Abstract
Methylation of certain lysine residues in the N-terminal tails of core histone proteins in nucleosome is of fundamental importance in the regulation of chromatin structure and gene expression. Such histone modification is catalyzed by protein lysine methyltransferases (PKMTs). PKMTs contain a conserved SET domain in almost all of the cases and may transfer one to three methyl groups from S-adenosyl-l-methionine (AdoMet) to the ε-amino group of the target lysine residue. Here, quantum mechanical/molecular mechanical molecular dynamics and free-energy simulations are performed on human PKMT SET7/9 and its mutants to understand two outstanding questions for the reaction catalyzed by PKMTs: the mechanism for deprotonation of positively charged methyl lysine (lysine) and origin of product specificity. The results of the simulations suggest that Tyr-335 (an absolute conserved residue in PKMTs) may play the role as the general base for the deprotonation after dissociation of AdoHcy (S-adenosyl-l-homocysteine) and before binding of AdoMet. It is shown that conformational changes could bring Y335 to the target methyl lysine (lysine) for proton abstraction. This mechanism provides an explanation why methyl transfers could be catalyzed by PKMTs processively. The free-energy profiles for methyl transfers are reported and analyzed for wild type and certain mutants (Y305F and Y335F) and the active-site interactions that are of importance for the enzyme's function are discussed. The results of the simulations provide important insights into the catalytic process and lead to a better understanding of experimental observations concerning the origin of product specificity for PKMTs.
Keywords: enzyme catalysis, quantum mechanical/molecular mechanical molecular dynamics simulations, potential of mean force
Histone lysine methylation is believed to be part of a histone code (1) and can govern many important biological processes, including transcription silencing and activation, DNA repair, cell cycle control, and DNA methylation (2–4). Several lysine residues on histone proteins have been identified to be the sites of methylation by PKMTs, including histone H3 Lys-4 (or simply H3-K4), H3-K9, H3-K27, H3-K36, H3-K79, and H4-K20 (for reviews, see refs. 1 and 3). Different SET domain protein lysine methyltransferases (PKMTs) may have different product specificities in the sense that they direct different degrees of methylation on the lysine residues (e.g., mono-, di-, or trimethylation), leading to different outcomes and signaling properties (1). The crystal structures of several PKMT complexes have been determined in recent years (5–20). These structures demonstrated that the SET domain is flanked by distinctive domains at the N and C termini. The substrate and S-adenosyl-l-methionine (AdoMet) are located on the opposite surfaces, with the target lysine side chain accessing the active site and the cofactor through a narrow channel in the SET domain.
Although PKMTs have been the subject of extensive experimental investigations, there are still considerable uncertainties concerning detailed catalytic process and origins of substrate and product specificities. One outstanding question that is of crucial importance for understanding the action of PKMTs concerns how positively charged lysine is deprotonated before its methylation. This deprotonation is absolutely required, because the basic form of the target lysine is the nucleophile, and the lone pair of electrons on its Nζ is the site where the methyl group is to be added (6, 13). Moreover, for the PKMTs that produce di- and trimethylated lysine products the multiple rounds of methylation are believed to proceed processively without the release of the intermediates from the active sites. Thus, after each round of methylation, the lysine side chain becomes positively charged (see Fig. 1A) and needs to be deprotonated again for the next round of methylation. So far, no functional groups/residues have been clearly identified to play this crucial role of the general base (6, 13). The mechanisms by which the enzymes (or bulk solvent) are able to perform the initial and multiple rounds of deprotonation to prepare for methylation are still unknown and have been the subject of considerable speculations (6, 10–14, 18). For instance, it was thought previously that an absolutely conserved Tyr residue in the SET domain (e.g., Tyr-335 in SET7/9, Tyr-283 in DIM-5, Tyr-287 in Rubisco LSMT, Tyr-336 in SET8 or Pr-SET7) might deprotonate the target lysine as well as methyl lysine (10, 12, 18). However, this possibility was later ruled out (14) based on the x-ray structures of the PKMT complexes (8–16), which showed that this tyrosine residue donates a conserved hydrogen bond to a backbone carboxyl oxygen (e.g., from Ala-295 in SET7/9 and Ile-240 in Dim-5) and is too far away from the target lysine or methyl lysine to perform the deprotonation function. Other active-site residues (e.g., Tyr-245 in SET7/9 and Tyr-178 in Dim-5) were also proposed as the possible general base (11,12). One major problem for the proposals based solely on the active-site residues is the lack of the ability to explain how the multiple rounds of deprotonation could occur (see above). In other words, it would be difficult to imagine that a general base would be able to deprotonate the methyl lysine and lysine repeatedly for multiple methyl additions. A recent hypothesis (15) seems to be able to overcome this problem by suggesting that bulk solvent might play a role in the direct deprotonation of lysine or methyl lysine. Although this mechanism is of interest, questions remain as to why PKMTs choose to use bulk solvent to perform this crucial step of the reaction and what is the role of the key residues in this process.
