Abstract
The goal of this study was to create an in vitro cell culture system that captures essential features of the in vivo erythroid micronucleus (MN) genotoxicity assay, thus enabling increased throughput and controlled studies of the hematopoietic DNA damage response. We show that adult bone marrow (BM) cultures respond to erythropoietin, the principal hormone that stimulates erythropoiesis, with physiological erythropoietic proliferation, differentiation, and enucleation. We then show that this in vitro erythropoietic system clearly signals exposure to genotoxicants through erythroid MN formation. Furthermore, we determined that DNA repair-deficient (MGMT−/−) BM displayed sensitivity to genotoxic exposure in vivo compared with WT BM and that this phenotypic response was reflected in erythropoietic cultures. These findings suggest that this in vitro erythroid MN assay is capable of screening for genotoxicity on BM in a physiologically reflective manner. Finally, responses to genotoxicants during erythroid differentiation varied with exposure time, demonstrating that this system can be used to study the effect of DNA damage at specific developmental stages.
Keywords: in vitro toxicity screens, genotoxicity, hematopoiesis, DNA damage and repair
Normal erythropoiesis in the adult human generates ≈1011 new RBCs in the bone marrow (BM) each day through the proliferation and differentiation of erythroid progenitors that descend from hematopoietic stem cells (1). In the presence of Epo, a single erythroid progenitor (known as a colony-forming unit erythroid or CFU-E) divides approximately five times over 2–3 days to produce ≈30 progeny that enucleate and form nascent erythrocytes, also known as reticulocytes or polychromatic erythrocytes (PCEs) (2).
If a cell suffers DNA damage while proliferating and differentiating, the biological outcomes are cell death, cell survival with mutations, or cell survival without mutations, depending on whether and how the cell repairs the damage. Some genotoxic agents, such as ionizing radiation, are direct-acting clastogens that create double-strand DNA breaks by direct scission. Other DNA-damaging agents generate DNA strand breaks as replication and repair intermediates, and these breaks can induce homologous or nonhomologous DNA recombination events; in addition, damaged centromeres or spindle components can trigger aneugenic events. Whether genetic fragmentation occurs because of clastogenic or aneugenic mechanisms, the result after erythropoietic growth is the same: cells that suffer sufficient DNA damage undergo enucleation but form PCEs that contain “micronuclei,” small fragments of nuclear membrane-encapsulated DNA (3). The spleen clears damaged erythrocytes, so normally <1% of circulating erythrocytes contain spontaneous micronuclei arising from background levels of DNA damage.
The hematopoietic system is highly sensitive to genotoxic agents, in part because hematopoietic cells undergo rapid division. The BM is often the dose-limiting tissue in chemotherapy and radiotherapy regimens, and therapy-related anemia, cytopenia, and leukemia are common side effects. These serious morbidities have prompted efforts to increase hematopoietic DNA repair capacity through gene therapy in patients receiving chemotherapeutic agents and to improve preclinical screening of new drugs for genotoxic effects (4–9). Although leukemias and cytopenias arise in white cell populations, erythrocyte populations serve as a useful system to measure DNA damage because normal and micronucleated PCEs are easily recognized and scored by microscopy, flow cytometry, and laser scanning cytometry (10–13). The assay system described here tests agents by using terminally differentiating erythrocyte populations to indicate general hematopoietic genotoxicity.
Efforts to predict the effect of drugs on human hematopoiesis, or to develop therapeutic approaches to mitigate these effects, are hindered by a lack of readily accessible model systems that duplicate the highly orchestrated set of cellular sensing, signaling, and repair responses that DNA damage triggers in exposed hematopoietic cells. Some repair pathways require sequential activity by multiple enzymes in a series of reactions that transiently decrease the stability of the double helix before the completion of repair. Reducing the level of one repair enzyme within the pathway can thus, paradoxically, increase resistance to genotoxic agents (14). The situation is further complicated by the diversity of expression levels of various DNA repair-related genes during different stages of hematopoietic differentiation (15, 16). Not surprisingly, then, model organisms and cell lines offer only modest predictive power for hematopoietic genotoxicity, and human genotoxicity in general (17–23).
Currently, one of the most robust tests for genotoxicity is the in vivo micronucleus (MN) assay, which is conducted by using an improved version of the method first described by Schmid and colleagues (24, 25). Close to 70% of known human carcinogens are detected by the in vivo MN test; accordingly, regulatory authorities around the world require a preclinical in vivo cytogenetic test, such as the MN assay (3, 26). At 24–48 h after injection of a test substance into a rodent, the BM is harvested and stained to distinguish PCEs from older normochromatic erythrocytes (NCEs) and to reveal the presence of micronuclei; a total of 2,000 PCEs from each animal are sampled to score MN frequency. Despite recent advances in the automated detection and enumeration of micronculeated PCEs that eliminate microscopy and increase assay throughput, this assay is still applied late in preclinical drug development because of the large numbers of animals required to encompass multiple doses and replicates for each agent tested (10–13).
We recently developed a flow cytometry protocol for analyzing erythropoietic differentiation in vivo in a quantitative and step-by-step fashion (27, 28). This analytical procedure was used to establish in vitro conditions that stimulate Ter-119− fetal liver erythroid progenitor cells to undergo terminal proliferation, erythroid differentiation, and enucleation in a physiologic manner. Using this system we have analyzed the role of several signal-transduction pathways activated by the Epo receptor (28, 29). Fetal erythropoiesis in the liver is very similar to adult erythropoiesis in the BM, although the CFU-E frequency in BM is much lower than that in fetal liver. Although adult BM-derived cells can form erythroid colonies in vitro, the fidelity of the in vitro process compared with physiological erythropoiesis in adult BM had not been examined.
