached a maximal level at stage 13–14 elongating spermatids in which rTspy protein was also at its maximal level (Fig. 5K). These observations suggested that rTspy was expressed at a period of spermatogenesis in which the core histones were being replaced by protamines and exported to the cytoplasm of maturing spermatids. The interaction between the rTspy and core histones (Fig. 3) raised the possibility that it might serve a role in the processing of the discarded core histones during spermiogenesis. Although the hTSPY is primarily located in the spermatogonia [7], our recent study showed that it could also be expressed in spermatids of adult testis [18]. The fact that it also interacts with core histones suggests that it might serve similar function(s) as that of the rat Tspy protein in spermiogenesis.
Figure 5.
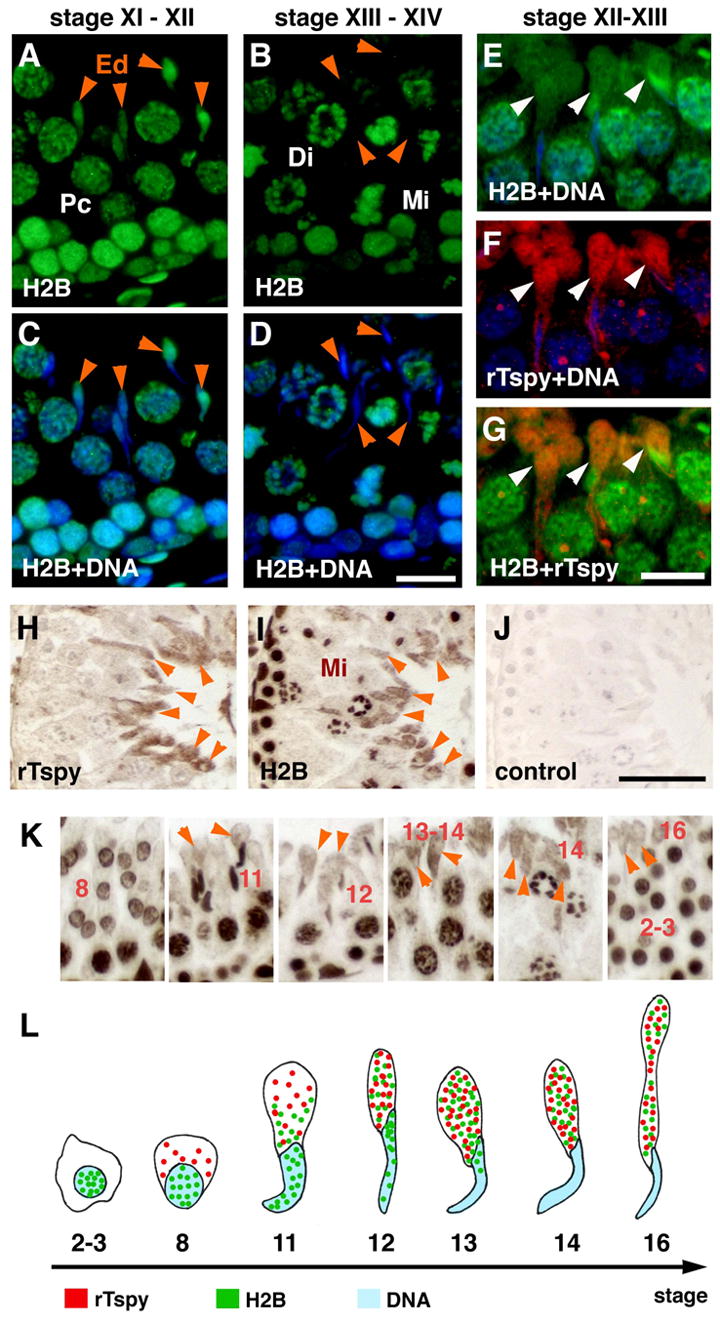
Co-localization of histone H2B and rTspy on elongated spermatids of adult rat testis. (A–D) Immunofluorescence of seminiferous tubules at stage XI–XII (A and C) and stage XIII–XIV (B and D) using anti-H2B antibody. DNA was stained with DAPI. C and D represented the merged images of H2B (green) and DNA (blue). Histone H2B was excluded from nuclei of elongated spermatids at stage XII–XIV. (E–G) Immunofluorescence of seminiferous tubules at stage XII–XIII doubly immunostained with individually labeled H2B and rTspy antibodies. The merged images of H2B (green) and DNA (blue) (E), rTspy (red) and DNA (blue) (F), and H2B (green) and rTspy (red) (G), respectively. The cytoplasm of elongating spermatids was positive for both H2B and rTspy, and was represented as yellow/orange fluorescent signals in the merged image (arrowheads in G). Abbreviations are same as in Figure 2. (H) Rat testis immunostained by anti-rTspy antiserum. Cytoplasm of elongated spermatids was positively stained (arrowheads). (I) Adjacent section of H that was immunostained by anti-H2B antibody. The cytoplasm of elongating spermatids (arrowheads), and the nuclei of other cells were stained. Arrowheads indicate the identical cells between H and I. (J) Image of the rat testis processed with the secondary antibody alone for negative control. Scale bars = 50 μm in A–D; 25 μm in E–G; 50 μm in H–J. (K) Immunstaining of histone H2B in various stages of spermatids. Arabic numbers indicate the stages of respective differentiating spermatids. Orange arrowheads point to the immuno-reactive cytoplasm of various spermatids. (L) A diagram illustrating the expression and localization of rTspy (red dots) and histone H2B (green dots) in adult rat testis. Arabic numbers indicate the stages of spermatids. The locations of DNA/nuclear were represented by blue color.
