Abstract
Sec1/Munc18 proteins (SM proteins) bind to soluble NSF attachment protein receptors (SNAREs) and play an essential role in membrane fusion. Divergent modes of regulation have been proposed for different SM proteins indicating that they can either promote or inhibit SNARE assembly. This is in part because of discrete modes of binding that have been described for various SM/SNARE complexes. One mode suggests that SM proteins bind only to Syntaxins (Stx) preventing SNARE assembly, whereas in another they facilitate SNARE assembly and bind to SNARE complexes. The mammalian cell surface SM protein Munc18c binds to an N-peptide in Stx4, and this is compatible with its interaction with SNARE complexes. Here we describe the crystal structure of Munc18c in complex with the Stx4 N-peptide. This structure shows remarkable similarity with a yeast complex indicating that the mode of binding, which can accommodate SNARE complexes, is highly conserved throughout evolution. Modeling reveals the presence of the N-peptide binding mode in most but not all yeast and mammalian SM/Stx pairs, suggesting that it has coevolved to fulfill a specific regulatory function. It is unlikely that the N-peptide interaction alone accounts for the specificity in SM/SNARE binding, implicating other contact surfaces in this function. Together with other data, our results support a sequential two-state model for SM/SNARE binding involving an initial interaction via the Stx N-peptide, which somehow facilitates a second, more comprehensive interaction comprising other contact surfaces in both proteins.
Keywords: crystallography, protein:protein interactions, vesicle trafficking
Intracellular trafficking relies on membrane fusion, a tightly regulated process that requires the formation of a coiled coil complex between helical motifs originating from soluble NSF attachment protein receptor (SNARE) proteins on vesicle and target membranes (1–3). Sec1/Munc18 proteins (SM proteins) play a fundamental role in this process by interacting with SNARE protein(s) and regulating the formation of SNARE complexes (4, 5). SM proteins have been identified in organisms from yeast to humans, and loss-of-function mutants lead to severe impairment in vesicle fusion (6).
Despite their obvious importance, the nature of SNARE regulation by SM proteins remains controversial, in that both positive and negative regulatory roles have been reported (6, 7). Possibly the most compelling evidence in support of negative regulation is the observation that the mammalian Syntaxin1a (Stx1a) SNARE protein involved in synaptic neurotransmission exists in both an “open” (SNARE complex-compatible) and a “closed” (SNARE complex-incompatible) conformation (8, 9). The crystal structure of Stx1a in complex with the SM protein Munc18-1 (Munc18a, nSec1) reveals that Munc18-1 embraces a closed conformation preventing Stx1a from forming the SNARE complex required for vesicle fusion (10). On the other hand, deletion of Munc18-1 results in impaired exocytosis in mouse neurons and chromaffin cells (11, 12), suggesting that it plays a positive regulatory role.
The case for a positive role is strengthened by observations in the yeast endoplasmic reticulum/Golgi system involving the SM protein Sly1p and its cognate Stx Sed5p. Sly1p binds to monomeric Sed5p as well as higher order SNARE complexes. Moreover, these interactions are facilitated by a short N-terminal peptide in Sed5p, which fits into a hydrophobic pocket in Sly1p. These data are consistent with a positive regulatory role for Sly1p in SNARE assembly and emphasize the importance of the N-peptide interaction (13–15).
In mammals, vesicle fusion with the plasma membrane is highly evolved and involves multiple SNARE complexes and SM proteins (6). There is a high degree of specificity such that the SM proteins Munc18-1 and Munc18b (Munc18-2) bind to Stxs 1–3 but not to Stx4, whereas Munc18c binds only to Stxs 2 and 4 (16). Although the function of Munc18-1 is confined to regulated exocytosis in neurons and neuroendocrine cells, Munc18c regulates vesicle transport to the plasma membrane in a range of cell types and has a key role in the insulin-stimulated movement of GLUT4 glucose transporters to the cell surface in muscle and adipocytes (17). The SNARE complex associated with this process involves VAMP2 (synaptobrevin) on intracellular GLUT4 storage vesicles and Stx4 and SNAP23 on the plasma membrane. We showed that Munc18c interacts with Stx4 alone (18) and with Stx4 SNARE complexes but not with VAMP2 or SNAP23 alone (19). Importantly, formation of the Munc18c/Stx4 heterodimer requires an N-terminal peptide in Stx4 similar to that described for yeast Sed5p (14, 20). Preformation of the Munc18c/Stx4 heterodimer promotes assembly of the SNARE ternary complex, indicating that Munc18c plays a positive regulatory role in membrane fusion (19). In the Sly1p/Sed5p structure, key interactions were found between Sed5p:Phe-10 and Sly1p:Leu-137. We predicted a similar binding mode for Stx4 and Munc18c and proposed that Phe-119 in Munc18c interacts with Leu-8 in Stx4. This was supported by site-directed mutagenesis (19).
We have now determined the crystal structure of the Munc18c/Stx4 N-peptide complex. The binding mode resembles that reported for Sly1p/Sed5p, indicating a conserved molecular mechanism. Based on comparison with the Munc18-1 structure and Stx4 and Stx1a N-peptide sequences, a similar N-peptide binding mode can be expected to form between Munc18-1 and Stx1a, consistent with recent reports (21, 22). We also used a structural bioinformatics approach to predict the presence or absence of an N-peptide binding mode in all yeast and mammalian SM/Stx pairs.