The overarching goal of this study was to determine whether erythropoietic cultures, derived from adult BM, can capture essential features of the in vivo MN assay, thus enabling step-by-step examination of the effects of genotoxic agents at different developmental stages while providing a platform for high-throughput in vitro screening of drugs and toxic environmental agents by using the BM of a single animal. To this end, we demonstrate that the lineage-marker-negative (Lin−) population of adult mouse BM undergoes erythropoietic differentiation according to established in vivo patterns and that erythrocytes generated in vitro undergo MN formation when exposed to direct-acting genotoxic agents. Further, we show that this in vitro system reflects a phenotypic sensitivity that we observed in DNA repair-deficient mice in vivo. Although this system removes the erythroid MN assay from the whole animal, and thus isolates promutagens from liver metabolism, it is well established that incubation with liver microsomes can restore some measure of metabolic activation to in vitro genotoxicity screens (30).
Results
Stimulation of Terminal Erythropoiesis in Lin− BM Cells.
Flow cytometric analysis of erythroid differentiation is achieved by double-staining for the transferrin receptor (CD71), which is dramatically up-regulated during the first two divisions of CFU-Es, and Ter-119, which is stably incorporated into the membrane of cells during late stages of erythroid differentiation (31). Erythropoietic progenitors (CD71−/Ter-119−) undergo several divisions to produce progeny that first express CD71, and then Ter-119, before CD71 is lost from the cell surface during reticulocyte maturation. It had been shown that this sequence of Epo-induced events can occur in vitro by using Ter-119− fetal liver cells (28). Here, we show that Lin− BM cells from adult mice also undergo Epo-induced differentiation according to this pattern.
The Lin− fraction of mouse BM was isolated by immunomagnetic negative selection, plated on fibronectin-coated culture dishes, and stimulated with Epo for the first 24 h of culture. Lin− BM cells represent ≈1.25% of cells in the marrow and comprise a mixture of hematopoietic and mesenchymal stem and progenitor cells. Analysis by flow cytometry indicated that a significant fraction of Lin− cells had begun erythroid differentiation (i.e., up-regulated CD71) after 1 day of culture with Epo (Fig. 1). By day 2, the majority of cells express CD71 and Ter-119, and, by day 3, the characteristic late-erythroid loss of CD71, reported for both in vivo fetal liver and cultured Ter-119− fetal liver, had begun (27, 28). Cytology confirmed that a substantial fraction of the Lin− BM population (nucleated cells lacking hemoglobin) had differentiated into enucleated, hemoglobinized reticulocytes over the course of 2–3 days (Fig. 1). A fraction of Lin− BM cells were unresponsive to Epo (Fig. 1), and it is likely that most of these cells are not erythropoeitic and do not survive under our culture conditions.
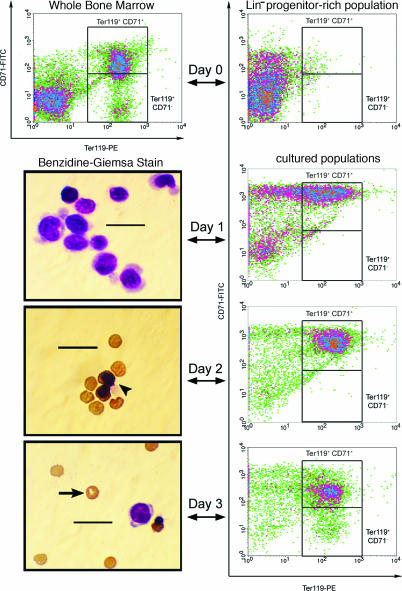
Fig. 1.
Terminal erythropoiesis stimulated in Lin− BM cells over 3 days in culture. BM cells were stained with biotinylated α-Lin mAbs, and the Lin+ fraction of the population was subsequently removed to obtain a progenitor-rich population (Lin− BM). These Lin− cells were then cultured in vitro for 3 days on fibronectin-coated plates in medium containing serum. Epo was included in the medium for the first day of culture, and then the medium was changed, and Epo was removed. The differentiation profile of the cultured cells was examined by both flow cytometry and benzidine–Giemsa stain after each day in culture, and a representative micrograph of these stained populations is presented from each day of erythropoieitc culture. After the third day of culture, flow cytometry indicated that the majority of the resulting population had acquired a late erythroid surface phenotype (Ter-119+). Furthermore, benizidine–Giemsa staining revealed that many cells in the harvested population had enucleated and expressed hemoglobin. The arrowhead indicates a hemoglobin+ normoblast, and the arrow indicates an enucleated reticulocyte. (Scale bars, 20 μm.)
Quantitative studies on the response of Lin− BM cultures after stimulus with Epo and other erythropoietic factors were then conducted to define the PCE yield per Lin− BM cell and to investigate whether adjustments to the culture protocol could provide increased yield while maintaining physiologic terminal erythropoiesis. A quantitative, cytology-based study revealed that an ≈8-fold increase in PCE yield could be achieved (up to 5.8 ± 0.9 PCEs per Lin− cell, J.S., H.F.L., and L.G.G., manuscript in preparation). Accordingly, all genotoxicity studies were conducted by using this improved erythropoietic culture protocol, which includes Epo, SCF, dexamethasone, and IGF1 in the culture medium for the first 24 h of culture. Erythropoiesis continued normally using the improved culture method, although expression of Ter-119 was slightly delayed [compare supporting information (SI) Fig. 5 with Fig. 1), and resulting PCE yields were sufficient to conduct hundreds of genotoxicity assays on the BM of a single mouse (SI Fig. 6).
