Abstract
Proline-rich Gla protein 2 (PRGP2) is one of four known vertebrate transmembrane γ-carboxyglutamic acid (Gla) proteins. Members of this protein family are broadly expressed in fetal and adult human tissues and share a common architecture consisting of a predicted propeptide and Gla domain, a single-pass transmembrane segment, and tandem Pro/Leu-Pro-Xaa-Tyr (PY) motifs near their C termini. Using a methodology developed for the regulated expression of enzymatically biotinylated proteins in mammalian cells, we demonstrate that PRGP2 undergoes γ-glutamyl carboxylation in a manner that is both dependent upon the presence of a proteolytically cleavable propeptide and sensitive to warfarin, a vitamin K antagonist that is widely used as an antithrombotic agent. When expressed at physiologically relevant levels, the majority of PRGP2 is present in the γ-glutamyl carboxylated, propeptide-cleaved (mature) form. We additionally demonstrate, by Western blotting and flow cytometry, that mature PRGP2 is predominantly located on the cell surface with the Gla domain exposed extracellularly. In a yeast two-hybrid screen that used the C-terminal cytoplasmic region of PRGP2 as bait, we identified the WW domain-containing transcriptional coactivator Yes-associated protein (YAP) as a binding partner for PRGP2. In GST pull-down experiments, both PRGP2 PY motifs and both YAP WW domains were essential for complex formation, as were residues proximal to the core sequence of the first PY motif. These findings suggest that PRGP2 may be involved in a signal transduction pathway, the impairment of which may be an unintended consequence of warfarin therapy.
Keywords: Gla domain, warfarin, WW domain
Vitamin K is a cofactor for the posttranslational conversion of glutamic acid residues to γ-carboxyglutamic acid (Gla) residues, a reaction that occurs in the endoplasmic reticulum (ER) (1, 2). The vitamin K-dependent coagulation factors each possess an N-terminal Gla domain, a protein module of ≈45 residues that contains between 9 and 13 Gla residues. The coordination of calcium ions by Gla residues within the Gla domain occurs at physiological plasma calcium concentration and maintains the Gla domain in a conformation that is capable of binding to phospholipid surfaces, specifically those that contain the anionic aminophospholipid, phosphatidylserine (PS) (3–5). PS is sequestered on the inner leaflet of the plasma membrane under normal cellular homeostasis but is expressed extracellularly in response to intracellular calcium mobilization, most notably during apoptosis and platelet activation (6–8). In the former case, PS exposure marks cells for efficient clearance by the reticuloendothelial system (7). In the latter, PS exposure on the surface of activated platelets provides a substrate for the deposition and proteolytic activation of vitamin K-dependent coagulation factors (6).
At least two distinct enzymatic activities are required for the γ-glutamyl carboxylation of Gla domain-containing proteins. The first of these, γ-glutamyl carboxylase, facilitates carboxyl transfer to Glu residues and the concomitant oxidation of the reduced hydroquinone form of the vitamin K cofactor (9, 10). γ-Glutamyl carboxylase recognizes substrates bearing an N-terminal propeptide that is removed by limited proteolysis after γ-glutamyl carboxylation (11–13). A second enzyme, vitamin K 2,3-epoxide reductase (VKOR) is required for the regeneration of reduced vitamin K, and it is this activity that is antagonized by warfarin-type anticoagulants (14, 15).
Despite the central role that the Gla domain plays in blood coagulation, several lines of evidence indicate nonhemostatic functions for the domain. For example, the anticoagulant cofactor protein S forms a complex with C4b-binding protein that negatively regulates complement on the surface of apoptotic cells, and the binding of this complex to apoptotic cells is mediated by the Gla domain of protein S (16–18). Gas6, a paralog of protein S, promotes survival and proliferation of various types of cells by activating members of the Axl subfamily of receptor tyrosine kinases. These activities are abrogated when Gas6 is produced in the presence of warfarin, highlighting the indispensable nature of γ-glutamyl carboxylation (19–22). Additional evidence that Gla domains perform functions unrelated to hemostasis is provided by the recent identification of Gla domain proteins in ascidians, marine invertebrates that lack a blood coagulation cascade (23–25).
The transmembrane Gla proteins constitute the most recently identified family of vertebrate Gla domain proteins, and include proline-rich Gla proteins (PRGPs) 1 and 2, and transmembrane Gla proteins (TMGs) 3 and 4 (26, 27). In contrast to the vitamin K-dependent coagulation factors, members of this protein family exhibit broad extrahepatic tissue distribution, and are predicted to be type I integral membrane proteins. The C-terminal regions of these proteins each contain two tandem repeats of a Pro/Leu-Pro-Xaa-Tyr (PY) motif, the minimal sequence recognized by type I WW domains (28–30). The WW domain is a protein module of ≈40 residues, first identified in Yes-associated protein (YAP) (31), that has subsequently been identified in a variety of signaling and cytoskeletal proteins, including dystrophin, Nedd4-like ubiquitin ligases, the tumor suppressor WWOX, and several others.
Results
Posttranslational Processing of PRGP2 Expressed in CHO Tet-On Cells.