The TSPY/SET/NAP1 domain of rTspy regulated/restricted the intracellular localization of rTspy in COS7 cells
To further explore the interaction between rTspy and histones, rTspy of various lengths and/or EGFP-testis type H2B fusion protein (EGFP-TH2B) were either singly or doubly transfected to COS7 cells, and analyzed for the subcellular locations of their respective proteins by immunofluorescence. The full-length rTspy was preferentially localized in cytoplasm consisting with the location of the endogenous rTspy protein in elongating spermatids (Fig. 6C). Interestingly, the rTspy(1–96), harboring the histone H2B binding site but not the TSPY/SET/NAP1 domain (Fig. 4), was distributed evenly in both nucleus and cytoplasm of transfected cells (Fig. 6A). In cells co-transfected with rTspy(1–96) and EGFP-TH2B, we found that rTspy(1–96) was well co-localized with the EGFP-TH2B on selected areas within the nuclei (Fig. 6D–6F and Supplemental Fig. S2 for control). Although some cells transfected with a slightly longer construct expressing the rTspy(1–162) including 12 amino acid of TSPY/SET/NAP1 domain, might show some cytoplasmic locations of the gene products, most of the rTspy(1–162) still remained in nuclei, as compared with the full-length rTspy (Fig. 6B and 6H). Hence, the TSPY/SET/NAP1 domain of rTspy could be important on restricting/regulating the cytoplasmic localization of the full-length rTspy. These observations support the hypothesis that rTspy interacts with the displaced histones in the cytoplasm of elongating spermatids.
Figure 6.
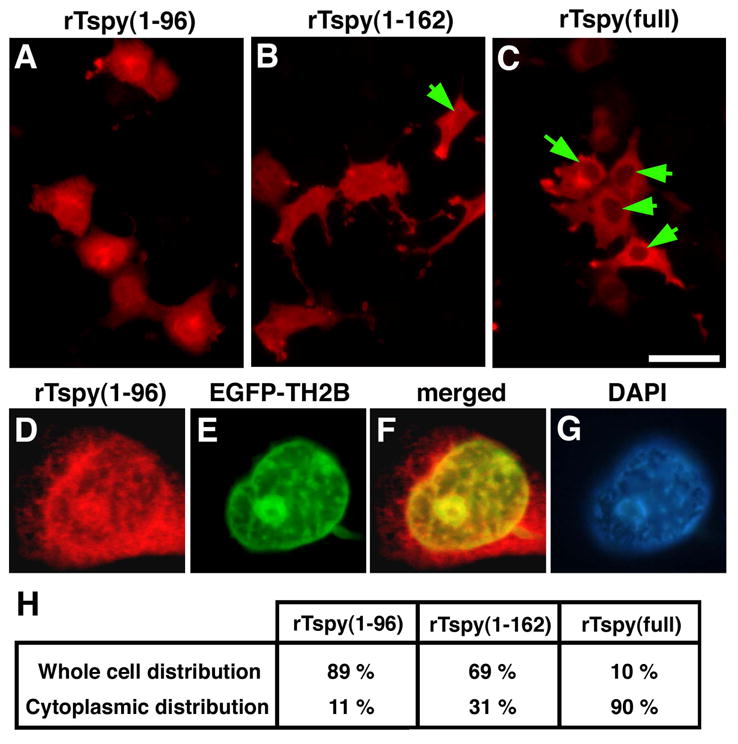
Intracellular localization of rTspy proteins of various lengths in COS7 cells. (A–C) Immunofluorescent images of COS7 cells singly transfected with pCS-rTspy(1–96) (A), pCS-rTspy(1–162) (B) or pCS-rTspy(full-length) (C) using anti-rTspy antiserum (red). Arrows indicate less fluorescence at the nuclei than the cytoplasm of transfected cells. Scale bar= 50μm. (D–G) Immunofluorescent images showing intranuclear localization of rTspy(1–96) (D) and EGFP-TH2B fusion protein (E) in the COS7 cell co-transfected with pCS-rTspy(1–96) and pEGFP-TH2B. The signal of EGFP-TH2B was enhanced by anti-EGFP antibody. The dots of rTpsy(1–96) (red) co-localized well with those of EGFP-TH2B (green), as represented in yellowish color in the merged image (F). DNA was visualized by DAPI staining (G). (H) Percentage of COS7 cells that displayed whole cell distribution (equal intensity between nuclei and cytoplasm) and preferentially cytoplasmic distribution of respective rTspy variants. For each construct, ~ 300 cells were randomly counted.