Detection of Genotoxic Exposure: In Vitro Erythroid MN Assay.
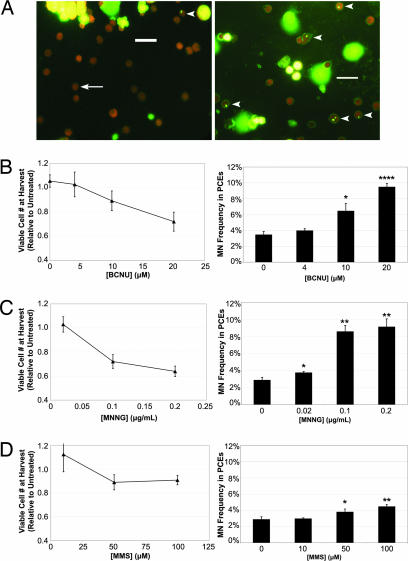
We used as model genotoxicants three mutagenic alkylating agents that test positive in the in vivo MN assay: BCNU (1,3-bis[2-chloroethyl]-1-nitrosourea), MNNG (N-methyl-N*-nitro-N-nitrosoguanidine), and MMS (methylmethane sulfonate). Cultures treated with genotoxicant for 1 h starting 23 h after seeding progressed normally through erythropoiesis (SI Fig. 7). Micronuclei appeared in both treated cultures and controls (Fig. 2A) but with greater frequency in treated cultures (Fig. 2 B–D), as expected based on the known ability of these agents to increase MN frequency above basal levels in vivo. Both the control and treated MN frequencies observed in vitro were higher than the levels typically observed in vivo, which might be related to the relatively high rate of erythropoietic growth. Indeed, increasing erythropoietic rates in vivo through bleeding or exogenous Epo expression increases erythroid MN frequencies and can sensitize the in vivo assay to genotoxic exposure (32, 33). Cell viability at harvest was >90% for all cultures (data not shown), but viable cell yields were lower for treated vs. control controls (Fig. 2 B–D). Cell proliferation and death were not explicitly measured, thus reductions in viable cell yields cannot be specifically attributed to cytotoxic or cytostatic mechanisms. However, these data demonstrate that genotoxic exposure can be detected by using this erythropoietic culture system.
Fig. 2.
Detection of genotoxicity through in vitro erythropoiesis. Purified Lin− cells were cultured in vitro for 1 day on fibronectin-coated plates in medium containing serum, Epo, SCF, dexamethasone, and IGF1. Genotoxic alkylating agents were introduced into the culture media 23 h after seeding. One hour later, the media were changed, and all soluble growth factors and alkylating agents were removed. Populations were then cultured for 2 additional days in media with serum. At harvest, the cells were removed from culture, and genotoxic effects were quantified through viable cell counts and differential cell counts. (A) Representative micrographs are provided from both treated (Right) and untreated (Left) cultures. The arrow indicates a normally enucleated PCE, and the arrowheads indicate micronucleated PCEs. (Scale bars, 20 μm.) (B) The response of this culture system to BCNU treatment is quantified in terms of relative viable cell numbers and MN frequencies. On the left, relative viable cell numbers are plotted vs. BCNU concentration. On the right, MN frequency is plotted vs. BCNU concentration. (C) The response of this culture system to MNNG treatment is quantified in terms of relative viable cell numbers and MN frequencies. (D) The response of this culture system to MMS treatment is quantified in terms of relative viable cell numbers and MN frequencies. Data are presented as the mean of three independent cultures ±SD. ∗ indicates a significant difference (P < 0.05) from untreated control cultures. ∗∗ indicates a significant difference (P < 0.01) from the untreated control cultures. ∗∗∗∗ indicates a significant difference (P < 0.0001) from the untreated control cultures.
MGMT−/− Mice Display Sensitivity to BCNU Exposure by the in Vivo MN Assay.
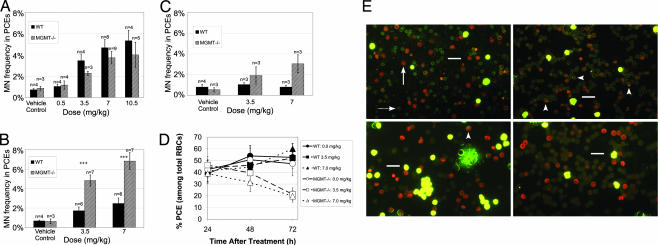
BCNU is an SN1 bifunctional alkylating agent that can form adducts at nucleophilic sites on DNA, including the O6 position of Guanine (34–36). After this initial addition reaction, the alkyl group on O6 of Guanine can react with the opposite Cytosine to form an interstrand cross-link; importantly, the O6 alkyl group can be removed by a DNA repair protein known as O6-methylguanine DNA methyltransferase (MGMT) (37, 38). The BM of MGMT−/− mice (on a C57BL/6J background) was previously shown to display sensitivity to BCNU-induced cytotoxicity (39). Here, MGMT−/− mice were treated with BCNU doses by i.p. injection and examined by the in vivo MN test. Counter to our expectations, the MN frequency in the MGMT−/− BM was lower than that observed in WT C57BL/6J BM 24 h after treatment at some BCNU doses (Fig. 3A). It seemed likely that the genotoxic response in the MGMT−/− marrow was delayed, possibly because proliferation slows in the presence of increased genomic instability. Therefore, we examined the marrow of MGMT−/− and WT mice 48 h after exposure, and, as initially expected, the MN frequency in the MGMT−/− marrow was drastically higher than that observed in WT marrow (Fig. 3B). Moreover, a decrease in the ratio of PCEs to total RBCs was also observed in MGMT−/− marrow 48 h after exposure (Fig. 3 D and E). The increased sensitivity of MGMT−/− BM to BCNU-induced MN formation and inhibition of PCE growth remained evident 72 h after exposure (Fig. 3 C–E). Taken together, these data indicate that MGMT−/− marrow is more sensitive to BCNU than WT marrow, which had largely recovered to resemble untreated marrow 48 h after treatment, both in terms of MN formation and as measured by erythroid cytotoxic/cytostatic effects (Fig. 3).