PRGP2 was expressed in CHO Tet-On cells by using plasmid pCBio, a vector designed for the doxycycline (dox)-regulated expression of enzymatically biotinylated proteins in mammalian cells (32). In this system, the target protein bearing a C-terminal biotin acceptor peptide (BioTag) is coexpressed on a bicistronic message with Escherichia coli biotin ligase (BirA) in response to dox added to the culture medium. The target protein then undergoes quantitative BirA-dependent site-specific biotinylation within the cytoplasm, facilitating its detection with a sensitivity that is superior to antibody-based reagents. PRGP2 constructs (Fig. 1A) included wild-type PRGP2, as well as variants with mutations that would be expected to abolish propeptide recognition by γ-glutamyl carboxylase [F(-19)A], prevent the proteolytic removal of the propeptide [R(-1)A], and result in deletion of the signal peptide, propeptide and extracellular domain of the protein (ΔEcto). Transiently transfected cells were cultured in the presence of either 5 μM vitamin K (Fig. 1B, odd-numbered lanes) or 5 μM warfarin (Fig. 1B, even-numbered lanes), and analyzed by Western blotting. Detection of total biotinylated PRGP2 with streptavidin-HRP (lanes 1–10) revealed four major protein species. A major band of ≈30 kDa and a minor band of ≈32 kDa were present in all lanes for full-length PRGP2 constructs, regardless of whether cells had been cultured with vitamin K or warfarin. Expression of the ΔEcto construct resulted in intense 12-kDa bands for both vitamin K- and warfarin-treated cells. Faint bands corresponding to approximately the same molecular mass were observed in all other lanes except those for the negative expression control, and likely represent a minor form of the protein that is cleaved proximal to its transmembrane region. Most notably, a 24-kDa band was present in the lane for vitamin K-treated cells expressing wild-type PRGP2 (lane 3), but absent in the lane for similarly transfected cells that had been treated with warfarin (lane 4). Biotinylated PRGP2 was isolated from cell lysates by adsorption to immobilized avidin and analyzed by Western blotting with a monoclonal antibody specific for Gla residues (lanes 11–20). The 24-kDa band corresponding to that in lane 3 was detected with the anti-Gla antibody in the lane for vitamin K-treated cells expressing wild-type PRGP2 (lane 13) but not in the lane for similarly transfected cells that had been treated with warfarin (lane 14), confirming that this band represents the vitamin K-dependent, carboxylated form of PRGP2 (hereafter referred to as “mature” PRGP2). Mutation F(-19)A, which corresponds to a mutation in factor IX that abolished its γ-glutamyl carboxylation (33), similarly affected PRGP2 (lane 15). A 30-kDa band was detected with the anti-Gla antibody in the lysate from cells that had been transfected with the R(-1)A mutant and treated with vitamin K (lane 17), but not in the lysate from similarly transfected cells that had been treated with warfarin (lane 18). This finding indicates that the 30-kDa band represents a precursor form of PRGP2 that retains the propeptide (pro-PRGP2), and that the R(-1)A mutation impairs its proteolytic removal. The 32-kDa band might therefore represent a form of the protein that additionally retains the signal peptide (prepro-PRGP2). The absence of a Gla-positive 30-kDa band corresponding to vitamin K-treated cells that had been transfected with wild-type PRGP2 (lane 13) indicates that carboxylation and propeptide removal are concerted, sequentially ordered processes, and that the former step is rate-limiting.
Fig. 1.
Expression of PRGP2 in CHO Tet-On cells. (A) Four constructs of PRGP2 were expressed, each with a C-terminal BioTag to facilitate detection and isolation. The wild-type (WT) construct spans the entire length of prepro-PRGP2 (residues −49 to 153). Propeptide mutant F(-19)A is analogous to mutations in other vitamin K-dependent proteins that prevent recognition and glutamyl carboxylation by γ-glutamyl carboxylase. Propeptide mutant R(-1)A was designed to prevent the proteolytic removal of the propeptide after γ-glutamyl carboxylation. A truncated form of PRGP2 that lacks the prepro leader sequence, the Gla domain, and nearly all of the juxtramembrane linker (Δ-Ecto) spans residues 54–153. SP, signal peptide; PP, propeptide; Gla, Gla domain; TM, transmembrane; PY, Leu/Pro-Pro-Xaa-Tyr motif; BT, BioTag. (B) CHO Tet-On cells were transiently transfected with the constructs indicated and cultured in the presence of either 5 μM vitamin K (odd-numbered lanes) or 5 μM warfarin (even-numbered lanes) for 24 h before lysis. Cell lysates were analyzed by Western blotting (lanes 1–10) with either streptavidin-HRP (SA-HRP; Upper) or with an antibody to E. coli biotin ligase (anti-BirA; Lower). Alternatively, biotinylated PRGP2 constructs were selectively enriched on immobilized avidin and analyzed by Western blotting with a monoclonal antibody that recognizes Gla residues (lanes 11–20). (C) Clonal CHO Tet-On cells stably transfected with the wild-type PRGP2 construct were cultured in the presence of either 5 μM vitamin K or 5 μM warfarin, and the expression level was adjusted by varying the concentration of the inducer, doxycycline (dox), as indicated. Total biotinylated PRGP2 and glutamyl carboxylated PRGP2 were analyzed by Western blotting as in B. The positions of molecular mass standards (in kilodaltons) are shown to the right of each blot.