Results
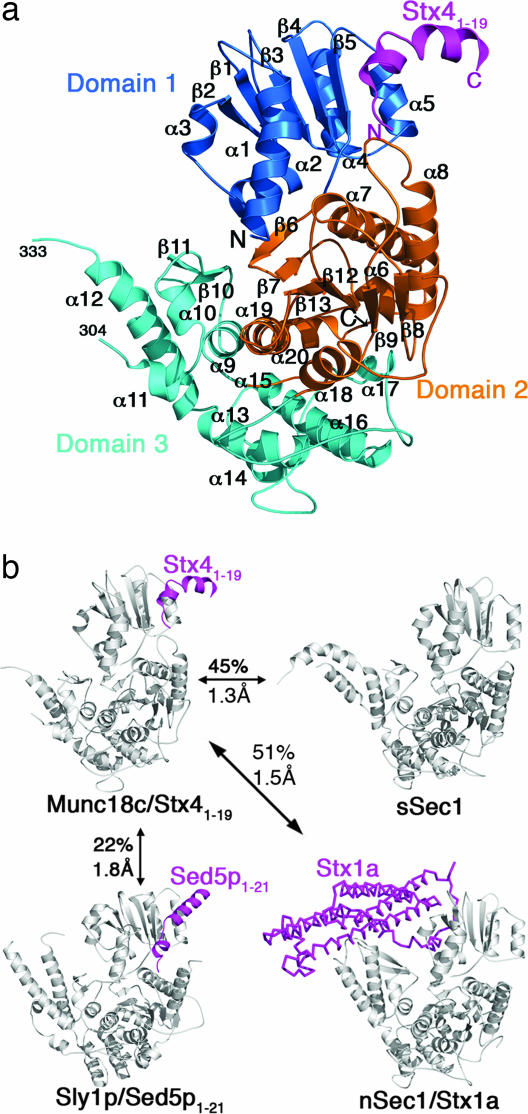
The structure of mouse Munc18c/Stx41–19 (Fig. 1 and Table 1) was solved by molecular replacement by using the squid Munc18-1 structure [sSec1, Protein Data Bank (PDB) ID code 1EPU (23)] as a model. The Munc18c structure resembles three other SM proteins that have been structurally characterized, rat Munc18-1 [from the complex with Stx1a, PDB ID code 1DN1 (10)], squid Munc18-1 [PDB ID code 1EPU (23)], and yeast Sly1p [PDB ID code 1MQS (20)] (Fig. 1). Of these, Munc18c is least similar to yeast Sly1p. Nevertheless, the two share the same Stx N-peptide binding mode.
Fig. 1.
Structure of Munc18c/Stx41–19 and comparison with other SM proteins. (a) The Munc18c/Stx41–19 complex showing domain 1 (residues 1–138) in dark blue, domain 2 (residues 139–250 and 476–584) in orange, and domain 3 (residues 251–475) in cyan. Stx4 residues 1–19 are shown in magenta. Helices are shown as coils, and strands are shown as arrows. N and C termini and secondary structure elements are labeled. (b) Comparison of mouse Munc18c/Stx41–19 with squid neuronal Munc18-1 [sSec1, PDB ID code 1EPU (23)], yeast Sly1p/Sed5p1–21 [PDB ID code 1MQS (20)], and rat neuronal Munc18-1/Stx1a (nSec1/Stx1a) [PDB ID code 1DN1 (10)] showing rmsd of the structures and the pairwise sequence identity.
Table 1.
X-ray data measurement and refinement statistics
| X-ray data measurement | |
| Resolution, Å | 30–3.15 (3.29–3.15) |
| Rmerge | 0.065 (0.560) |
| I/σI | 15.0 (3.3) |
| Completeness, % | 99.9 (100.0) |
| Redundancy | 6.3 (6.3) |
| Structure refinement | |
| Resolution, Å | 12–3.15 (3.23–3.15) |
| No. of reflections total/test set | 26,463/1,421 |
| Rfactor, %/Rfree, % | 25.2 (31.7)/28.0 (32.3) |
| rmsd from ideal bond lengths, Å | 0.008 |
| rmsd from ideal bond angles, ° | 1.4 |
| Ramachandran plot,* | 94.5/0.8 |
Values in parentheses represent data for the highest-resolution shell.
*Percentage of residues in most favored and additionally allowed regions/disallowed region.
N-Peptide Binding Mode Is Conserved from Yeast to Mammals.
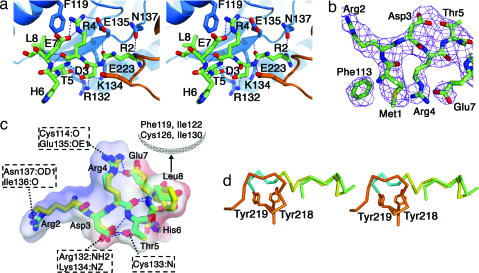
Bound Stx4 N-peptide is predominantly helical (Fig. 1), although residues 1–4 are extended and form two antiparallel hydrogen bonds to Munc18c β5. The bound conformation and interactions formed by the Stx4 N-peptide with Munc18c are strikingly similar to those of the Sed5p N-peptide with Sly1p [Figs. 1–3 and supporting information (SI) Table 2]. In both cases, the SM proteins interact with a short segment of the ≈30-residue peptide (the first eight and 10 residues of the Stx4 and Sed5p N-peptides, respectively). Indeed, the majority of the interactions involve just two residues, a highly conserved arginine in a DRT motif (Arg-4 of Stx4 and Arg-6 of Sed5p) and a hydrophobic residue (Leu-8 of Stx4 and Phe-10 of Sed5p) (Fig. 2).