Fig. 3.
MGMT−/− mice exhibit sensitivity to MN formation after in vivo exposure to BCNU: dynamic quantification of the frequency of PCEs and micronucleated PCEs in BM. WT and MGMT−/− male mice aged 6–8 weeks were treated with either BCNU or vehicle control (PBS/10% EtOH) by i.p. injection. After 24, 48, or 72 h, the treated animals were killed, the BM was flushed from the femurs, and slides were prepared and stained with acridine orange for differential cell counting. Data are presented as the mean ±SD. (A) The MN frequency in BM PCEs was quantified 24 h after exposure to BCNU. (B) The MN frequency in BM PCEs was quantified 48 h after exposure to BCNU. +++ indicates a significant difference (P < 0.001) from WT mice. (C) The MN frequency in BM PCEs was quantified 72 h after exposure to BCNU. (D) The PCE frequency in total BM RBCs was quantified at various times after exposure to BCNU. At 48 h after exposure to BCNU, the BM of MGMT−/− mice was found to contain a reduced frequency of PCEs, and 72 h after exposure, this effect had become even more pronounced. (E) Representative micrographs are shown for BM from the two genotypes examined at various times after treatment with either BCNU (3.5 mg/kg) or vehicle control. The top left micrograph (WT mice 48 h after treatment with vehicle control) is characteristic of untreated, WT BM, with a PCE/RBC ratio between 40% and 60% and a low MN frequency. The arrow indicates a PCE, and the dashed arrow indicates an NCE. The bottom left micrograph is representative of WT BM 48 h after treatment with BCNU (3.5 mg/kg), which has largely recovered to resemble untreated BM (the top left micrograph). The arrowhead indicates a micronucleated PCE. The top right micrograph represents MGMT−/− BM 48 h after treatment with BCNU, which contains an elevated MN frequency among PCEs and a reduced PCE/RBC frequency. The bottom right micrograph illustrates that the decreased PCE/RBC ratio becomes even more pronounced in MGMT−/− BM 72 h after treatment with BCNU. (Scale bars, 20 μm.)
In Vitro Erythroid MN Assay Reflects the in Vivo Phenotype of MGMT−/− Mice.
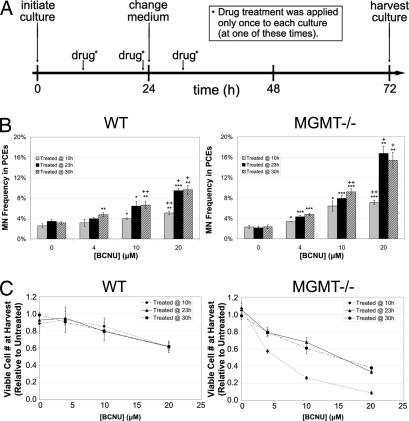
MGMT−/− Lin− BM was used in our erythropoietic culture system to test whether an in vitro genotoxicity screen could reflect the DNA repair capacity of primary BM. In the absence of genotoxic exposure, erythropoietic differentiation in MGMT−/− cultures was indistinguishable from that of WT cultures (SI Fig. 8 vs. SI Fig. 5). After BCNU addition, erythropoiesis in MGMT−/− cultures continued normally before leading to significant increases in MN frequency as compared with WT cultures (Fig. 4A). Furthermore, reduced PCE yields were observed in the MGMT−/− cultures, meaning that this assay can detect the increased sensitivity of MGMT−/− erythroid progenitors to BCNU both by cytotoxic/cytostatic signals and by increased production of micronucleated progeny (Fig. 4B).
Fig. 4.
Similar to in vivo responses, Lin− BM from MGMT−/− mice exhibits sensitivity to MN formation and decreased PCE yields when treated with BCNU during erythropoietic culture. (A) Purified Lin− cells from MGMT−/− and WT mice were cultured in vitro for 1 day on fibronectin-coated plates in medium containing serum, Epo, SCF, dexamethasone, and IGF1. After 1 day, the medium was changed to one containing serum without other hormones. Populations were then cultured for 2 additional days before harvest at 72 h. BCNU was introduced into the culture media at various times (10, 23, or 30 h) after seeding. At harvest, the cells were removed from culture, and genotoxic effects were quantified through viable cell counts and MN enumeration. (B) The MN response of Lin− bone marrow from WT and MGMT−/− mice to BCNU in this erythropoietic culture system is quantified. MN frequency is plotted vs. BCNU concentration, and data are presented as the mean of three independent cultures ±SD. + indicates a significant difference (P < 0.05) between WT and MGMT−/− cultures. ++ indicates a significant difference (P < 0.01) between WT and MGMT−/− cultures. ∗ indicates a significant difference (P < 0.05) from the vehicle control. ∗∗ indicates a significant difference (P < 0.01) from the vehicle control. ∗∗∗ indicates a significant difference (P < 0.001) from the vehicle control. (C) The effect of BCNU exposure on the viable cell number of Lin− bone marrow from WT and MGMT−/− mice after erythropoietic culture is quantified. Relative viable cell number is plotted vs. BCNU concentration, and data are presented as the mean of three independent cultures ±SD.