Discussion
The Tspy gene is evolutionarily conserved and located on the Y chromosome of all mammalian species examined [1, 2, 4, 5]. However, beside the TSPY/SET/NAP1 domain, it diverges considerably along the evolutionary tree. The human TSPY gene is tandemly repeated in 20.4-kb repeat units that encompass ~0.7 MB DNA on the short arm of the Y chromosome and constitute the largest block of functional repeated genes in the human genome [41]. Such repetitiveness of the gene has been conserved among some mammalian species, such as bovine and equine genomes [1]. However, in the rodents, including species of the Apodemus, Mus and Rattus, the functional Tspy gene is a single-copy gene [42]. In particular, the presence of nonsense mutations along its putative coding sequence in the laboratory mice, i.e. Mus musculus and Mus musculus domesticus, suggests that this gene might be undergoing a decaying process(es) on the Y chromosome [6, 15]. The rodent Tspy gene, in addition to its sequence divergence with the human TSPY gene, exhibits an expression pattern, as reported here, different from that of the human TSPY or bovine Tspy genes. The preferential expression of the human TSPY in embryonic germ cells and spermatogonia in adult testis suggests that it might exert a critical effect(s) on germ cell proliferation and meiotic division [1, 7]. On the other hand, the preferential spermatid expression of the rat Tspy in adult testis suggests that the rodent Tspy gene might play an important role in the spermiogenesis process. Our recent demonstration of detectable expression of the human TSPY in spermatids of adult testis [18] indicates that it might also play similar role as the rodent Tspy in spermatozoa maturation. Hence, it is reasonable to assume that the human TSPY may play a variety of roles in germ cell proliferation, meiotic division, and spermiogenesis. The rodent, i.e. the rat, Tspy could have lost its role in germ cell proliferation and meiotic division, but retains that in spermiogenesis.
Currently the exact mechanism(s) of such differential expression between the human TSPY and the rat Tspy gene is uncertain. Preliminary sequence analysis of the putative rat Tspy promoter region revealed that it is significantly different from that of the human TSPY gene. The rat Tspy promoter contains a CRE element located at 50-bp upstream of the transcription start site (Supplemental Fig. S1). Such element is absent in the human TSPY gene promoter. The transcription factor, CREM, binds to the CRE element and mediates postmeiotic transcription of many spermatid-specific genes [43], including protamine 2, TP1 and the Y-located Ssty1 [43–46]. The promoters of many of these spermatid-specific genes, including the rat Tspy, do not harbor any TATA box, but contains the CRE element (Supplemental Fig. S1). Mice homozygous for a CREM null allele showed an arrest of their spermatogenesis at the haploid germ cell stage [47]. The promoter for the human TSPY gene is also TATA-less, but is rich in CpG sequences. The human TSPY promoter is subjected to methylation that regulates its expression in certain cancer cells [48]. Despite such differences between the promoters of the rat Tspy and human TSPY, the exact mechanism(s) of their differential expression patterns cannot be unambiguously defined since various transgenic mouse studies showed variation of transgene expression patterns by the human TSPY promoter [18, 49, 50]. Transgenic mice harboring a human 2.4-kb TSPY promoter directed reporter gene showed a preferential transgene-expression in spermatids of the testis, similar to the rTspy expression pattern in the present study [18]. Another study suggested that the SV40 large T antigen oncogene directed by a 1.3-kb human TSPY promoter was capable of inducing tumor formation in the pituitary gland of the host [49]. Other transgenic mice harboring multiple copies of a 8.2-kb human TSPY DNA showed a spermatogonia/spermatocytes specific expression of the transgene, similar to the human TSPY expression pattern [50]. Although the respective transgene expression could be influenced by the length of the promoter, integration sites and genetic background of the hosts, it is conceivable that a combination of species-specific cis and trans-elements are important for the differential expression of the endogenous hTSPY and rTspy in respective types of germ cells.