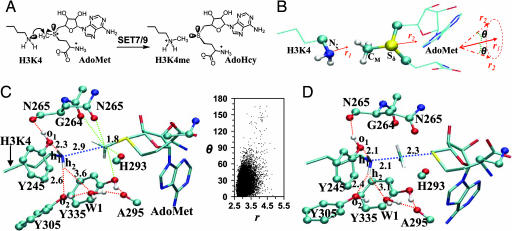
Fig. 1.
The methyl transfer reaction catalyzed by SET7/9 and active site structures obtained from the simulations. (A) A methyl group is transferred from AdoMet to H3-K4, leading to the formation of a monomethylated lysine product (H3-K4me). (B) The relative orientation of AdoMet and H3-K4 in the reactant complex. The efficiency of the methyl transfer depends on the values of r(CM… Nζ) and θ in the reactant complexes. θ is defined as the angle between the two vectors r1 and r2. Here, r1 is the direction of the lone pair of electrons on Nζ, and r2 is the vector pointing from CM to Sδ. (C) An average active-site structure obtained from the QM/MM MD simulations of SET7/9 complexed with AdoMet and a histone H3-K4 peptide along with two-dimensional plot of r(CM… Nζ) and θ distributions from the 1-ns MD simulations. SET7/9 is shown in balls and sticks, and AdoMet and H3-K4 are in sticks. The distances are in angstroms. The free-energy changes as functions of r(CM… Nζ) and θ obtained based on the distributions are given in SI Fig. 5A. (D) The average active-site structure in the area near the transition state of monomethylation in wild type obtained based on the free-energy simulations (see below).
Here, the results of quantum mechanical/molecular mechanical (QM/MM) molecular dynamics (MD) and free-energy simulations are reported for the first and second methyl transfers in wild type and different mutants (i.e., in the generation of mono- and dimethylated lysine products from the reactant complexes containing the target lysine and monomethylated lysine, respectively) for understanding the deprotonation process during histone methylation catalyzed by SET7/9 and the origin of product specificity.
Results
Product Specificity.
The average active-site structures of the reactant complex and near transition state for monomethylation (Fig. 1 A and B) in wild type are plotted in Fig. 1 C and D, respectively. The average structure for the product complex (data not shown) is very close to the x-ray structure after 1-ns simulations with an average deviation of only 0.1 Å for key active-site interactions, suggesting that the models and methods used in this study are meaningful. Fig. 1C shows that the methyl group of AdoMet is well aligned with the lone pair of electrons on Nζ in the reactant complex, consistent with the earlier studies (9, 21). Thus, the enzyme is able to stabilize the active conformations for monomethylation. Two of the interactions that contribute to the formation of the reactive structure are the hydrogen bonds between o1 of Tyr-245 and h1 of the ε-amino group and between o2 of Tyr-305 and h2. These interactions help to fix the direction of the lone pair of electrons toward the methyl group of AdoMet. It is of interest to note that these two hydrogen bonds along with the one involving W1 become stronger as the system changes from the reactant complex to transition state (Fig. 1D). This strengthening of the interactions may play a role in the transition state stabilization for the methyl transfer. The importance of the hydrogen bond involving Tyr-245 was demonstrated by mutagenesis studies (8, 9, 12), which showed that replacement of Tyr-245 by Ala or Phe led to a significant reduction of the activity on a H3-K4 peptide. In addition, there are several C H… O hydrogen bonds that might play a role in the methyl transfer (13, 17). The ability of SET7/9 to stabilize the reactive conformations for the unmodified lysine in the substrate and AdoMet is further demonstrated by the r(CM… Nζ) and θ distributions plotted in Fig. 1C as well as the corresponding free-energy changes [see supporting information (SI) Fig. 5A]. Indeed, there is a large population for the structures with relatively short r(CM… Nζ) distances (≈2.9 Å) and small θ angles (≈0°). The formation of such active conformations has been suggested to play an important role for the catalysis (5–21).