In the study shown in Fig. 4, BCNU was introduced into erythropoietic cultures at various times after seeding to determine the time at which these differentiating populations are most sensitive to MN formation in response to genotoxic exposure. In MGMT−/− BM cultures, BCNU treatment at earlier times exacerbated decreases in viable cell numbers, whereas BCNU treatment at later times resulted in further increases in micronculeated PCE frequencies; when WT BM cultures were treated 4 h after culture initiation, a similar trend was observed (compare SI Fig. 9 to Fig. 4 WT data). These observations lead us to hypothesize that unrepaired genetic damage to Epo-responsive progenitors (approximately CFU-E stage) during the first day of culture results in a greater degree of cell death/senescence, whereas genetic damage to the erythroid cells present later in culture, if not repaired, is more likely to yield micronucleated PCEs.
Discussion
Many drugs and environmental agents can damage DNA, and understanding and predicting the outcomes of exposure to DNA-damaging agents remains an important challenge in pharmaceutical development and in environmental health sciences. The complex interplay between the numerous cellular pathways that influence damage and repair determines the final biological consequences of exposure. The outcomes we observed for MN formation in mice treated with BCNU (Fig. 3) underscore this complexity: mice lacking the DNA repair enzyme MGMT appear to be slightly less sensitive than WT mice 24 h after exposure, but exhibit a clear sensitivity to genetic alkylation 48 and 72 h after exposure. Chromosome aberrations arising through the processing of the O6 alkyl guanine lesion require a second round of DNA replication to be expressed, and this might contribute to the late appearance of micronuclei in MGMT−/− BM (40, 41). Furthermore, our dynamic treatment studies (Fig. 4) and in vitro growth measurements (SI Fig. 6) show that differentiating erythroid populations are extremely sensitive to MN formation while they undergo the final 2–3 divisions before enucleation.
As a first step toward capturing a complex physiological response in vitro, we established that Lin− cells from adult mouse BM undergo normal erythropoiesis in culture, as assessed by flow cytometry and cytology. In this culture system all cells are Epo-stimulated during the first day of culture, and erythrocytes near the CFU-E stage of development then undergo metasynchronous development. The trends we observed in vivo and in vitro suggest that MGMT functions in erythroid progenitors to mitigate the effects of BCNU, and responses in this in vitro assay system lead us to hypothesize that unrepaired damage leads to different outcomes (decreased reticulocyte yield vs. increased MN frequency) depending on the stage of erythropoiesis.
When SCF, dexamethasone, and IGF1 were added along with Epo, the fraction of cells at each stage of differentiation (as assessed by flow cytometry) was different from that in Epo-only cultures (Fig. 1 vs. SI Fig. 5), in a manner consistent with proliferation of mature burst-forming units erythroid (BFU-Es) stimulated by SCF and Epo. Because SCF and Epo are withdrawn 24 h after seeding, these cells might then arrest near the CFU-E stage. Therefore, it might be possible to examine the effect of chronic or early genotoxic exposure during erythropoiesis by reintroducing Epo to cultures late in the culture period. Indeed, colony assays conducted on populations harvested from these cultures at day 3 revealed that these populations contain CFU-Es (data not shown).
As the correlation between the in vivo and in vitro responses of MGMT−/− marrow illustrates, this in vitro erythropoietic system is capable of detecting the DNA-repair capacity of primary BM donors, and thus, if extended to human cells, may allow individual patient testing, or testing panels of human cells both to predict average human responses and to better understand the basic biology of the human hematopoietic response to genotoxic agents. Further, the levels of key molecules in DNA damage-response pathways can be manipulated more easily in culture than in vivo (e.g., via RNAi), allowing the effects of pathway components to be explored. Finally, this genotoxicity screen conducts the erythroid MN assay on adult tissue in vitro to provide increased throughput over the existing in vivo erythroid MN assay, which tests a single agent (at a single dose) per animal.
Materials and Methods
Cells.
BM cells were isolated from the hind legs of C57BL/6J mice aged 6–8 weeks (The Jackson Laboratory, Bar Harbor, ME) and were mechanically dissociated by pipetting in Iscove's modified Dulbecco's medium (IMDM)/4% FBS. Single-cell suspensions were prepared by passing dissociated cells through 70-μm cell strainers. BM cells with diameter >6 μm were counted by using a Coulter particle counter Z1 (Beckman Coulter, Fullerton, CA).
Erythropoietic Culture.