During mammalian spermiogenesis, dynamic chromatin remodeling occurs in germ cell nuclei. Histones are replaced by protamines, and the chromatin is packed into highly condensed sperm nucleus [45, 51]. Various studies suggest that certain factors (e.g. histone chaperones tNASP and CIA-II) are involved in the replacement of histones during spermiogenesis [51–53]. The general role of histone chaperones has been postulated to associate with histones and facilitate their interactions with other molecules without being components of the final reaction product and regulating histone metabolism [54]. NAP1, another histone chaperone, shares the TSPY/SET/NAP1 domain with rTspy, binds to the core histones H2A and H2B, and assembles nucleosome or modulates chromatin structure [33–35]. Further, SET/TAFIβ has been demonstrated to interact with sperm basic proteins, and decondenses the sperm chromatin in the fertilized egg [55]. In the present study, we demonstrated that the rTspy protein was capable of binding core histones, H2A, H2B, H3 and H4, but preferentially in the cytoplasm of elongating spermatids (Fig. 2). Although its cytoplasmic location precludes it to have any role in nucleosome assembly, as ascribed for SET/TAF1β, its interactions with the core histones at the period of spermiogenesis when the core histones are being replaced by protamines suggest that the rTspy could be an important chaperone for these sequestered core histones. We hypothesize that rTspy is involved in the metabolism of core histones, i.e. sequestrating the exported histones from nucleus and targeting them for degradation, in elongating spermatids.
The apparent decay of the rodent Tspy gene on their Y chromosome, i.e. gene inactivation in the laboratory mouse and spermatid-restricted expression in others, suggests that functions assigned to those of higher organisms, e.g. human and bovine, in germ cell proliferation and meiotic division will most likely be performed by other genes in the rodent genomes, as illustrated in the human Y-located HSFY and autosomal HSFY-like system, discussed above. Conceivably, the autosomal Tspy-like (Tspyl) or other Y-located gene(s) would have assumed function(s) of TSPY in the rodent germ cell biology. Alternatively, other genes on the Y chromosome, e.g. Sry and Eif2s3y, have been postulated to serve vital function for spermatogenesis and are expressed as late as the round spermatid stage [23, 25]. The Eif2s3y (also termed as Eif2γ) gene encodes a translation initiation factor and is essential for spermatogenic cells to proceed to the round spermatid stage in the mouse. The human homologous gene, EIF2γ, however, is not located on the Y-chromosome [25, 41], suggesting the reverse situation in which Y-located gene on the rodents is being replaced by an autosomal gene in humans. These, as well as numerous Tspy-like genes identified on the X chromosome and autosomes of the mouse, rat and human [26, 27], might be capable of substituting the prescribed function(s) of TSPY in germ cells at different developmental stages. Interestingly, mutations of the human TSPY-L1 gene on chromosome 6 have been demonstrated to be responsible for to a certain sudden infant death syndrome that is also associated with dysgenesis of the testes (SIDDT) [56]. The mutation introduced a stop codon upstream of the coding sequence for the TSPY/SET/NAP1 domain, thereby truncating an essential interactive domain common among members of this protein family. Patients with SIDDT show various abnormalities in the nervous system as well as testicular development, suggesting a likely role for TSPY-L1 in testicular function. Further studies on the relationship between the Y-located TSPY/Tspy and numerous autosomal and X-located Tspy-like genes will be significant in providing insights on this family of proteins, whose members are associated with each other primarily by their conserved cyclin B binding TSPY/SET/NAP1 domain.
Supplementary Material
Acknowledgments
Y-F. C. Lau is a Research Career Scientist of Department of Veterans Affairs.
Footnotes
This work was partially supported by the Department of Defense Prostate Cancer Research Program (DAMD-17-03-1-0081) and National Institutes of Health (1RO1HD038117).