H… O hydrogen bonds that might play a role in the methyl transfer (13, 17). The ability of SET7/9 to stabilize the reactive conformations for the unmodified lysine in the substrate and AdoMet is further demonstrated by the r(CM… Nζ) and θ distributions plotted in Fig. 1C as well as the corresponding free-energy changes [see supporting information (SI) Fig. 5A]. Indeed, there is a large population for the structures with relatively short r(CM… Nζ) distances (≈2.9 Å) and small θ angles (≈0°). The formation of such active conformations has been suggested to play an important role for the catalysis (5–21).
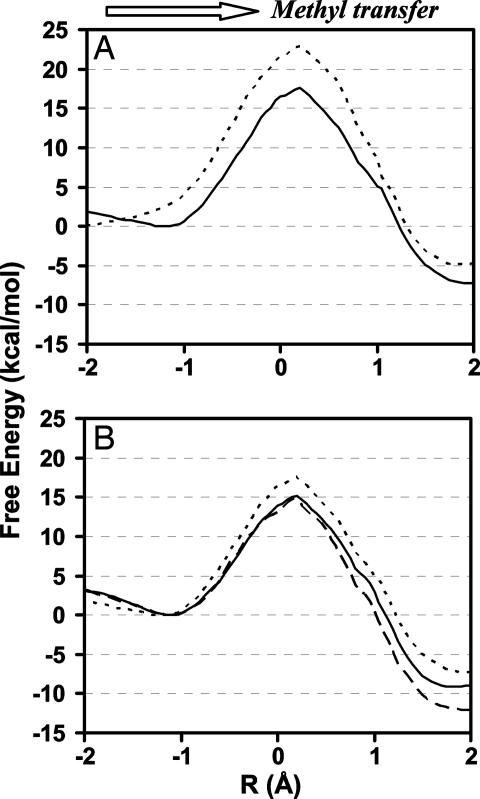
The free-energy profiles for the first and second methyl transfers as a function of the reaction coordinate are plotted in Fig. 2A for wild type and Fig. 2B for Y305F and Y335F, respectively. As can be seen from Fig. 2A, the free-energy barrier for monomethylation is ≈17–18 kcal/mol for wild type, and this barrier becomes 5 kcal/mol higher for the second methyl transfer to H3-K4me. Thus, for the wild-type enzyme, the methyl transfer from AdoMet to H3-K4me is considerably more difficult than monomethylation of lysine. Presumably, the product of monomethylation would already be released before the second methyl group could be added to H3-K4me. The results of the simulations agree with the findings (6, 8, 9, 11, 21) that SET7/9 is mainly a monomethyltransferase. The free-energy profiles for Y305F in Fig. 2B demonstrate that, unlike wild type, the free-energy barriers for both methyl transfers from AdoMet to H3-K4 and from AdoMet to H3-K4me are rather low. The results of the free-energy simulations are therefore consistent with the experimental observations that the Y305→F mutation resulted in an increase in activity of SET7/9 on histones and that Y305F is a dimethyltransferase (11). Interestingly, the free-energy barrier for the methyl transfer from AdoMet to the target lysine in Y335F is very similar to that in Y305F, even though the effects of the mutations observed experimentally are different; the role of Y335 will be discussed later (see below).
Fig. 2.
The free-energy changes for the first methyl transfer from AdoMet to H3-K4 and the second methyl transfer from AdoMet to H3-K4me as a function of the reaction coordinate [R = r(CM… Sδ) − r(CM… Nζ)] in wild type and certain mutants. (A) Wild type. Solid line, the first methyl transfer; dotted line, the second methyl transfer. (B) Y305F and Y335F. Solid line, the first methyl transfer in Y305F; dotted line, the second methyl transfer in Y305F; dashed line, the first methyl transfer in Y335F.