Total BM cells were labeled with biotin-conjugated α-Lin Abs, consisting of α-CD3e, α-CD11b, α-CD45R/B220, α-Ly6G/Ly6C, and α-TER-119 Abs (2 μl of each Ab/106 cells; BD Pharmingen, San Diego, CA), and Lin− cells were purified through a 0.3-in StemSep negative selection column as per the manufacturer's instructions (StemCell Technologies, Vancouver, BC, Canada). Purified cells were seeded in fibronectin-coated (2 μg/cm2) tissue culture treated polystyrene wells (BD Discovery Labware, Bedford, MA) at a cell density of 105 per ml. On the first day, the purified cells were cultured in IMDM containing basal supplements consisting of: 15% FBS, 1% detoxified BSA, 200 μg/ml holotransferrin (Sigma, St Louis, MO), 10 μg/ml recombinant human insulin (Sigma), 2 mM l-glutamine, 10−4 M β-mercaptoethanol, 50 units/ml penicillin G, and 50 μg/ml streptomycin; as well as soluble erythropoietic factors. The previously described erythropoietic medium, including Epo (Amgen, Thousand Oaks, CA) at 2 units/ml along with the basal supplements, was used in the cultures presented in Fig. 1 (28). An improved erythropoietic medium formulation, including Epo at 10 units/ml, SCF (R & D Systems, Minneapolis, MN) at 10 ng/ml, dexamethasone (Sigma) at 10 μM, and IGF1 (R & D Systems) at 100 ng/ml, along with basal supplements, was used in all other cultures. For all cultures, the media was replaced with erythroid-differentiation medium (EDM) (IMDM with 20% FBS, 2 mM l-glutamine, and 10−4 M β-mercaptoethanol) after 1 day of culture. At harvest, suspended cells were removed from culture wells by pipetting, and then the culture well was incubated in PBS/10% FBS/5 mM EDTA at 37°C for 5 min to dissociate adherent cells. Dissociated cells were then removed from a culture well by pipetting and combined with the suspended cell fraction from the same culture well for analysis.
Immunostaining and Flow Cytometry.
BM-derived cells were immunostained at 4°C in PBS/4% FBS. Cells were incubated with phycoerythrin-conjugated α-Ter119 (1:200; BD Pharmingen) and fluorescein isothiocyanante-conjugated α-CD71 (1:200; BD Pharmingen) Abs for 15 min and were then washed in PBS/4% FBS. Flow cytometry was carried out on a FACSCalibur (Becton Dickinson, Franklin Lakes, NJ). Flow cytometry plots and region statistics were acquired by using CellQuest Pro (BD Biosciences, San Jose, CA).
Cytospin Preparation and Cytological Staining.
Approximately 2 × 104 cells per culture were centrifuged onto slides for 3 min at 800 rpm (Cytospin 3; Thermo Shandon, Pittsburgh, PA) and air dried. For benzidine–Giemsa staining, cells were fixed in −20°C methanol for 2 min and stained with 3,3′-diaminobenzidine and Giemsa stains according to the manufacturer's recommendations (Sigma). For acridine orange staining, cells were fixed in 25°C methanol for 10 min and stained in acridine orange (Fisher Scientific, Hanover Park, IL) at a concentration of 20 μg/ml in staining buffer (19 mM NaH2PO4 and 81 mM Na2HPO4) for 10 min at 4°C. After acridine orange staining, slides were protected from light, washed for 10 min in 4°C staining buffer, air dried, and stored at 4°C until microscopic examination and scoring was complete.
In Vivo Genotoxic Treatment and Cytology.
BCNU (Sigma) was first dissolved in cold absolute ethanol (EtOH), and mice were treated with BCNU by i.p. injection using a vehicle of PBS/10% EtOH. At the appropriate time after dosage (24, 48, or 72 h), the mice were killed by using CO2, and the BM was removed from the femurs and tibiae of the hind legs. A single cell suspension was generated by mechanical dissociation, and this suspension was then passed through a cellulose column to enrich erythrocytes. This enriched erythroid population was then spread on a slide, fixed in methanol, and stained with acridine orange for cytological examination.
Genotoxic Treatment of Erythropoietic Cultures.
Lin− BM was cultured in 500 μl of medium per culture well according to the method described for erythropoietic culture. Alkylating agents were added to the culture medium at various times after seeding and, if added during the first day of culture, were removed when the medium was exchanged for EDM. When treating 30 h after seeding, BCNU was not removed from the cultures. BCNU was first dissolved in 4°C EtOH to make a 10 mM solution. This 10 mM solution was diluted in 4°C IMDM to produce an IMDM/0.4 mM BCNU/4% EtOH solution. The required volume of this 0.4 mM BCNU solution (0–25 μl) was then added to each culture, along with a compensatory volume of vehicle (IMDM/4% EtOH), such that each culture was exposed to the targeted concentration of BCNU (0–20 μM) and an equal concentration of EtOH (0.2% by volume). A MNNG (Aldrich, Milwaukee, WI) stock solution [1 mg/ml in 0.1M sodium acetate buffer (pH 5.1)] was mixed with 4°C IMDM to yield two solutions of different concentration (2 μg/ml and 10 μg/ml). To deliver 0.02 μg/ml treatments to cultures, 5 μl of 2 μg/ml MNNG was added to the medium; to deliver 0.10 μg/ml or 0.20 μg/ml treatments to cultures, 5 μl or 10 μl of 10 μg/ml MNNG was added to the medium, respectively. MMS (Aldrich) was mixed with 4°C IMDM to yield two solutions of different concentration (0.5 mM and 5 mM). To deliver 10 μM treatments to cultures, 10 μl of 0.5 mM MMS was added to the medium; to deliver 50 μM or 100 μM treatments to cultures, 5 μl or 10 μl of 5 mM MMS were added to the medium, respectively. All alkylating agent solutions were prepared immediately before treatment to minimize degradation and decreased reactivity.
Viable Cell Counting.
Viable cell counts, based on Trypan blue exclusion, were conducted by using a ViCellXR viable cell counter (Beckman Coulter, Miami, FL) according to the manufacturer's instructions. The instrument was set to include cells with diameters from 6–50 μm in analyses. The relative number of viable cells in treated cultures was determined by normalizing the mean of viable cell counts from a set of treated cultures to the mean of viable cell counts from the set of untreated cultures.
Histological Imaging and Quantification.