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Vogel T, Dechend F, Manz E, Jung C, Jakubiczka S, Fehr S, Schmidtke J, Schnieders F. Organization and expression of bovine TSPY. Mamm Genome. 1997;8:491–496. doi: 10.1007/s003359900482. [DOI] [PubMed] [Google Scholar]
- 2.Arnemann J, Epplen JT, Cooke HJ, Sauermann U, Engel W, Schmidtke J. A human Y-chromosomal DNA sequence expressed in testicular tissue. Nucleic Acids Res. 1987;15:8713–8724. doi: 10.1093/nar/15.21.8713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang JS, Yang-Feng TL, Muller U, Mohandas TK, de Jong PJ, Lau YF. Molecular isolation and characterization of an expressed gene from the human Y chromosome. Hum Mol Genet. 1992;1:717–726. doi: 10.1093/hmg/1.9.717. [DOI] [PubMed] [Google Scholar]
- 4.Kim HS, Hirai H, Takenaka O. Molecular features of the TSPY gene of gibbons and Old World monkeys. Chromosome Res. 1996;4:500–506. doi: 10.1007/BF02261777. [DOI] [PubMed] [Google Scholar]
- 5.Dechend F, Schubert S, Nanda I, Vogel T, Schmid M, Schmidtke J. Organization and expression of rat Tspy. Cytogenet Cell Genet. 1998;83:270–274. doi: 10.1159/000015169. [DOI] [PubMed] [Google Scholar]
- 6.Schubert S, Dechend F, Skawran B, Krawczak M, Schmidtke J. Molecular evolution of the murine tspy genes. Cytogenet Cell Genet. 2000;91:239–242. doi: 10.1159/000056852. [DOI] [PubMed] [Google Scholar]
- 7.Schnieders F, Dork T, Arnemann J, Vogel T, Werner M, Schmidtke J. Testis-specific protein, Y-encoded (TSPY) expression in testicular tissues. Hum Mol Genet. 1996;5:1801–1807. doi: 10.1093/hmg/5.11.1801. [DOI] [PubMed] [Google Scholar]
- 8.Lau YF. Gonadoblastoma, testicular and prostate cancers, and the TSPY gene. Am J Hum Genet. 1999;64:921–927. doi: 10.1086/302353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lau Y, Chou P, Iezzoni J, Alonzo J, Komuves L. Expression of a candidate gene for the gonadoblastoma locus in gonadoblastoma and testicular seminoma. Cytogenet Cell Genet. 2000;91:160–164. doi: 10.1159/000056838. [DOI] [PubMed] [Google Scholar]
- 10.Lau YF, Zhang J. Expression analysis of thirty one Y chromosome genes in human prostate cancer. Mol Carcinog. 2000;27:308–321. doi: 10.1002/(sici)1098-2744(200004)27:4<308::aid-mc9>3.0.co;2-r. [DOI] [PubMed] [Google Scholar]
- 11.Repping S, van Daalen SK, Brown LG, Korver CM, Lange J, Marszalek JD, Pyntikova T, van der Veen F, Skaletsky H, Page DC, Rozen S. High mutation rates have driven extensive structural polymorphism among human Y chromosomes. Nat Genet. 2006 doi: 10.1038/ng1754. [DOI] [PubMed] [Google Scholar]
- 12.Page DC. Hypothesis: a Y-chromosomal gene causes gonadoblastoma in dysgenetic gonads. Development. 1987;101(Suppl):151–155. doi: 10.1242/dev.101.Supplement.151. [DOI] [PubMed] [Google Scholar]
- 13.Salo P, Kaariainen H, Petrovic V, Peltomaki P, Page DC, de la Chapelle A. Molecular mapping of the putative gonadoblastoma locus on the Y chromosome. Genes Chromosomes Cancer. 1995;14:210–214. doi: 10.1002/gcc.2870140309. [DOI] [PubMed] [Google Scholar]
- 14.Tsuchiya K, Reijo R, Page DC, Disteche CM. Gonadoblastoma: molecular definition of the susceptibility region on the Y chromosome. Am J Hum Genet. 1995;57:1400–1407. [PMC free article] [PubMed] [Google Scholar]
- 15.Schubert S, Dechend F, Skawran B, Kunze B, Winking H, Weile C, Romer I, Hemberger M, Fundele R, Sharma T, Schmidtke J. Silencing of the Y-chromosomal gene tspy during murine evolution. Mamm Genome. 2000;11:288–291. doi: 10.1007/s003350010054. [DOI] [PubMed] [Google Scholar]