A similar conclusion can be drawn from the results of experiments using stably tranfected CHO Tet-On cells (Fig. 1C). Here, a clonal population of stably selected cells was cultured in the presence of varying concentrations of the inducer, dox. As in the previous experiment, PRGP2 expression was monitored by Western blotting with streptavidin-HRP (Upper), and the extent of carboxylation of avidin-adsorbed PRGP2 was monitored by Western blotting with the anti-Gla antibody (Lower). For cells grown in the presence of vitamin K, the intensity of 24-kDa streptavidin-positive band (mature PRGP2) reached a maximum between 100 and 200 ng/ml dox, paralleling the dox-dependence of the 24-kDa Gla-positive band. At lower expression levels, mature PRGP2 predominates, whereas at higher expression levels, Gla-negative pro-PRGP2 is generated at increasing levels. In contrast to the results of the transient transfection experiments described above, an additional 27-kDa band appears at higher expression levels in both vitamin K- and warfarin-treated cells. Whereas the nature of this protein species remains uncertain, it is not recognized by the anti-Gla antibody, nor does it react with antibodies to phosphotyrosine, phosphoserine, or phosphothreonine (data not shown). Taken together, the results of both transient and stable expression experiments are consistent with a model in which the host cell's intrinsic carboxylation machinery operates effectively at low levels of PRGP2 expression, but is overwhelmed at higher, presumably supraphysiological expression levels. Moreover, the glutamyl carboxylation of PRGP2 seems to be both necessary and sufficient to direct the efficient proteolytic removal of the propeptide.
Localization of PRGP2 on the Cell Surface.
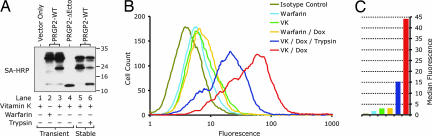
Sequence elements within PRGP2, namely a predicted signal peptide and a largely hydrophobic 26 residue segment, suggest a topological orientation in which the N-terminal Gla domain of mature PRGP2 is exposed to the ER lumen and the C terminus is directed to the cytoplasm. Although PRGP2 lacks any discernible motifs that would specify retention within the ER or the Golgi apparatus, it was not immediately obvious whether PRGP2 was localized to the plasma membrane or restricted to intracellular organelles. The results of a preliminary experiment, in which expression of a PRGP2-GFP fusion protein was placed under control of a strong CMV promoter and monitored by confocal fluorescence microscopy, suggested a punctate perinuclear expression pattern (data not shown). Given the unexpected nature of this result, we investigated the susceptibility of PRGP2 to trypsin-mediated proteolysis in intact CHO Tet-On cells by Western blotting (Fig. 2A). In these experiments, the overall expression level of PRGP2 was adjusted, by titrating the dox concentration, such that the levels of pro-PRGP2 (30 kDa) and mature PRGP2 (24 kDa) were approximately equal. Treatment of PRGP2-expressing cells with trypsin before cell lysis resulted in the selective diminution of the 24-kDa band and the emergence of a 12-kDa band corresponding to the putative juxtamembrane cleavage product (lane 6). In contrast, the intensities of the 30-kDa band (pro-PRGP2) and the 27-kDa band, both of which were previously shown to represent noncarboxylated species, were only slightly diminished upon trypsin treatment. These findings indicate that mature PRGP2 is predominantly located on the cell surface, whereas incompletely processed precursor forms of the protein are not.
Fig. 2.
Localization of recombinant PRGP2 on the surface of CHO Tet-On cells. (A) Western blotting of biotinylated PRGP2 constructs produced by transient transfection (lanes 1–4) and stable transfection (lanes 5 and 6) of CHO Tet-On cells. Stably transfected cells were either left untreated (lane 5) or treated with trypsin (lane 6) before Western blotting. Biotinylated proteins were detected with streptavidin-HRP (SA-HRP). The positions of molecular mass standards (in kilodaltons) are shown to the right of the blot. (B) Analysis of cell surface expression of glutamyl carboxylated PRGP2 by flow cytometry using an anti-Gla monoclonal antibody. (C) Graphical representation of median fluorescence values derived from the histogram shown in B. The data depicted represent raw median fluorescent values minus the baseline median fluorescent value from cells cultured in the presence of dox and vitamin K but not treated with streptavidin-FITC before flow cytometry. Bar colors correspond to those in B.
The surface exposure of carboxylated PRGP2 in stably transfected CHO Tet-On cells was further investigated by flow cytometry by using the anti-Gla monoclonal antibody (Fig. 2 B and C). In the absence of the inducer (dox), surface-exposed Gla was observed at a level slightly above that seen for the isotype control antibody regardless of whether the cells were cultured in the presence of warfarin (light blue) or vitamin K (green). This phenomenon could be attributed either to low-level leaky expression of PRGP2 before induction or to the presence of endogenous cell surface Gla proteins in the host cell line. The amount of cell surface Gla in cells that were induced with dox in the presence of warfarin (yellow) was virtually identical to noninduced cells. However, dox-mediated induction of PRGP2 expression in the presence of vitamin K resulted in a dramatic increase in cell surface Gla (red) that was significantly reduced when cells were treated with trypsin before analysis (dark blue).
These findings allow several inferences to be made regarding the biosynthesis, topology, and localization of PRGP2. First, when expressed at relatively low levels in the presence of vitamin K, PRGP2 is preferentially converted to its mature form and deposited on the cell surface with its N-terminal Gla domain exposed extracellularly. Second, inhibition of carboxylation, either by mutation of the propeptide or by warfarin treatment, abrogates the proteolytic removal of the propeptide. Third, expression of PRGP2 at high levels overwhelms the host cell's intrinsic carboxylation system, resulting in the production of uncarboxylated pro-PRGP2 that is largely retained within the cell.
PRGP2 Associates with YAP.