Slides were examined by using a Labophot microscope (Nikon, Garden City, NY) and representative micrographs were acquired by using a DSC-P93A Cyber-Shot digital camera (Sony, San Diego, CA). Micrographs of benzidine–Giemsa stained cells were acquired by using a ×100 oil-immersion objective and bright-field illumination, whereas micrographs of acridine orange stained cells were acquired by using a ×40 oil-immersion objective and fluorescence (100-W Hg lamp excitation). Cytological slides were examined blind, and differential cell counting was used to enumerate relevant cell types and thus quantify the frequency of MN-PCEs among PCEs (>2,000 PCEs scored per slide), and the frequency of PCEs among RBCs (>1,000 RBCs scored per slide).
Statistics.
To determine the statistical significance of mean comparisons, distributions were first checked for normality by using the Shapiro–Wilk test [using JMP5 (SAS Institute, Cary, NC)]. Normal data sets were then subjected to two-tailed Welch's t tests to determine P values by using the data analysis tool in Excel (Microsoft, Redmond, WA).
Supplementary Material
Acknowledgments
We thank Wes Overton, Sophia Kamran, and Catherine Moroski for technical assistance; Drs. Lisiane Meira and Dharini Shah for guidance in genotoxicity studies; and Glenn Paradis for expert technical advice in flow-cytometry studies. The animals used in this study were treated and housed in accordance with approved guidelines and supervised by Massachusetts Institute of Technology (MIT)'s animal care committee. This work was supported by the Cambridge–MIT Institute (J.S.), a grant from Amgen (to L.G.G.), National Institutes of Health Grants ES02109 and CA75576 (to L.D.S.), and National Science Foundation Grant EEC9843342 (to L.G.G.).
Abbreviations
- BM
bone marrow
- Epo
erythropoietin
- MN
micronucleus
- PCE
polychromatic erythrocyte
- CFU-E
colony-forming unit erythroid
- Lin−
lineage-marker negative
- SCF
stem cell factor
- IGF1
insulin-like growth factor 1
- MGMT
O6-methylguanine DNA methyltransferase
- BCNU
1,3-bis(2-chloroethyl)-1-nitrosourea
- MNNG
N-methyl-N*-nitro-N-nitrosoguanadine
- MMS
methylmethane sulfonate.
Note Added in Proof.
Near the completion of these studies we realized that some of the MGMT−/− mice used in the experiments were heterozygous for another DNA-repair gene (3-methyladenine DNA glycosylase). We have established that this mutation has no effect on the production of micronucleated PCEs after treatment with BCNU, both in a previously published report (14) and in unpublished data (J.S. and L.D.S.); accordingly, only the relevant MGMT genotype is indicated.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0701829104/DC1.
References
- 1.Zon LI. Hematopoiesis. New York: Oxford Univ Press; 2001. [Google Scholar]
- 2.Lodish HF. Molecular Cell Biology. New York: Freeman; 2003. [Google Scholar]
- 3.Heddle JA, Cimino MC, Hayashi M, Romagna F, Shelby MD, Tucker JD, Vanparys P, MacGregor JT. Environ Mol Mutagen. 1991;18:277–291. doi: 10.1002/em.2850180414. [DOI] [PubMed] [Google Scholar]
- 4.Roth RB, Samson LD. Mutat Res. 2000;462:107–120. doi: 10.1016/s1383-5742(00)00021-1. [DOI] [PubMed] [Google Scholar]
- 5.Cai S, Hartwell JR, Cooper RJ, Juliar BE, Kreklau E, Abonour R, Goebel WS, Pollok KE. Mol Ther. 2006;13:1016–1026. doi: 10.1016/j.ymthe.2005.11.017. [DOI] [PubMed] [Google Scholar]
- 6.Kreklau EL, Pollok KE, Bailey BJ, Liu N, Hartwell JR, Williams DA, Erickson LC. Mol Cancer Ther. 2003;2:1321–1329. [PubMed] [Google Scholar]
- 7.Pollok KE, Hartwell JR, Braber A, Cooper RJ, Jansen M, Ragg S, Bailey BJ, Erickson LC, Kreklau EL, Williams DA. Hum Gene Ther. 2003;14:1703–1714. doi: 10.1089/104303403322611728. [DOI] [PubMed] [Google Scholar]
- 8.Woolford LB, Southgate TD, Margison GP, Milsom MD, Fairbairn LJ. J Gene Med. 2006;8:29–34. doi: 10.1002/jgm.816. [DOI] [PubMed] [Google Scholar]
- 9.Cornetta K, Croop J, Dropcho E, Abonour R, Kieran MW, Kreissman S, Reeves L, Erickson LC, Williams DA. Cancer Gene Ther. 2006;13:886–895. doi: 10.1038/sj.cgt.7700963. [DOI] [PubMed] [Google Scholar]
- 10.Dertinger SD, Torous DK, Tometsko KR. Mutat Res. 1996;371:283–292. doi: 10.1016/s0165-1218(96)90117-2. [DOI] [PubMed] [Google Scholar]