- 16.Mazeyrat S, Mitchell MJ. Rodent Y chromosome TSPY gene is functional in rat and non-functional in mouse. Hum Mol Genet. 1998;7:557–562. doi: 10.1093/hmg/7.3.557. [DOI] [PubMed] [Google Scholar]
- 17.Kinoshita K, Shinka T, Sato Y, Kurahashi H, Kowa H, Chen G, Umeno M, Toida K, Kiyokage E, Nakano T, Ito S, Nakahori Y. Expression analysis of a mouse orthologue of HSFY, a candidate for the azoospermic factor on the human Y chromosome. J Med Invest. 2006;53:117–122. doi: 10.2152/jmi.53.117. [DOI] [PubMed] [Google Scholar]
- 18.Kido T, Lau YF. A Cre gene directed by a human TSPY promoter is specific for germ cells and neurons. Genesis. 2005;42:263–275. doi: 10.1002/gene.20147. [DOI] [PubMed] [Google Scholar]
- 19.Conlon RA, Rossant J. Exogenous retinoic acid rapidly induces anterior ectopic expression of murine Hox-2 genes in vivo. Development. 1992;116:357–368. doi: 10.1242/dev.116.2.357. [DOI] [PubMed] [Google Scholar]
- 20.Turner DL, Weintraub H. Expression of achaete-scute homolog 3 in Xenopus embryos converts ectodermal cells to a neural fate. Genes Dev. 1994;8:1434–1447. doi: 10.1101/gad.8.12.1434. [DOI] [PubMed] [Google Scholar]
- 21.Rupp RA, Snider L, Weintraub H. Xenopus embryos regulate the nuclear localization of XMyoD. Genes Dev. 1994;8:1311–1323. doi: 10.1101/gad.8.11.1311. [DOI] [PubMed] [Google Scholar]
- 22.Oh HJ, Li Y, Lau YF. Sry Associates with the Heterochromatin Protein 1 Complex by Interacting with a KRAB Domain Protein. Biol Reprod. 2005;72:407–415. doi: 10.1095/biolreprod.104.034447. [DOI] [PubMed] [Google Scholar]
- 23.Toure A, Grigoriev V, Mahadevaiah SK, Rattigan A, Ojarikre OA, Burgoyne PS. A protein encoded by a member of the multicopy Ssty gene family located on the long arm of the mouse Y chromosome is expressed during sperm development. Genomics. 2004;83:140–147. doi: 10.1016/s0888-7543(03)00216-7. [DOI] [PubMed] [Google Scholar]
- 24.Conway SJ, Mahadevaiah SK, Darling SM, Capel B, Rattigan AM, Burgoyne PS. Y353/B: a candidate multiple-copy spermiogenesis gene on the mouse Y chromosome. Mamm Genome. 1994;5:203–210. doi: 10.1007/BF00360546. [DOI] [PubMed] [Google Scholar]
- 25.Mazeyrat S, Saut N, Grigoriev V, Mahadevaiah SK, Ojarikre OA, Rattigan A, Bishop C, Eicher EM, Mitchell MJ, Burgoyne PS. A Y-encoded subunit of the translation initiation factor Eif2 is essential for mouse spermatogenesis. Nat Genet. 2001;29:49–53. doi: 10.1038/ng717. [DOI] [PubMed] [Google Scholar]
- 26.Vogel T, Dittrich O, Mehraein Y, Dechend F, Schnieders F, Schmidtke J. Murine and human TSPYL genes: novel members of the TSPY-SET-NAP1L1 family. Cytogenet Cell Genet. 1998;81:265–270. doi: 10.1159/000015042. [DOI] [PubMed] [Google Scholar]
- 27.Ozbun LL, Martinez A, Angdisen J, Umphress S, Kang Y, Wang M, You M, Jakowlew SB. Differentially expressed nucleolar TGF-beta1 target (DENTT) in mouse development. Dev Dyn. 2003;226:491–511. doi: 10.1002/dvdy.10257. [DOI] [PubMed] [Google Scholar]
- 28.Russell L, Ettlin R, Sinha Hikim A, ED C. Histological and Histopathological Evaluation of the Testis. Cache River Press; Vienna, IL: 1990. [Google Scholar]
- 29.Marchler-Bauer A, Bryant SH. CD-Search: protein domain annotations on the fly. Nucleic Acids Res. 2004;32:W327–331. doi: 10.1093/nar/gkh454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ozbun LL, You L, Kiang S, Angdisen J, Martinez A, Jakowlew SB. Identification of differentially expressed nucleolar TGF-beta1 target (DENTT) in human lung cancer cells that is a new member of the TSPY/SET/NAP-1 superfamily. Genomics. 2001;73:179–193. doi: 10.1006/geno.2001.6505. [DOI] [PubMed] [Google Scholar]