Fig. 2.
Interaction between Stx4 N-peptide and Munc18c. (a) Stereo diagram showing the bound N-peptide (green) and residues of Munc18c (blue, domain 1; orange, domain 2). Hydrogen bond interactions are shown as dotted lines. (b) Electron density of the Stx4 N-peptide (2Fo − Fc contoured at 1σ). (c) Superimposition of SM-bound conformations of Stx4 (green) and Sed5p (yellow) N-peptides. Intramolecular hydrogen bonds (blue dotted lines) and Munc18c-interacting residues are indicated (hydrogen bond partners for Arg-2, Asp-3, Arg-4, and Thr-5 and hydrophobic interacting partners for Leu-8). The electrostatic surface for the Stx4 peptide is also shown, revealing a basic face (Arg-2 and Arg-4) and an acidic face (Asp-3 and Glu-7) that are complementary to the Munc18c binding surface (Fig. 3). (d) Stereo diagram of the α8 loop region of Munc18c (orange) and the Stx4 (green) and Sed5p (yellow and blue) N-peptides, showing the overlap between the loop and the nonnative residues of Sed5p (blue).
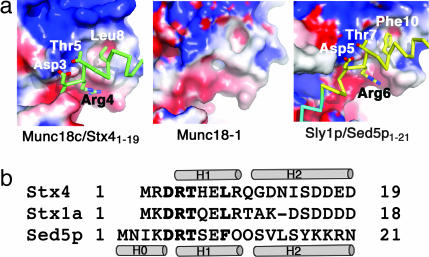
Fig. 3.
A structurally conserved N-peptide binding site in Munc18c, Sly1p, and Munc18-1. (a) Electrostatic surfaces are shown for the structures of Munc18c, Munc18-1, and Sly1p in the N-peptide binding region (colored blue to red from +5 to −5 KbT/e). An acidic groove (red), basic patch (blue), and hydrophobic pocket (white) characterize the N-peptide binding sites of Munc18c and Sly1p, and these features are also present in Munc18-1. The main chain of the N-peptides of Stx4 (green) and Sed5p (yellow and blue) and the side chains of the DRT motif and hydrophobic residues are shown. (b) Sequence alignment of the N-peptides of mouse Stx4, rat Stx1a, and yeast Sed5p is shown with the DRT motif and hydrophobic residue indicated in bold. Helices are shown as cylinders for Stx4 and Sed5p.
The conservation in binding mode for these two Stx residues is exemplified by the arginine of the DRT motif (Fig. 2). The side chain of Stx4 Arg-4 interacts with Munc18c Glu-135 (side chain) and Cys-114 (main chain oxygen) and forms an intramolecular interaction with Stx4 Glu-7 (side chain). A β-strand interaction is formed between the main chain nitrogen of Arg-4 and the main chain oxygen of Cys-133 (not shown in Fig. 2). Finally, the main chain oxygen of Arg-4 forms an intramolecular helical hydrogen bond with the main chain nitrogen of Leu-8. All five interactions are mirrored in the structure of the Sly1p/Sed5p N-peptide complex, although Glu-135 of Munc18c is replaced by Asp-158 in Sly1p. By contrast, the role of the flanking residues in the DRT motif appears to be mainly structural; these residues form intramolecular interactions (Fig. 2) that are conserved in the Sly1p/Sed5p N-peptide complex.
The N-peptide hydrophobic residue fits into an SM protein hydrophobic pocket in both complexes (Fig. 3). The Sly1p pocket is formed from Leu-137, Leu-140, Val-144, Ile-153, and Val-156. Mutation of Sed5p Phe-10 to Ala prevents binding to Sly1p (24). Similarly, mutation of Stx4 Leu-8 to Lys or Munc18c Phe-119 to Glu disrupts binding of Munc18c to Stx4 (19), and we proposed that Leu-8 replaces Phe-10 of Sed5p. Our prediction is now proven to be correct; the Munc18c/Stx41–19 structure shows that Leu-8 binds to a hydrophobic pocket consisting of Munc18c residues Phe-119, Ile-122, Cys-126, Ile-130, and Cys-133.
Despite the similar binding mode, there is a difference between the structures of Munc18c and Sly1p at the N-peptide binding site. This occurs at a region of Sly1p, where five nonnative residues of the Sed5p N-peptide are bound; residues GlyAlaMetAlaGly (numbered −4, −3, −2, −1, and 0 in the structure) are remnants of the cloning and proteolysis procedure used to produce the Sed5p peptide (20). In Munc18c, the α8 loop (residues 222–231) connecting α8 and β9 (SI Fig. 5) of domain 2 occupies the position equivalent to the nonnative Sed5p residues (Fig. 2). The α8 loop forms a hydrogen bond and van der Waals interactions with the Stx4 N-peptide. The electron density for the loop is well defined in Munc18c, whereas the equivalent region in Sly1p (residues 245–256) is disordered. Evidence supporting an important role for this loop in Munc18c is that phosphorylation of Tyr-219, a buried residue on α8, causes dissociation of the Munc18c/Stx4 complex (25). Based on our structure, it is likely that Tyr-219 phosphorylation disrupts the loop and its interactions with the N-peptide.
Munc18-1 Has an N-Peptide Binding Site.