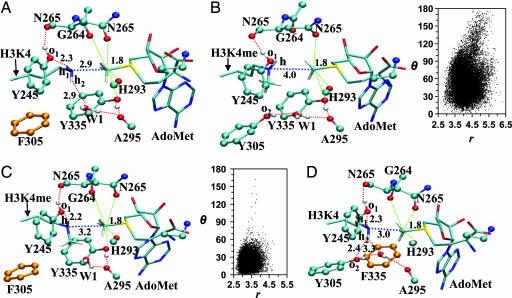
Some average active-site structures for the reactant complexes are given in Fig. 3. Fig. 3A shows that, for the monomethylation of H3-K4 in Y305F, an active-site water (W1) replaces Y305 for stabilizing the active conformation of the ε-amino group. It is interesting to note that the interactions involving the both W1 and Y245 become stronger in Y305F as the system changes from the reactant complex (Fig. 3A) to transition state (see SI Fig. 6A), and these changes are more profound than those observed for wild type (see above). The strengthening of these interactions may play a role in reducing the free-energy barrier for monomethylation. To determine the origin of the relatively low (high) free-energy barrier for the methyl transfer from AdoMet to H3-K4me in Y305F (wild type), the active-site structures of the reactant complexes for wild type and Y305F are plotted in Fig. 3 B and C, respectively, along with the r(CM… Nζ) and θ distributions. The structures in Fig. 3 B and C show that the distance between CM and Nζ is considerably longer in wild type than in Y305F for dimethylation probably because of steric repulsions that prevent K4me and AdoMet to approach each other in wild type. Fig. 3B shows that, for wild type, the r(CM… Nζ) and θ distributions in the reactant complex for the second methyl transfer are quite different from those of monomethylation (Fig. 1C). Indeed, the distributions are much broader and the possibility for the system to sample the structures with r(CM… Nζ) < 2.9 Å and θ < 15° becomes rather small, in agreement with an earlier study (21). Fig. 3C shows that, for Y305F, the active site is able to adopt the structures with a good alignment of the electron lone pair of H3-K4me and the methyl group of AdoMet, although there is only one hydrogen bond interacting with H3-K4me. This is probably due in part to the existence of the hydrophobic interaction between the methyl group of H3-K4me and the ring of Tyr-335.
Fig. 3.
The average structures of the reactant complexes of Y305F and Y335F from the simulations. (A) Y305F complexed with AdoMet and H3-K4. (B) Wild type complexed with AdoMet and H3-K4me along with the r(CM… Nζ) and θ distributions. (C) Y305F complexed with AdoMet and H3-K4me along with the r(CM… Nζ) and θ distributions. (D) Y335F complexed with AdoMet and H3-K4.
Deprotonation of Methyl Lysine: The Role of Tyr-335.
Tyr-335 is an absolute conserved residue for PKMTs, and the replacement of this Tyr was found to abolish the activity of the enzymes (8, 10, 12). For this reason, it is rather surprising to see from Fig. 2B that the free-energy barrier for the methyl transfer does not increase at all as a result of the Y335→F mutation. Consistent with the results of the free-energy simulations, Fig. 3D shows that the methyl group of AdoMet is well aligned with the lone pair of electrons on Nζ in the Y335F reactant complex. Because other experimental observations have been well reproduced by the simulations, it appears that the interactions involving this Tyr may indeed have little effect on the methyl transfer. One possible reason for this residue to be crucial for the catalysis could be due to its role in binding of AdoMet and/or peptide. However, previous experimental binding studies (8) showed that mutation of Tyr-335 does not alter the binding affinity for the cofactor and has only limited effect on the peptide binding. Alternatively, Tyr-335 could act as the general base for deprotonation of the target lysine and methyl lysine (see above).