- 31.Altman R, Kellogg D. Control of mitotic events by Nap1 and the Gin4 kinase. J Cell Biol. 1997;138:119–130. doi: 10.1083/jcb.138.1.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kellogg DR, Kikuchi A, Fujii-Nakata T, Turck CW, Murray AW. Members of the NAP/SET family of proteins interact specifically with B-type cyclins. J Cell Biol. 1995;130:661–673. doi: 10.1083/jcb.130.3.661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ishimi Y, Kikuchi A. Identification and molecular cloning of yeast homolog of nucleosome assembly protein I which facilitates nucleosome assembly in vitro. J Biol Chem. 1991;266:7025–7029. [PubMed] [Google Scholar]
- 34.Mosammaparast N, Ewart CS, Pemberton LF. A role for nucleosome assembly protein 1 in the nuclear transport of histones H2A and H2B. Embo J. 2002;21:6527–6538. doi: 10.1093/emboj/cdf647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Park YJ, Chodaparambil JV, Bao Y, McBryant SJ, Luger K. Nucleosome assembly protein 1 exchanges histone H2A-H2B dimers and assists nucleosome sliding. J Biol Chem. 2005;280:1817–1825. doi: 10.1074/jbc.M411347200. [DOI] [PubMed] [Google Scholar]
- 36.Kutney SN, Hong R, Macfarlan T, Chakravarti D. A signaling role of histone-binding proteins and INHAT subunits pp32 and Set/TAF-Ibeta in integrating chromatin hypoacetylation and transcriptional repression. J Biol Chem. 2004;279:30850–30855. doi: 10.1074/jbc.M404969200. [DOI] [PubMed] [Google Scholar]
- 37.Schneider R, Bannister AJ, Weise C, Kouzarides T. Direct binding of INHAT to H3 tails disrupted by modifications. J Biol Chem. 2004;279:23859–23862. doi: 10.1074/jbc.C400151200. [DOI] [PubMed] [Google Scholar]
- 38.Seo SB, Macfarlan T, McNamara P, Hong R, Mukai Y, Heo S, Chakravarti D. Regulation of histone acetylation and transcription by nuclear protein pp32, a subunit of the INHAT complex. J Biol Chem. 2002;277:14005–14010. doi: 10.1074/jbc.M112455200. [DOI] [PubMed] [Google Scholar]
- 39.Okuwaki M, Nagata K. Template activating factor-I remodels the chromatin structure and stimulates transcription from the chromatin template. J Biol Chem. 1998;273:34511–34518. doi: 10.1074/jbc.273.51.34511. [DOI] [PubMed] [Google Scholar]
- 40.Seo SB, McNamara P, Heo S, Turner A, Lane WS, Chakravarti D. Regulation of histone acetylation and transcription by INHAT, a human cellular complex containing the set oncoprotein. Cell. 2001;104:119–130. doi: 10.1016/s0092-8674(01)00196-9. [DOI] [PubMed] [Google Scholar]
- 41.Skaletsky H, Kuroda-Kawaguchi T, Minx PJ, Cordum HS, Hillier L, Brown LG, Repping S, Pyntikova T, Ali J, Bieri T, Chinwalla A, Delehaunty A, Delehaunty K, Du H, Fewell G, Fulton L, Fulton R, Graves T, Hou SF, Latrielle P, Leonard S, Mardis E, Maupin R, McPherson J, Miner T, Nash W, Nguyen C, Ozersky P, Pepin K, Rock S, Rohlfing T, Scott K, Schultz B, Strong C, Tin-Wollam A, Yang SP, Waterston RH, Wilson RK, Rozen S, Page DC. The male-specific region of the human Y chromosome is a mosaic of discrete sequence classes. Nature. 2003;423:825–837. doi: 10.1038/nature01722. [DOI] [PubMed] [Google Scholar]
- 42.Vogel T, Boettger-Tong H, Nanda I, Dechend F, Agulnik AI, Bishop CE, Schmid M, Schmidtke J. A murine TSPY. Chromosome Res. 1998;6:35–40. doi: 10.1023/a:1009214307764. [DOI] [PubMed] [Google Scholar]
- 43.Monaco L, Kotaja N, Fienga G, Hogeveen K, Kolthur US, Kimmins S, Brancorsini S, Macho B, Sassone-Corsi P. Specialized rules of gene transcription in male germ cells: the CREM paradigm. Int J Androl. 2004;27:322–327. doi: 10.1111/j.1365-2605.2004.00494.x. [DOI] [PubMed] [Google Scholar]