The remarkably similar binding modes of Munc18c and Sly1p for their respective Stx N-peptides prompted us to ask whether an N-peptide binding mode is likely in Munc18-1. Munc18c shares a higher sequence identity and structural similarity with Munc18-1 than with Sly1p (Fig. 1). Furthermore, evidence is emerging that Munc18-1 can interact with SNARE complexes (21, 22, 26, 27) and that the Stx1a N terminus is important for this interaction (21, 22). We surmised that an N-peptide interaction can occur in Munc18-1 and that this would explain the requirement for the N-peptide in SNARE complex interactions with Munc18-1.
Comparison between the N-peptide binding sites of Munc18c and Sly1p and the equivalent region in Munc18-1 (Fig. 3) revealed that the three features that characterize the Munc18c and Sly1p N-peptide binding site (an acidic groove, a neighboring basic region, and a hydrophobic pocket) are all present in the Munc18-1 crystal structure. Furthermore, the key residues of the Stx4 N-peptide (Arg-4 in a DRT motif, and Leu-8) are conserved in Stx1a (Fig. 3). Also, the Munc18c residues contacted by Arg-4 and Leu-8 are conserved in Munc18-1. Hence, the features necessary for the N-peptide binding mode are present in Stx1a and Munc18-1, and this likely explains how Munc18-1 binds to SNARE complexes.
Which SM/Stx Pairs Have an N-Peptide Binding Mode?
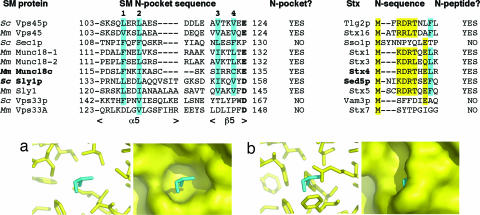
The Munc18c/Stx41–19 and Sly1p/Sed5p1–21 structures allow delineation of the features of the N-peptide binding mode. The major interaction involves a hydrophobic residue (Leu or Phe) present in an Stx N-terminal motif corresponding to MX0–2(R/K)DRTX(e/q)(L/F). In Stx3 the motif is slightly different, in that Leu replaces Thr. Stx16 has RRLT rather than (R/K)DRT, and this N-motif is known to be required for binding to the SM protein Vps45 (24).
The hydrophobic residue (L/F) of the N-peptide binds in a hydrophobic pocket (N-pocket) on the surface of the SM protein. In Munc18c, the N-pocket is formed by the hydrophobic residues F119, I122, C126, I130, and C133, which align with the Sly1p residues lining its N-pocket, L137, L140, V144, I153, and V156 (SI Fig. 5). Sequence alignment of this region for all yeast and mammalian SM proteins reveals those SM proteins likely to have a hydrophobic N-pocket (Fig. 4). In this analysis we did not include C126 of Munc18c or V144 of Sly1p because the differing loop lengths make alignment of loop residues problematic. We therefore confined our analysis to the four hydrophobic residues (sites 1, 2, 3, and 4) that are located in α5 and β5. We found that, in addition to Sly1p and Munc18c, an N-pocket is likely to be present in yeast and mammalian Vps45 (L, L, V, V and V, L, V, V, respectively), in mammalian Munc18-1 and Munc18-2 (F, L, I, L and F, L, V, L, respectively) and mammalian Sly1 (L, I, I, V).
Fig. 4.
SM proteins predicted to have a hydrophobic N-pocket have cognate Stxs with an N-peptide sequence motif. A sequence alignment of the N-pocket of SM proteins from yeast (S. cerevisiae, Sc) and mammals (M. musculus, Mm) with the proteins involved in the two N-peptide complexes that have been structurally characterized shown in bold. Hydrophobic residues that align with sites 1, 2, 3, and 4 of the N-pockets of Munc18c and Sly1p are highlighted in cyan. The SM protein acidic residue that interacts with the DRT motif of the N-peptide is shown in bold. All SM proteins except Sec1p, Vps33p, Vps33A, and Vps33B (data not shown) are predicted to have hydrophobic N-pockets that could interact with the N-peptide hydrophobic residue of cognate Stxs. All Stxs except the cognate Stxs of Sec1p, Vps33p, and Vps33A/B have an N-peptide motif (shown in yellow with the hydrophobic residue in cyan). For the Stx N-peptides, alignment of the N-terminal sequences of Sso1p and Vam3p is based on the sequence alignment in ref. 50. (a) Munc18c residues from the N-pocket (yellow) that interact with the hydrophobic residue of the Stx4 N-peptide Leu-8 (cyan). The surface of the hydrophobic N-pocket is also shown. (b) The same region is shown for yeast Sec1p, predicted from the structure of Munc18c and the sequence alignment above, showing that the hydrophobic residues at sites 1–4 of Sec1p fill the pocket.
Yeast Sec1p, on the other hand, does not possess this N-pocket. It has four hydrophobic residues at sites 1–4, but sites 1–3 are F rather than L, I, or V, and these three aromatic side chains fill the space that would comprise the N-pocket (Fig. 4). Furthermore, yeast Sec1p does not have the conserved acidic residue that interacts with the N-peptide DRT motif (Glu-135 in Munc18c and Asp-158 in Sly1p), and there is no predicted N-peptide motif in Sso1p. These findings indicate that the SM/Stx pair of yeast Sec1p/Sso1p does not utilize the N-peptide binding mode, in agreement with recent data on this system (28).