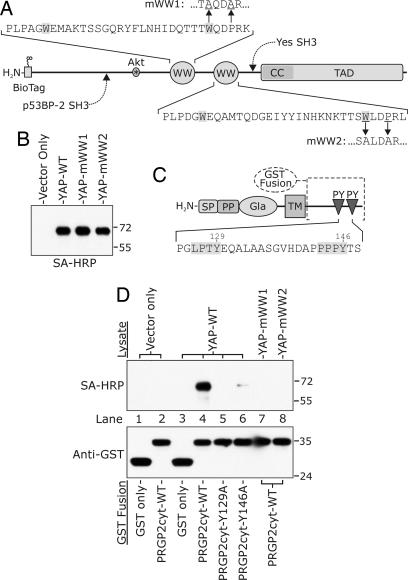
A yeast two-hybrid screen of a human kidney cDNA library, using the cytoplasmic tail of PRGP2 (residues 87–153) as bait, allowed the identification of YAP as a potential PRGP2-binding protein. A schematic representation of YAP (Fig. 3A) illustrates several key features of the molecule that are essential for its function, including binding sites for the SH3 domains of p53-binding protein 2 (p53BP-2) and Yes kinase, and an Akt phosphorylation site that mediates the association of YAP with 14-3-3 (34). In the yeast two-hybrid screen, we isolated a major variant of YAP that contains two type I WW domains, protein modules that bind to PY motifs and act in concert with the C-terminal transcriptional activation domain (TAD) to promote signal transduction and regulate transcription (35–39).
Fig. 3.
Analysis of PRGP2–YAP interactions by GST pull-down assay. (A) Schematic representation of YAP constructs used in the assay. The diagram indicates the relative positions of the binding sites for the SH3 domains of p53-binding protein 2 (p53BP-2) and Yes kinase, as well as the locations of the Akt phosphorylation site, tandem WW domains, the coiled-coil region (CC), and the transcriptional activation domain (TAD). Conserved Trp residues that are characteristic of WW domains are highlighted. Mutations designed to disrupt the binding of WW domains 1 and 2 to PY motifs (mWW1 and mWW2, respectively) have been described (35). (B) Western blotting with streptavidin-HRP (SA-HRP) to detect biotinylated YAP variants in the lysates of transiently transfected CHO Tet-On cells. (C) Schematic representation of GST-PRGP2 cytoplasmic domain fusions used in the assay. GST was fused to the cytoplasmic region of PRGP2 (residues 87–153) as indicated with the dashed line. Mutations designed to impair binding of PRGP2 PY motifs to WW domains (Y129A and Y146) are indicated. (D) Interactions between GST constructs and biotinylated YAP variants were assessed by Western blotting of immobilized GST fusions with SA-HRP to detect associated biotinylated YAP variants (Upper). The same material was blotted and detected with an anti-GST antibody (Lower) to verify the presence of GST or GST fusion proteins. The positions of molecular mass standards (in kilodaltons) are shown to the right of each blot in B and D.
To confirm the association of PRGP2 and YAP, the entire coding sequence of human YAP was ligated into plasmid pNBio, a mammalian expression vector that directs the production and enzymatic biotinylation of cytoplasmic proteins bearing an N-terminal biotin-acceptor peptide (32). Biotinylated wild-type YAP and two variants in which each of the WW domains had been independently mutated to prevent association with PY motifs (mWW1 and mWW2) were transiently expressed in CHO Tet-On cells (Fig. 3B). The association of these YAP constructs with the wild-type PRGP2 cytoplasmic domain, as well as variants in which the Tyr residues within each PY motif had been independently mutated to Ala residues (Y129A and Y146A, Fig. 3C), was assessed by using a GST pull-down approach (Fig. 3D). Nonspecific binding of GST to YAP was not observed, nor was binding of the GST-PRGP2 cytoplasmic domain fusion to streptavidin-reactive components in cells that had been transfected with the empty vector. An intense streptavidin-reactive band was observed for the association of wild-type YAP with the wild-type cytoplasmic domain of PRGP2 (lane 4). The interaction with wild-type YAP was abolished by mutation of the first PY motif of PRGP2 (Y129A; lane 5), and very nearly so by mutation of the second (Y146A; lane 6), indicating that both PY motifs of PRGP2 are essential for formation of a tight complex with YAP, and that this complex is bidentate with respect to both proteins.
Defining an Extended Noncanonical PY Motif.
We used a similar GST pull-down strategy to identify residues within the first putative PY motif of PRGP2 that contribute to its association with YAP (Fig. 4). Mutation of the two residues preceding the LPTY sequence to Ala (Pro124A and Gly125A) did not disrupt YAP binding (lanes 4 and 5). As expected from a number of previous studies that have clearly established the core PY motif, mutation of the corresponding key residues to Ala (Leu126A, Pro127A and Tyr129A; lanes 6, 7, and 9) abrogated binding to wild-type YAP. Unexpectedly, substitution of Thr-128 (corresponding to the unrestricted Xaa residue in the Leu/Pro-Pro-Xaa-Tyr core motif) also abolished binding to YAP, as no band corresponding to biotinylated YAP was observed even upon prolonged exposure of the blot to film (lane 8). In addition, mutation of residues downstream of the core motif (E130A and Q131A) significantly impaired binding to YAP (lanes 10 and 11), with bands corresponding to biotinylated YAP visible only upon extended exposure of the blot.
Fig. 4.