- 11.Dertinger SD, Torous DK, Tometsko KR. Mutat Res. 1997;390:257–262. doi: 10.1016/s1383-5718(97)00022-3. [DOI] [PubMed] [Google Scholar]
- 12.Romagna F, Staniforth CD. Mutat Res. 1989;213:91–104. doi: 10.1016/0027-5107(89)90035-3. [DOI] [PubMed] [Google Scholar]
- 13.Styles JA, Clark H, Festing MF, Rew DA. Cytometry. 2001;44:153–155. doi: 10.1002/1097-0320(20010601)44:2<153::aid-cyto1095>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 14.Roth RB, Samson LD. Cancer Res. 2002;62:656–660. [PubMed] [Google Scholar]
- 15.Bracker TU, Giebel B, Spanholtz J, Sorg UR, Klein-Hitpass L, Moritz T, Thomale J. Stem Cells. 2006;24:722–730. doi: 10.1634/stemcells.2005-0227. [DOI] [PubMed] [Google Scholar]
- 16.Gerson SL, Miller K, Berger NA. J Clin Invest. 1985;76:2106–2114. doi: 10.1172/JCI112215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ames BN, Durston WE, Yamasaki E, Lee FD. Proc Natl Acad Sci USA. 1973;70:2281–2285. doi: 10.1073/pnas.70.8.2281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ames BN, McCann J, Yamasaki E. Mutat Res. 1975;31:347–364. doi: 10.1016/0165-1161(75)90046-1. [DOI] [PubMed] [Google Scholar]
- 19.Fenech M. Mutat Res. 2000;455:81–95. doi: 10.1016/s0027-5107(00)00065-8. [DOI] [PubMed] [Google Scholar]
- 20.Fenech M, Morley AA. Mutat Res. 1985;147:29–36. doi: 10.1016/0165-1161(85)90015-9. [DOI] [PubMed] [Google Scholar]
- 21.Sancar A, Lindsey-Boltz LA, Unsal-Kacmaz K, Linn S. Annu Rev Biochem. 2004;73:39–85. doi: 10.1146/annurev.biochem.73.011303.073723. [DOI] [PubMed] [Google Scholar]
- 22.Tennant RW, Margolin BH, Shelby MD, Zeiger E, Haseman JK, Spalding J, Caspary W, Resnick M, Stasiewicz S, Anderson B, et al. Science. 1987;236:933–941. doi: 10.1126/science.3554512. [DOI] [PubMed] [Google Scholar]
- 23.Zeiger E. Cancer Res. 1987;47:1287–1296. [PubMed] [Google Scholar]
- 24.Schmid W. Agents Actions. 1973;3:77–85. doi: 10.1007/BF01986538. [DOI] [PubMed] [Google Scholar]
- 25.Schmid W. Mutat Res. 1975;31:9–15. doi: 10.1016/0165-1161(75)90058-8. [DOI] [PubMed] [Google Scholar]
- 26.Morita T, Asano N, Awogi T, Sasaki YF, Sato S, Shimada H, Sutou S, Suzuki T, Wakata A, Sofuni T, Hayashi M. Mutat Res. 1997;389:3–122. doi: 10.1016/s1383-5718(96)00070-8. [DOI] [PubMed] [Google Scholar]
- 27.Socolovsky M, Nam H, Fleming MD, Haase VH, Brugnara C, Lodish HF. Blood. 2001;98:3261–3273. doi: 10.1182/blood.v98.12.3261. [DOI] [PubMed] [Google Scholar]
- 28.Zhang J, Socolovsky M, Gross AW, Lodish HF. Blood. 2003;102:3938–3946. doi: 10.1182/blood-2003-05-1479. [DOI] [PubMed] [Google Scholar]
- 29.Zhang J, Lodish HF. Blood. 2004;104:1679–1687. doi: 10.1182/blood-2004-04-1362. [DOI] [PubMed] [Google Scholar]
- 30.Suling WJ, Rice LS, Shannon WM. J Natl Cancer Inst. 1983;70:767–769. [PubMed] [Google Scholar]
- 31.Kina T, Ikuta K, Takayama E, Wada K, Majumdar AS, Weissman IL, Katsura Y. Br J Haematol. 2000;109:280–287. doi: 10.1046/j.1365-2141.2000.02037.x. [DOI] [PubMed] [Google Scholar]
- 32.Hirai O, Miyamae Y, Fujino Y, Izumi H, Miyamoto A, Noguchi H. Mutat Res. 1991;264:109–114. doi: 10.1016/0165-7992(91)90125-n. [DOI] [PubMed] [Google Scholar]
- 33.Yajima N, Kurata Y, Imai E, Sawai T, Takeshita Y. Mutagenesis. 1993;8:231–236. doi: 10.1093/mutage/8.3.231. [DOI] [PubMed] [Google Scholar]
- 34.Singer B, Bodell WJ, Cleaver JE, Thomas GH, Rajewsky MF, Thon W. Nature. 1978;276:85–88. doi: 10.1038/276085a0. [DOI] [PubMed] [Google Scholar]
- 35.Bodell WJ. Chem Res Toxicol. 1999;12:965–970. doi: 10.1021/tx980200c. [DOI] [PubMed] [Google Scholar]
- 36.Ludlum DB. Mutat Res. 1990;233:117–126. doi: 10.1016/0027-5107(90)90156-x. [DOI] [PubMed] [Google Scholar]
- 37.Kohn KW. Cancer Res. 1977;37:1450–1454. [PubMed] [Google Scholar]
- 38.Samson L, Cairns J. Nature. 1977;267:281–283. doi: 10.1038/267281a0. [DOI] [PubMed] [Google Scholar]
- 39.Glassner BJ, Weeda G, Allan JM, Broekhof JL, Carls NH, Donker I, Engelward BP, Hampson RJ, Hersmus R, Hickman MJ, et al. Mutagenesis. 1999;14:339–347. doi: 10.1093/mutage/14.3.339. [DOI] [PubMed] [Google Scholar]
- 40.Galloway SM. Environ Mol Mutagen. 1994;24(23) Suppl:44–53. doi: 10.1002/em.2850230612. [DOI] [PubMed] [Google Scholar]
- 41.Kaina B, Fritz G, Coquerelle T. Environ Mol Mutagen. 1993;22:283–292. doi: 10.1002/em.2850220418. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.