Other Stxs lacking an N-peptide motif are Vam3p and Stx7. Vps33p, the cognate SM protein of Vam3p, may have a partial hydrophobic pocket (F, V, T, P), but the Pro residue at site 4 in Vps33 sequences suggests structural differences in this region compared with other SM proteins. In any case, it is clear that mammalian Vps33A does not have a hydrophobic pocket because sites 1 and 3 have acidic rather than hydrophobic residues.
In summary, SM proteins that have predicted hydrophobic N-pockets all have partner Stxs with a putative N-peptide motif (Vps45p/Tlg2p, Vps45/Stx16, Munc18-1/Stx1, Munc18-2/Stx3, Munc18c/Stx4, Sly1p/Sed5p, and Sly1/Stx5). Conversely, SM proteins that do not have a hydrophobic N-pocket have cognate Stxs that lack an N-peptide motif (Sec1p/Sso1p, Vps33p/Vam3p, and Vps33/Stx7). This indicates that the N-peptide binding mode may have coevolved in SM and Stx proteins.
Discussion
The foremost question in understanding the biological role of SM/SNARE complexes in vesicle transport is, how can we reconcile the different binding modes that have been described? Sly1p (14, 20, 29), Vps45p (24, 30), and Munc18c (19) bind to an N-peptide on their cognate Stxs, and this interaction can accommodate binding to monomeric Stxs as well as SNARE complexes. Evidence is emerging to show that Munc18-1 also binds SNARE complexes (21, 22, 26, 27), as well as monomeric Stx1a. The interaction between Munc18-1 and the SNARE complex relies on the Stx1a N-peptide (21, 22). We suggest that the presence of an N-terminal hexahistidine tag in Stx1a used to generate the Munc18-1/Stx1a crystal structure may have precluded the N-peptide interaction with Munc18-1 (10).
In contrast to Munc18-1 and Munc18c, the yeast cell surface SM protein Sec1p binds only to SNARE complexes in vivo and in vitro, and this interaction does not require an N-peptide in Sso1p (28, 31, 32). Furthermore, the SM protein Vps33p is part of a large complex (HOPS or VpsC) that interacts with Vam3p (33, 34), but a direct Vps33p/Vam3p interaction is unlikely (N. Bryant and D.E.J., unpublished data).
The structure of the Munc18c/Stx4 N-peptide complex reveals a high degree of conservation with yeast Sly1p/Sed5p. The strict conservation of this binding mode from yeast to mammals provides compelling evidence that an N-peptide interaction is important and probably serves an evolutionarily conserved function in some but not all vesicle transport processes.
It is becoming clear that the universal feature of SM proteins is their ability to interact with SNARE complexes. In the case of Munc18-1 and Sly1p, the N-peptide is required for this interaction (21, 22), and it has been proposed that the N-peptide facilitates SNARE assembly (21). This seems unlikely because Sec1p/Sso1p lack the hydrophobic N-pocket/N-peptide, but Sec1p nevertheless binds to SNARE complexes but not to monomeric Sso1p (28, 31, 32). We have shown that the N-peptide is absolutely required for the interaction of Munc18c with monomeric Stx4 (19), and similar results were found for Sly1p/Sed5p (14) and Vps45p/Tlg2p (24). Therefore, it seems more likely that the function of the N-peptide is to enable a direct interaction between the SM protein and monomeric Stx. Munc18-1 can interact with a closed conformation of Stx1, and this does not require the Stx1 N-peptide (10), but recent work suggests that a second Munc18-1/Stx1 binding mode occurs, possibly involving an open conformation of the Stx, that does require the N-peptide (35). It is noteworthy that the stimulation of SNARE-mediated membrane fusion by Munc18-1 occurs only when the SM protein is preincubated with t-SNARE and v-SNARE vesicles for several hours at 4°C (21). This was interpreted to mean that preincubation exposes a transient SNARE complex, but we favor the view that preincubation allows a transient SM/Stx complex to form, involving the N-peptide interaction. In vivo this transient N-peptide interaction is likely to be precisely regulated by extrinsic factors such as phosphorylation or lipids.
The Munc18c/Stx41–19 and Sly1p/Sed5p1–21 (20) structures indicate that the N-peptide interactions are insufficient to explain the high degree of specificity between the SM and Stx partners. Furthermore, domains 1 of Munc18-1, Munc18-2, or Munc18c can all interact with Stx1a (36), supporting the notion that specificity requires binding interactions beyond those involved in N-peptide binding. Indeed, there is evidence that Stx4 binding sites exist elsewhere on the surface of Munc18c. For example, a peptide corresponding to a loop not involved in N-peptide binding can block Stx4 binding to Munc18c (37). A more comprehensive binding interaction must occur to explain the specificity of the interaction. One explanation for how this might happen is that binding of the N-peptide to the SM protein enables a second more comprehensive binding mode, perhaps through induction of conformational changes in Stx or the SM protein.
It is tempting to speculate that this second interaction might involve a closed to open transition of those Stxs that adopt a closed conformation, or binding of an open conformation in those that do not. This would explain the specificity of the SM/Stx interaction, the ability of some SM proteins to bind a closed fusion-incompetent Stx, and the ability of SM proteins to stimulate SNARE complex formation and membrane fusion. Binding of the closed conformation of Stxs might also imply a checkpoint function for SM proteins that is consistent with data showing that in some cases overexpression of SM proteins inhibits SNARE assembly and in vitro data showing that under certain conditions SM proteins are inhibitory (6, 7). It also agrees with our findings in adipocytes that all of the Munc18c is bound to monomeric Stx4 with no detectable interaction with SNARE complexes (D.E.J., unpublished data). Conversely, Sec1p binds only to SNARE complexes in yeast and not to Sso1p alone (28, 31, 32). This would indicate that yeast and mammalian cell surface SM proteins have evolved to regulate the assembly point at different stages of the membrane fusion process, early in the case of Munc18c and late in the case of Sec1p. The added layer of regulation imparted by Munc18c, mediated by the N-peptide, may represent a key regulatory node in the vesicle transport process as exemplified by the presence of an insulin-regulated tyrosine phosphorylation site at the N-peptide location.