Mapping of an extended noncanonical PY motif in the PRGP2 cytoplasmic domain. Pull-down assays were performed as in Fig. 3. GST fusion proteins consisted of a series of single point mutations within the region encompassing the first PY motif of PRGP2. Only biotinylated wild-type (WT) YAP was used as the target protein. Bound biotinylated YAP was detected with streptavidin-HRP (SA-HRP) by using brief (Top) and extended (Middle) exposure times. The same material was blotted and detected with an anti-GST antibody (Bottom) to verify the presence of GST or GST fusion proteins. The positions of molecular mass standards (in kilodaltons) are shown to the right of the blot.
Discussion
The results presented here establish that PRGP2 undergoes γ-glutamyl carboxylation in a manner that is inhibited by warfarin. The propeptide is efficiently and quantitatively removed from the carboxylated protein, and the resulting mature protein is directed to the plasma membrane with the Gla domain exposed extracellularly and the C-terminal region retained within the cytoplasm. When expressed at levels that exceed the capacity of the host cell's intrinsic carboxylation system, PRGP2 is produced in an immature des-Gla proPRGP2 form. This immature PRGP2 is largely resistant to trypsin-mediated proteolysis in intact cells, indicating that it is predominantly retained within the cell. This finding additionally suggests that the previously observed punctuate perinuclear localization of the PRGP2-GFP chimera is likely to have resulted from the intracellular retention of unprocessed biosynthetic intermediates of PRGP2.
The cytoplasmic tail of PRGP2 seemed to bind specifically to YAP, because it did not bind to WWOX, a WW domain-containing tumor suppressor (data not shown). However, it should be noted that PRGP2 was previously identified as a potential binding partner for the ubiquitin ligase Nedd4 (40). In this study, the second PY motif of PRGP2 (Ala-Pro-Pro-Pro-Pro-Tyr-Thr-Ser; core motif underlined) did not seem to be involved in binding to Nedd4, highlighting the potential importance of the first PY motif for this interaction. In the present study, we characterized the first PY motif as a noncanonical variant (Leu-Pro-Thr-Tyr-Glu-Gln) in which residues C-terminal to the core PY sequence confer additional binding specificity, a property that has been observed in recent structural studies (41–43). We additionally demonstrated that mutation of the Thr residue (occupying the Xaa position of the core motif) to Ala abolished binding to YAP. This finding contrasts with the conclusion, derived from extensive peptide binding experiments and structural studies, that the Xaa position is variable owing to the solvent exposure of its side chain. Interestingly, this position is occupied by either a Thr or Ser residue, not only in all known vertebrate orthologs of PRPG2, but also in its paralogs (PRGP1, TMG3, and TMG4), raising the possibility that phosphorylation may regulate formation of the bidentate PRGP2-YAP complex.
YAP was originally identified based on its ability to bind to the SH3 domain of Yes kinase, and has been shown to associate with the SH3 domains of numerous proteins involved in intracellular signaling pathways (31, 44, 45). In addition, YAP has been extensively characterized as a transcriptional coactivator that forms productive complexes with numerous PY motif-containing transcription factors including p73, PEBP2, TEAD2, and the cytoplasmic domain of ErbB4 (35–39). The phosphorylation of YAP by Akt has been shown to promote the cytoplasmic association of YAP with 14-3-3, thereby preventing translocation to the nucleus and abrogating the p73-mediated proapoptotic up-regulation of Bax (34). Based on these observations, at least three distinct outcomes can be proposed for the association of YAP with PRGP2. First, YAP may serve as an adaptor to recruit SH3 domain-containing signaling molecules to the cytoplasmic domain of PRGP2. Second, as in the case of ErbB4, the cytoplasmic region of PRGP2 may be liberated by regulated intramembrane proteolysis and form a transcriptionally active complex with YAP. Finally, PRGP2 could serve to sequester YAP and thereby attenuate its activity as a transcriptional coactivator. Although the physiological significance of the PRGP2-YAP complex is presently unclear, the results presented here set the stage for the functional characterization of PRGP2 as well as other members of the vertebrate transmembrane Gla protein family.
Materials and Methods
DNA Constructs for Protein Expression in CHO Tet-On Cells.
The full-length PRGP2 coding sequence and variants thereof were amplified by PCR from the PRGP2 cDNA (GenBank accession no. AF009243). Point mutations were introduced by standard two-step overlap extension PCR (46). For all PRGP2 constructs, the forward primer contained a terminal BamHI site and a consensus Kozak translation initiation sequence, and the reverse primer contained an in-frame stop codon preceding a NotI site. Amplimers were ligated between the BamHI and NotI sites of plasmid pCBio (GenBank accession no. DQ520291), a vector that directs the coexpression of E. coli biotin ligase (BirA) and target proteins bearing a C-terminal BioTag (32).
The wild-type human YAP cDNA as well as YAP cDNAs in which each WW domain had been mutated to abolish binding to PY motifs (35) were kindly provided by Marius Sudol (Weis Center for Research, Danville, PA). Full-length wild-type and mutant YAP coding sequences were amplified by PCR with a forward primer that contained a terminal BglII site and a reverse primer that contained a stop codon and a terminal NotI site. Amplimers were ligated between the BamHI and NotI sites of plasmid pNBio (GenBank accession no. DQ520290), a vector developed for the expression of target proteins bearing an N-terminal BioTag (32).
Cell Culture.