- 44.Beissbarth T, Borisevich I, Horlein A, Kenzelmann M, Hergenhahn M, Klewe-Nebenius A, Klaren R, Korn B, Schmid W, Vingron M, Schutz G. Analysis of CREM-dependent gene expression during mouse spermatogenesis. Mol Cell Endocrinol. 2003;212:29–39. doi: 10.1016/j.mce.2003.09.023. [DOI] [PubMed] [Google Scholar]
- 45.Sassone-Corsi P. Unique chromatin remodeling and transcriptional regulation in spermatogenesis. Science. 2002;296:2176–2178. doi: 10.1126/science.1070963. [DOI] [PubMed] [Google Scholar]
- 46.Martianov I, Fimia GM, Dierich A, Parvinen M, Sassone-Corsi P, Davidson I. Late arrest of spermiogenesis and germ cell apoptosis in mice lacking the TBP-like TLF/TRF2 gene. Mol Cell. 2001;7:509–515. doi: 10.1016/s1097-2765(01)00198-8. [DOI] [PubMed] [Google Scholar]
- 47.Blendy JA, Kaestner KH, Weinbauer GF, Nieschlag E, Schutz G. Severe impairment of spermatogenesis in mice lacking the CREM gene. Nature. 1996;380:162–165. doi: 10.1038/380162a0. [DOI] [PubMed] [Google Scholar]
- 48.Gallagher WM, Bergin OE, Rafferty M, Kelly ZD, Nolan IM, Fox EJ, Culhane AC, McArdle L, Fraga MF, Hughes L, Currid CA, O'Mahony F, Byrne A, Murphy AA, Moss C, McDonnell S, Stallings RL, Plumb JA, Esteller M, Brown R, Dervan PA, Easty DJ. Multiple markers for melanoma progression regulated by DNA methylation: insights from transcriptomic studies. Carcinogenesis. 2005 doi: 10.1093/carcin/bgi152. [DOI] [PubMed] [Google Scholar]
- 49.Tascou S, Trappe R, Nayernia K, Jarry H, Konig F, Schulz-Schaeffer W, Saeger W, Meinhardt A, Engel W, Schmidtke J, Burfeind P. TSPY-LTA transgenic mice develop endocrine tumors of the pituitary and adrenal gland. Mol Cell Endocrinol. 2003;200:9–18. doi: 10.1016/s0303-7207(02)00426-4. [DOI] [PubMed] [Google Scholar]
- 50.Schubert S, Skawran B, Dechend F, Nayernia K, Meinhardt A, Nanda I, Schmid M, Engel W, Schmidtke J. Generation and characterization of a transgenic mouse with a functional human TSPY. Biol Reprod. 2003;69:968–975. doi: 10.1095/biolreprod.103.016501. [DOI] [PubMed] [Google Scholar]
- 51.Govin J, Caron C, Lestrat C, Rousseaux S, Khochbin S. The role of histones in chromatin remodelling during mammalian spermiogenesis. Eur J Biochem. 2004;271:3459–3469. doi: 10.1111/j.1432-1033.2004.04266.x. [DOI] [PubMed] [Google Scholar]
- 52.Umehara T, Horikoshi M. Transcription initiation factor IID-interactive histone chaperone CIA-II implicated in mammalian spermatogenesis. J Biol Chem. 2003;278:35660–35667. doi: 10.1074/jbc.M303549200. [DOI] [PubMed] [Google Scholar]
- 53.Lee YH, O'Rand MG. Ultrastructural localization of a nuclear autoantigenic sperm protein in spermatogenic cells and spermatozoa. Anat Rec. 1993;236:442–448. doi: 10.1002/ar.1092360304. [DOI] [PubMed] [Google Scholar]
- 54.Loyola A, Almouzni G. Histone chaperones, a supporting role in the limelight. Biochim Biophys Acta. 2004;1677:3–11. doi: 10.1016/j.bbaexp.2003.09.012. [DOI] [PubMed] [Google Scholar]
- 55.Matsumoto K, Nagata K, Miyaji-Yamaguchi M, Kikuchi A, Tsujimoto M. Sperm chromatin decondensation by template activating factor I through direct interaction with basic proteins. Mol Cell Biol. 1999;19:6940–6952. doi: 10.1128/mcb.19.10.6940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Puffenberger EG, Hu-Lince D, Parod JM, Craig DW, Dobrin SE, Conway AR, Donarum EA, Strauss KA, Dunckley T, Cardenas JF, Melmed KR, Wright CA, Liang W, Stafford P, Flynn CR, Morton DH, Stephan DA. Mapping of sudden infant death with dysgenesis of the testes syndrome (SIDDT) by a SNP genome scan and identification of TSPYL loss of function. Proc Natl Acad Sci U S A. 2004;101:11689–11694. doi: 10.1073/pnas.0401194101. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