In summary, our structural data have provided the basis for the prediction of which SM/Stx complexes are likely to use an N-peptide binding model and a model for the role of the N-peptide binding mode in SM regulation of SNARE-mediated membrane fusion. This prediction should now facilitate future studies to test this model.
Methods
Full-length Munc18c (mouse, residues 1–592) was expressed as an N-terminally 6×His-tagged fusion protein by using baculovirus-infected insect cells and purified as described previously (18). A peptide corresponding to residues 1–29 of Stx4 was synthesized chemically (19). Met-1 was included in the N-peptide for direct comparison with similar studies using N-peptides but may not be present in mature Stx4. The process of producing diffraction-quality crystals is reported in detail elsewhere (38). The crystals used to measure the diffraction data reported here were prepared from purified Munc18c (14 mg/ml in 25 mM Hepes, pH 7.0/150 mM NaCl/2 mM 2-mercaptoethanol) mixed with a 10-fold molar excess of the N-peptide. The crystals were grown by hanging drop vapor diffusion at 20°C using 2 μl of the Munc18c/Stx41–29 complex and 2 μl of reservoir (10–13% PEG3350/0.2 M Mg acetate/0.1 M MES, pH 6.5/50 mM MgCl2). Crystals appeared within 3 days and grew to full size within a week with typical dimensions of 0.2–0.3 mm3. Crystals were dehydrated with 25–30% PEG 3350 and cryoprotected with 15% ethylene glycol before data measurement. Data were measured at the Advanced Light Source beamline 8.3.1 at a wavelength of 1.115872 Å. The crystals belong to the cubic space group P213 with unit cell a = b = c = 170.4 Å, two molecules in the asymmetric unit, and a calculated solvent content of 54% (39). Data were processed with HKL2000 (40), and statistics are shown in Table 1.
The structure of Munc18c/Stx41–29 was determined by molecular replacement. The search model was squid Munc18-1 [PDB ID code 1EPU (20)], with nonconserved residues replaced by alanine and nonhomologous regions omitted. A molecular replacement solution was found by CNS (41) and PHASER (42, 43). The model was refined by using CNS with noncrystallographic symmetry restraints. Several rounds of torsion-angle simulated annealing and positional and group temperature factor refinements in CNS and model building in COOT (44) resulted in an Rfactor of 29.7% and an Rfree of 33.6% (12–3.15 Å). Refinement was then continued with TLS parameters in REFMAC5 (45). Unbiased 2Fo − Fc composite annealed omit maps were used during model building. The 2Fo − Fc and Fo − Fc electron density maps revealed density for the Stx4 N-peptide, although the side chains of Stx4 residues 9–14 and the main chain of residues 13–14 are poorly ordered. The structure of the complex was refined to an Rfactor of 25.2% with an Rfree of 28.0%. The final model includes Munc18c residues 8–584 with three loop regions not modeled because of disorder (272–278, 305–332, and 502–526). The peptide used in crystallization experiments corresponded to Stx4 residues 1–29, but density was present only for the first 19 residues (residues 20–29 were not modeled). Statistics for the final refined structure are given in Table 1. The quality and geometry of the model were evaluated by PROCHECK (46). Structural analysis was performed by using LSQMAN (47). Figures were made by using PyMol (48), and APBS (49) was used to generate electrostatic surfaces.
The following sequences were downloaded from the National Center for Biotechnology Information in January 2007 (accession codes are shown in parentheses) for use in the analysis of SM proteins and Stxs from yeast and mammals. Yeast (Saccharomyces cerevisiae): Sly1p (CAA86695), Sec1p (P30619), Vps45p (CAA96801), Vps33p (NP_013500), Sed5p (NP_013126), Sso1p (NP_013908), Sso2p (NP_015092), Tlg2p (NP_014624), and Vam3p (NP_014749); and mouse (Mus musculus): Munc18-1 (NP_084101), Munc18-2 (AAF71616), Munc18c (NP035634), Sly1 (NP_084101), Vps33A (NP_084205), Vps33B (NP_835171), Stx1 (O35526), Stx2 (Q00262), Stx3 (NP_689344), Stx4 (P70452), Stx5 (NP_062803), Stx7 (O70439), and Stx16 (AAH94436). The crystal structures of mouse Munc18c (this work), rat Munc18-1 (10), and yeast Sly1p (20) were superimposed by using COOT (43), and this was used to generate an initial structure-based sequence alignment for the N-pocket region of SM proteins. Other SM protein sequences were aligned by using ClustalW (www.ebi.ac.uk/clustalw). The results hold for all of the proteins used in this alignment, but we have not included discussion in the text for Vam33B because of the high sequence conservation with Vam33A. Similarly, the N-peptides of Sso2p and Stx2 are not discussed because these are similar to Sso1p and Stx3, respectively. The model of the yeast Sec1p N-pocket was derived from the structure of Munc18c by using COOT to mutate the N-pocket residues to those in Sec1p (based on the alignment in Fig. 4) and selecting side chain rotamers or torsion angles that minimized steric clashes.