The cultivation and transfection of CHO Tet-On cells (Clontech, Palo Alto, CA) with pCBio and pNBio expression constructs was performed as described (25, 32). For the expression of biotinylated PRGP2 constructs in transiently transfected cells, the culture medium was supplemented with 2 μg/ml dox, 25 μM d-biotin, and either 5 μM vitamin K1 (Abbott Laboratories, Abbott Park, IL) or 5 μM sodium warfarin (Sigma–Aldrich, St. Louis, MO), and cells were grown for 24 h before lysis. For the expression of biotinylated PRGP2 in stably transfected cells, cells were cultured for 24 h in the presence of 25 μM d-biotin, either 5 μM vitamin K1 or 5 μM sodium warfarin, and varying concentrations of dox as indicated in Fig. 1C. Alternatively, stably transfected cells were cultured for 24 h in the presence of 100 ng/ml dox, 25 μM d-biotin, and 5 μM vitamin K1. Cells were then either left untreated or treated with 0.25% trypsin/1 mM EDTA in Hanks' balanced salt solution (Invitrogen, Carlsbad, CA) for 30 min before Western blotting. Trypsin was inactivated by the addition of 2 mg/ml soybean trypsin inhibitor and 100 μM diisopropyl fluorophosphate.
Western Blotting.
Cell lysis, SDS/PAGE, electrophoretic blotting, detection of biotinylated proteins with streptavidin-HRP, and detection of Gla with an anti-Gla monoclonal antibody (American Diagnostica, Stamford, CT) were performed as described (25, 32).
Flow Cytometry.
CHO Tet-On cells that had stably transfected with the wild-type PRGP2 expression vector were plated at 106 cells per 10 cm plate and grown overnight. Expression of PRGP2 was induced for by the addition of 2 μg/ml dox to medium containing either 5 μM vitamin K1 or 5 μM sodium warfarin. After 24 h, cells were washed twice with PBS and then either detached from plates with 0.5 mM EDTA/PBS or treated for 30 min with 0.25% trypsin/1 mM EDTA. Trypsin activity was inhibited with soybean trypsin inhibitor and diisopropyl fluorophosphate as described above. Cells that had been detached from plates by either EDTA or by trypsin were suspended in PBS containing 1 mM EDTA and 2% BSA. After two washes with PBS, cells were resuspended in PBS/2% BSA/10 mM EDTA containing either the anti-Gla monoclonal antibody or an IgG2b isotype control antibody (Santa Cruz Biotechnology, Santa Cruz, CA) both at 5 μg/ml. After incubation with primary antibodies for 90 min at 4°C with gentle mixing, cells were washed three times with PBS, and resuspended in PBS/2% BSA/10 mM EDTA containing FITC-conjugated goat anti-mouse IgG2b (Santa Cruz Biotechnology) diluted 1:400 from the commercially supplied stock solution. Cells were washed twice with PBS containing 10 mM EDTA, fixed with 0.5% paraformaldehyde in PBS, and analyzed on a FACScan flow cytometer (Becton Dickinson, San Jose, CA).
Yeast Two-Hybrid Screening.
The bait plasmid used in yeast two-hybrid screening was generated by PCR amplification of region of the PRGP2 cDNA corresponding to its cytoplasmic domain (residues 87–153), and insertion of the resulting amplimer between the EcoRI and BamHI sites of plasmid pAS2-1 (Clontech; GenBank accession no. U30497). S. cerevisiae strain CG1945 was cotransfected by the lithium acetate method (47) with the bait plasmid and a human adult kidney cDNA-GAL4 activation domain library in plasmid pACT2 (Clontech; GenBank accession no. U29899). Histidine prototrophs were selected on plates containing triple dropout medium (lacking Leu, Trp, and His) supplemented with 5 mM 3-amino-1,2,4-triazole to suppress background colony growth. Library plasmid DNA was isolated by using the Yeastmaker plasmid isolation kit (Clontech) from clones that reverted to histidine auxotrophy upon cycloheximide counterselection for loss of the bait plasmid. Library plasmid inserts were identified by automated DNA sequencing with pACT2-specific oligonucleotide primers. Molecular interactions were confirmed by β-gal colony lift assay of S. cerevisiae strain Y187 (Clontech) that had been cotransfected with both bait and library plasmids.
GST Pull-Down Experiments.
The region of the PRGP2 cDNA corresponding to the cytoplasmic domain (residues 87–153) was amplified by using PCR primers that introduced SpeI and XhoI sites at the 5′ and 3′ termini, respectively. Alternatively, a series of Ala-encoding point mutations were introduced by overlap extension PCR (46) using the same terminal primers. The resulting amplimers were inserted between the SpeI and XhoI sites of pET41a (Novagen, Madison, WI). To generate a plasmid for the expression of GST without a C-terminal extension, a 244 base pair segment between the SpeI and XhoI sites of pET41a was excised and replaced with the sequence 5′-AGATCTGATATC-3′. E. coli BL21(DE3)-RIPL Codon Plus cells (Stratagene, La Jolla, CA) were transformed with these plasmids, and protein expression was induced with 0.5 mM isopropyl β-d-1-thiogalactopyranoside for 3 h. GST fusion proteins were purified on glutathione Sepharose 4B (GE Healthcare, Piscataway, NJ) according to the manufacturer's instructions. Ten microliters of glutathione resin with bound protein was added to 250 μl of CHO Tet-On cell lysate (5 μg of protein) and incubated at 4°C with gentle mixing for 45 min. The resin was washed three times with lysis buffer, and bound protein was eluted in SDS/PAGE loading buffer by heating at 95°C for 5 min. Detection of biotinylated YAP with streptavidin-HRP was performed as described (25, 32). GST fusion proteins were detected by Western blotting with polyclonal antibodies that had been affinity purified from sera of rabbits immunized with a GST-BirA fusion protein (32).