Supplementary Material
Acknowledgments
We thank Karl Byriel and the University of Queensland Macromolecular X-Ray Crystallography Facility; Tom Alber, James Holton, Jane Tanamachi, and George Meigs for help on beamline 8.3.1 at the Advanced Light Source, supported by the Director, Office of Science, Office of Basic Energy Sciences, of the U.S. Department of Energy under Contract DE-AC02-05CH11231; Nathan Cowieson and Harry Tong for assistance with data measurement; the Australian Synchrotron Research Program (funded by the Commonwealth of Australia under the Major National Research Facilities Program) for supporting access to synchrotrons; Linda Lua and Michelle Christie for assistance with protein production; Paul Alewood and Chris Armishaw (University of Queensland, Brisbane) for providing the Stx4 N-peptide; and Nia Bryant, Fred Meunier, and Begoña Heras for helpful discussions. This research was supported by the National Health and Medical Research Council of Australia.
Abbreviations
- SM protein
Sec1/Munc18 protein
- SNARE
soluble NSF attachment protein receptor
- Stx
Syntaxin
- PDB
Protein Data Bank.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors for Munc18c/Stx41–29 have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 2PJX).
This article contains supporting information online at www.pnas.org/cgi/content/full/0701124104/DC1.
References
- 1.Bennett MK, Garcia-Arraras JE, Elferink LA, Peterson K, Fleming AM, Hazuka CD, Scheller RH. Cell. 1993;74:863–873. doi: 10.1016/0092-8674(93)90466-4. [DOI] [PubMed] [Google Scholar]
- 2.Brunger AT. Q Rev Biophys. 2005;38:1–47. doi: 10.1017/S0033583505004051. [DOI] [PubMed] [Google Scholar]
- 3.Sutton RB, Fasshauer D, Jahn R, Brunger AT. Nature. 1998;395:347–353. doi: 10.1038/26412. [DOI] [PubMed] [Google Scholar]
- 4.Rizo J, Sudhof TC. Nat Rev Neurosci. 2002;3:641–653. doi: 10.1038/nrn898. [DOI] [PubMed] [Google Scholar]
- 5.Jahn R, Scheller RH. Nat Rev Mol Cell Biol. 2006;7:631–643. doi: 10.1038/nrm2002. [DOI] [PubMed] [Google Scholar]
- 6.Toonen RF, Verhage M. Trends Cell Biol. 2003;13:177–186. doi: 10.1016/s0962-8924(03)00031-x. [DOI] [PubMed] [Google Scholar]
- 7.Gallwitz D, Jahn R. Trends Biochem Sci. 2003;28:113–116. doi: 10.1016/S0968-0004(03)00028-8. [DOI] [PubMed] [Google Scholar]
- 8.Yang B, Steegmaier M, Gonzalez LC, Jr, Scheller RH. J Cell Biol. 2000;148:247–252. doi: 10.1083/jcb.148.2.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dulubova I, Sugita S, Hill S, Hosaka M, Fernandez I, Sudhof TC, Rizo J. EMBO J. 1999;18:4372–4382. doi: 10.1093/emboj/18.16.4372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Misura KM, Scheller RH, Weis WI. Nature. 2000;404:355–362. doi: 10.1038/35006120. [DOI] [PubMed] [Google Scholar]
- 11.Voets T, Toonen RF, Brian EC, de Wit H, Moser T, Rettig J, Sudhof TC, Neher E, Verhage M. Neuron. 2001;31:581–591. doi: 10.1016/s0896-6273(01)00391-9. [DOI] [PubMed] [Google Scholar]
- 12.Verhage M, Maia AS, Plomp JJ, Brussaard AB, Heeroma JH, Vermeer H, Toonen RF, Hammer RE, van den Berg TK, Missler M, et al. Science. 2000;287:864–869. doi: 10.1126/science.287.5454.864. [DOI] [PubMed] [Google Scholar]
- 13.Bracher A, Weissenhorn W. J Mol Biol. 2001;306:7–13. doi: 10.1006/jmbi.2000.4347. [DOI] [PubMed] [Google Scholar]
- 14.Yamaguchi T, Dulubova I, Min S-W, Chen X, Rizo J, Sudhof TC. Dev Cell. 2002;2:295–305. doi: 10.1016/s1534-5807(02)00125-9. [DOI] [PubMed] [Google Scholar]
- 15.Peng R, Gallwitz D. J Cell Biol. 2002;157:645–655. doi: 10.1083/jcb.200202006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tellam JT, Macaulay SL, McIntosh S, Hewish DR, Ward CW, James DE. J Biol Chem. 1997;272:6179–6186. doi: 10.1074/jbc.272.10.6179. [DOI] [PubMed] [Google Scholar]
- 17.Thurmond DC, Ceresa BP, Okada S, Elmendorf JS, Coker K, Pessin JE. J Biol Chem. 1998;273:33876–33883. doi: 10.1074/jbc.273.50.33876. [DOI] [PubMed] [Google Scholar]
- 18.Hu S-H, Gee CL, Latham CF, Rowlinson SW, Rova U, Jones A, Halliday JA, Bryant NJ, James DE, Martin JL. Protein Expression Purif. 2003;31:305–310. doi: 10.1016/s1046-5928(03)00197-9. [DOI] [PubMed] [Google Scholar]
- 19.Latham CF, Lopez JA, Hu S-H, Gee CL, Westbury E, Duncan Blair D, Armishaw C, Alewood PF, Bryant NJ, James DE, Martin JL. Traffic. 2006;7:1408–1419. doi: 10.1111/j.1600-0854.2006.00474.x. [DOI] [PubMed] [Google Scholar]
- 20.Bracher A, Weissenhorn W. EMBO J. 2002;21:6114–6124. doi: 10.1093/emboj/cdf608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shen J, Tareste DC, Paumet F, Rothman JE, Melia TJ. Cell. 2007;128:183–195. doi: 10.1016/j.cell.2006.12.016. [DOI] [PubMed] [Google Scholar]
- 22.Dulubova I, Khvotchev M, Liu S, Huryeva I, Sudhof TC, Rizo J. Proc Natl Acad Sci USA. 2007;104:2697–2702. doi: 10.1073/pnas.0611318104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bracher A, Perrakis A, Dresbach T, Betz H, Weissenhorn W. Structure (London) 2000;8:685–694. doi: 10.1016/s0969-2126(00)00156-8. [DOI] [PubMed] [Google Scholar]
- 24.Dulubova I, Yamaguchi T, Gao Y, Min S-W, Huryeva I, Sudhof TC, Rizo J. EMBO J. 2002;21:3620–3631. doi: 10.1093/emboj/cdf381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Oh E, Thurmond DC. J Biol Chem. 2006;281:17624–17634. doi: 10.1074/jbc.M601581200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Latham CF, Osborne SL, Cryle MJ, Meunier FA. J Neurochem. 2007;100:1543–1554. doi: 10.1111/j.1471-4159.2006.04286.x. [DOI] [PubMed] [Google Scholar]
- 27.Zilly FE, Sorensen JB, Jahn R, Lang T. PLoS Biol. 2006;4:1789–1792. doi: 10.1371/journal.pbio.0040330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Togneri J, Cheng YS, Munson M, Hughson FM, Carr CM. Proc Natl Acad Sci USA. 2006;103:17730–17735. doi: 10.1073/pnas.0605448103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Araç D, Dulubova I, Pei J, Huryeva I, Grishin NV, Rizo J. J Mol Biol. 2005;346:589–601. doi: 10.1016/j.jmb.2004.12.004. [DOI] [PubMed] [Google Scholar]
- 30.Carpp LN, Ciufo LF, Shanks SG, Boyd A, Bryant NJ. J Cell Biol. 2006;173:927–936. doi: 10.1083/jcb.200512024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Carr CM, Grote E, Munson M, Hughson FM, Novick PJ. J Cell Biol. 1999;146:333–344. doi: 10.1083/jcb.146.2.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Scott BL, Van Komen JS, Irshad H, Liu S, Wilson KA, McNew JA. J Cell Biol. 2004;167:75–85. doi: 10.1083/jcb.200405018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sato TK, Rehling P, Peterson MR, Emr SD. Mol Cell. 2000;6:661–671. doi: 10.1016/s1097-2765(00)00064-2. [DOI] [PubMed] [Google Scholar]
- 34.Seals DF, Eitzen G, Margolis N, Wickner WT, Price A. Proc Natl Acad Sci USA. 2000;97:9402–9407. doi: 10.1073/pnas.97.17.9402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rickman C, Medine CN, Bergmann A, Duncan RR. J Biol Chem. 2007;282:12097–12103. doi: 10.1074/jbc.M700227200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dulubova I, Yamaguchi T, Arac D, Li H, Huryeva I, Min SW, Rizo J, Sudhof TC. Proc Natl Acad Sci USA. 2003;100:32–37. doi: 10.1073/pnas.232701299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thurmond DC, Kanzaki M, Khan AH, Pessin JE. Mol Cell Biol. 2000;20:379–188. doi: 10.1128/mcb.20.1.379-388.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Latham CF, Hu S-H, Gee CL, Armishaw CJ, Alewood PF, James DE, Martin JL. Acta Crystallogr F. 2007 doi: 10.1107/S1744309107022361. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Matthews BW. J Mol Biol. 1968;33:491–497. doi: 10.1016/0022-2836(68)90205-2. [DOI] [PubMed] [Google Scholar]
- 40.Otwinowski Z, Minor W. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 41.Brünger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang J-S, Kuszewski J, Nilges M, Pannu NS, et al. Acta Crystallogr D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 42.Storoni LC, McCoy AJ, Read RJ. Acta Crystallogr D. 2004;60:432–438. doi: 10.1107/S0907444903028956. [DOI] [PubMed] [Google Scholar]
- 43.Read RJ. Acta Crystallogr D. 2001;57:1373–1382. doi: 10.1107/s0907444901012471. [DOI] [PubMed] [Google Scholar]
- 44.Emsley P, Cowtan K. Acta Crystallogr D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 45.Murshudov GN, Vagin AA, Dodson EJ. Acta Crystallogr D. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 46.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. J Appl Crystallogr. 1993;26:283–291. [Google Scholar]
- 47.Kleywegt GT. Acta Crystallogr D. 1996;52:842–857. doi: 10.1107/S0907444995016477. [DOI] [PubMed] [Google Scholar]
- 48.DeLano WL. The Pymol User's Manual. San Carlos, CA: DeLano Scientific; 2002. [Google Scholar]
- 49.Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Proc Natl Acad Sci. 2001;98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dulubova I, Yamaguchi T, Wang Y, Sudhof TC, Rizo J. Nat Struct Biol. 2001;8:258–264. doi: 10.1038/85012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.