Acknowledgments
We thank Dominic Chung, Greg Mize, and Mary Hu (University of Washington) for their helpful advice and assistance in the preparation of this manuscript, and Marius Sudol (Weis Center for Research, Danville, PA) for kindly providing human YAP cDNAs. This work was supported by National Institutes of Health Grants HL-16919 and HL-071599.
Abbreviations
- Gla
γ-carboxyglutamic acid
- dox
doxycycline
- PRGP
proline-rich Gla protein
- TMG
transmembrane Gla protein
- YAP
Yes-associated protein
- ER
endoplasmic reticulum.
Footnotes
The authors declare no conflict of interest.
References
- 1.Stenflo J, Fernlund P, Egan W, Roepstorff P. Proc Natl Acad Sci USA. 1974;71:2730–2733. doi: 10.1073/pnas.71.7.2730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Suttie JW. Annu Rev Biochem. 1985;54:459–477. doi: 10.1146/annurev.bi.54.070185.002331. [DOI] [PubMed] [Google Scholar]
- 3.Huang M, Rigby AC, Morelli X, Grant MA, Huang G, Furie B, Seaton B, Furie BC. Nat Struct Biol. 2003;10:751–756. doi: 10.1038/nsb971. [DOI] [PubMed] [Google Scholar]
- 4.Nelsestuen GL. J Biol Chem. 1976;251:5648–5656. [PubMed] [Google Scholar]
- 5.Soriano-Garcia M, Padmanabhan K, de Vos AM, Tulinsky A. Biochemistry. 1992;31:2554–2566. doi: 10.1021/bi00124a016. [DOI] [PubMed] [Google Scholar]
- 6.Bevers EM, Comfurius P, van Rijn JL, Hemker HC, Zwaal RF. Eur J Biochem. 1982;122:429–436. doi: 10.1111/j.1432-1033.1982.tb05898.x. [DOI] [PubMed] [Google Scholar]
- 7.Fadok VA, Voelker DR, Campbell PA, Cohen JJ, Bratton DL, Henson PM. J Immunol. 1992;148:2207–2216. [PubMed] [Google Scholar]
- 8.Op den Kamp JA. Annu Rev Biochem. 1979;48:47–71. doi: 10.1146/annurev.bi.48.070179.000403. [DOI] [PubMed] [Google Scholar]
- 9.Sadowski JA, Esmon CT, Suttie JW. J Biol Chem. 1976;251:2770–2776. [PubMed] [Google Scholar]
- 10.Wu SM, Cheung WF, Frazier D, Stafford DW. Science. 1991;254:1634–1636. doi: 10.1126/science.1749935. [DOI] [PubMed] [Google Scholar]
- 11.Foster DC, Rudinski MS, Schach BG, Berkner KL, Kumar AA, Hagen FS, Sprecher CA, Insley MY, Davie EW. Biochemistry. 1987;26:7003–7011. doi: 10.1021/bi00396a022. [DOI] [PubMed] [Google Scholar]
- 12.Jorgensen MJ, Cantor AB, Furie BC, Brown CL, Shoemaker CB, Furie B. Cell. 1987;48:185–191. doi: 10.1016/0092-8674(87)90422-3. [DOI] [PubMed] [Google Scholar]
- 13.Suttie JW, Hoskins JA, Engelke J, Hopfgartner A, Ehrlich H, Bang NU, Belagaje RM, Schoner B, Long GL. Proc Natl Acad Sci USA. 1987;84:634–637. doi: 10.1073/pnas.84.3.634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chu PH, Huang TY, Williams J, Stafford DW. Proc Natl Acad Sci USA. 2006;103:19308–19313. doi: 10.1073/pnas.0609401103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Whitlon DS, Sadowski JA, Suttie JW. Biochemistry. 1978;17:1371–1377. doi: 10.1021/bi00601a003. [DOI] [PubMed] [Google Scholar]
- 16.Dahlback B, Stenflo J. Proc Natl Acad Sci USA. 1981;78:2512–2516. doi: 10.1073/pnas.78.4.2512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Webb JH, Blom AM, Dahlback B. Blood Coagul Fibrinolysis. 2003;14:355–359. doi: 10.1097/00001721-200306000-00006. [DOI] [PubMed] [Google Scholar]
- 18.Webb JH, Blom AM, Dahlback B. J Immunol. 2002;169:2580–2586. doi: 10.4049/jimmunol.169.5.2580. [DOI] [PubMed] [Google Scholar]
- 19.Hafizi S, Dahlback B. FEBS J. 2006;273:5231–5244. doi: 10.1111/j.1742-4658.2006.05529.x. [DOI] [PubMed] [Google Scholar]
- 20.Hasanbasic I, Rajotte I, Blostein M. J Thromb Haemost. 2005;3:2790–2797. doi: 10.1111/j.1538-7836.2005.01662.x. [DOI] [PubMed] [Google Scholar]
- 21.Nakano T, Kawamoto K, Kishino J, Nomura K, Higashino K, Arita H. Biochem J. 1997;323:387–392. doi: 10.1042/bj3230387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stenhoff J, Dahlback B, Hafizi S. Biochem Biophys Res Commun. 2004;319:871–878. doi: 10.1016/j.bbrc.2004.05.070. [DOI] [PubMed] [Google Scholar]
- 23.Jiang Y, Doolittle RF. Proc Natl Acad Sci USA. 2003;100:7527–7532. doi: 10.1073/pnas.0932632100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang CP, Yagi K, Lin PJ, Jin DY, Makabe KW, Stafford DW. J Thromb Haemost. 2003;1:118–123. doi: 10.1046/j.1538-7836.2003.00069.x. [DOI] [PubMed] [Google Scholar]
- 25.Kulman JD, Harris JE, Nakazawa N, Ogasawara M, Satake M, Davie EW. Proc Natl Acad Sci USA. 2006;103:15794–15799. doi: 10.1073/pnas.0607543103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kulman JD, Harris JE, Haldeman BA, Davie EW. Proc Natl Acad Sci USA. 1997;94:9058–9062. doi: 10.1073/pnas.94.17.9058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kulman JD, Harris JE, Xie L, Davie EW. Proc Natl Acad Sci USA. 2001;98:1370–1375. doi: 10.1073/pnas.98.4.1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Macias MJ, Hyvonen M, Baraldi E, Schultz J, Sudol M, Saraste M, Oschkinat H. Nature. 1996;382:646–649. doi: 10.1038/382646a0. [DOI] [PubMed] [Google Scholar]
- 29.Pires JR, Taha-Nejad F, Toepert F, Ast T, Hoffmuller U, Schneider-Mergener J, Kuhne R, Macias MJ, Oschkinat H. J Mol Biol. 2001;314:1147–1156. doi: 10.1006/jmbi.2000.5199. [DOI] [PubMed] [Google Scholar]
- 30.Kasanov J, Pirozzi G, Uveges AJ, Kay BK. Chem Biol. 2001;8:231–241. doi: 10.1016/s1074-5521(01)00005-9. [DOI] [PubMed] [Google Scholar]
- 31.Sudol M, Bork P, Einbond A, Kastury K, Druck T, Negrini M, Huebner K, Lehman D. J Biol Chem. 1995;270:14733–14741. doi: 10.1074/jbc.270.24.14733. [DOI] [PubMed] [Google Scholar]
- 32.Kulman JD, Satake M, Harris JE. Protein Expression Purif. 2007;52:320–328. doi: 10.1016/j.pep.2006.09.011. [DOI] [PubMed] [Google Scholar]
- 33.Rabiet MJ, Jorgensen MJ, Furie B, Furie BC. J Biol Chem. 1987;262:14895–14898. [PubMed] [Google Scholar]
- 34.Basu S, Totty NF, Irwin MS, Sudol M, Downward J. Mol Cell. 2003;11:11–23. doi: 10.1016/s1097-2765(02)00776-1. [DOI] [PubMed] [Google Scholar]
- 35.Komuro A, Nagai M, Navin NE, Sudol M. J Biol Chem. 2003;278:33334–33341. doi: 10.1074/jbc.M305597200. [DOI] [PubMed] [Google Scholar]
- 36.Milewski RC, Chi NC, Li J, Brown C, Lu MM, Epstein JA. Development (Cambridge, UK) 2004;131:829–837. doi: 10.1242/dev.00975. [DOI] [PubMed] [Google Scholar]
- 37.Omerovic J, Puggioni EM, Napoletano S, Visco V, Fraioli R, Frati L, Gulino A, Alimandi M. Exp Cell Res. 2004;294:469–479. doi: 10.1016/j.yexcr.2003.12.002. [DOI] [PubMed] [Google Scholar]
- 38.Strano S, Munarriz E, Rossi M, Castagnoli L, Shaul Y, Sacchi A, Oren M, Sudol M, Cesareni G, Blandino G. J Biol Chem. 2001;276:15164–15173. doi: 10.1074/jbc.M010484200. [DOI] [PubMed] [Google Scholar]
- 39.Yagi R, Chen LF, Shigesada K, Murakami Y, Ito Y. EMBO J. 1999;18:2551–2562. doi: 10.1093/emboj/18.9.2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jolliffe CN, Harvey KF, Haines BP, Parasivam G, Kumar S. Biochem J. 2000;351:557–565. [PMC free article] [PubMed] [Google Scholar]
- 41.Chong PA, Lin H, Wrana JL, Forman-Kay JD. J Biol Chem. 2006;281:17069–17075. doi: 10.1074/jbc.M601493200. [DOI] [PubMed] [Google Scholar]
- 42.Kanelis V, Bruce MC, Skrynnikov NR, Rotin D, Forman-Kay JD. Structure (London) 2006;14:543–553. doi: 10.1016/j.str.2005.11.018. [DOI] [PubMed] [Google Scholar]
- 43.Kanelis V, Rotin D, Forman-Kay JD. Nat Struct Biol. 2001;8:407–412. doi: 10.1038/87562. [DOI] [PubMed] [Google Scholar]
- 44.Sudol M. Oncogene. 1994;9:2145–2152. [PubMed] [Google Scholar]
- 45.Espanel X, Sudol M. J Biol Chem. 2001;276:14514–14523. doi: 10.1074/jbc.M008568200. [DOI] [PubMed] [Google Scholar]
- 46.Ho SN, Hunt HD, Horton RM, Pullen JK, Pease LR. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- 47.Schiestl RH, Gietz RD. Curr Genet. 1989;16:339–346. doi: 10.1007/BF00340712. [DOI] [PubMed] [Google Scholar]