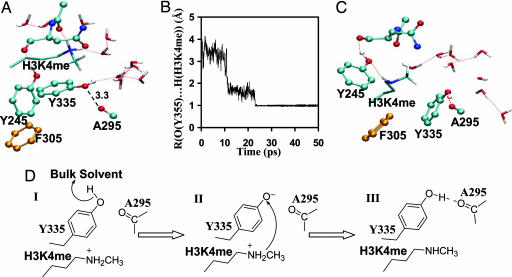
Y305F is a dimethyltransferase (as suggested by the both experimental and computational studies), and its product complex from monomethylation can therefore be used to examine the possibility of whether Tyr-335 might play a role in the deprotonation of positively charged H3-K4me. As in the x-ray structures of the PKMT complexes (8–16), the simulations showed that Tyr-335 donates a rather stable hydrogen bond to the backbone carboxyl oxygen of Ala-295 in all of the reactant and product complexes (Figs. 1C and 3 A–C). This would make it difficult to remove the proton from Tyr-335 and produce the negatively charged general base. However, the general base could be produced to perform its function after dissociation of S-adenosyl-l-homocysteine (AdoHcy) and before binding of the next AdoMet molecule. To see whether this process would be possible, AdoHcy in the Y305F product complex from monomethylation was removed to generate the state in which AdoHcy is dissociated from the enzyme and the binding of the next AdoMet molecule has not occurred yet. The MD simulations on the system showed that the conserved hydrogen bond between Tyr-335 and the backbone carboxyl oxygen was broken, and the hydroxyl group of Tyr-335 donated a hydrogen bond to bulk water molecules instead (Fig. 4A). The structural changes from the simulations seem to suggest that Tyr-335 could be deprotonated by bulk solvent and then act as the general base for deprotonation of the positively charged methyl lysine through conformational changes. To test this hypothesis, Tyr-335 was deprotonated manually and the QM/MM MD simulations were performed. Consistent with our hypothesis, the deprotonated Y335 underwent conformational transitions and performed the deprotonation function very efficiently (Fig. 4B). Fig. 4C shows the average structure after the deprotonation of positively charged methyl lysine in Y305F. As is evident from Fig. 4C, two of the key active-site hydrogen bonds between Tyr-245 and the ε-amino group and between Tyr-335 and the backbone carbonyl oxygen of Ala-295 were recovered during the simulations, providing additional support for the hypothesis. The similar results were also obtained for the deprotonation of positively charged lysine by Y335 for monomethylation before AdoMet binding in the wild-type enzyme (see SI Fig. 7).
Fig. 4.
Proton abstraction from H3-K4me by Y335 in the Y305F mutant. (A) The average structure of Y305F complexed with the positively charged H3-K4me without the cofactor. The conserved hydrogen bond between Tyr-335 and the backbone carboxyl oxygen of Ala-295 was broken, and the hydroxyl group of Tyr-335 donates a hydrogen bond to bulk water molecules instead. (B) The distance between the Tyr-335 oxygen and the proton of H3-K4me as a function of time during the MD simulations. This plot shows that, when Y335 is deprotonated, it undergoes conformational changes and abstracts a proton from the positively charged H3-K4me. (C) The average structure from the MD simulations after Tyr-335 abstracted the proton from H3-K4me. Two hydrogen bonds observed in the crystal structure involving Tyr-245 (with H3-K4me) and Tyr-335 (with the carbonyl oxygen of Ala-295) are recovered. (D) The mechanism of deprotonation of methyl lysine proposed from this study.
Discussion
SET7/9 can methylate multiple proteins, including histone H3 (8–9, 12), tumor suppressor p53 (19), and TBP-associated factor TAF10 (20). It has been widely accepted (6) that the action of the wild-type enzyme on histone H3 is mainly to add a single methyl group to Lys-4 (Fig. 1A). SET7/9 is therefore considered as a histone H3-K4 monomethyltransferase (6). The previous studies on SET7/9 have provided important insights into the origin of product specificity. The lone pair of electrons on Nζ of H3-K4 or H3-K4me is the site for methyl addition. Therefore, one would expect that a good alignment of the methyl group of AdoMet with the electron lone pair on Nζ in the reactant complex would lead to an efficient methyl transfer, whereas a bad alignment would make the methyl transfer less likely. This structure–reactivity relationship provides an explanation why SET7/9 is a monomethyltransferase. Indeed, the crystal structure of SET7/9 complexed with AdoHcy and a histone H3-K4me peptide showed that the K4me side chain in this product complex of monomethylation adopts a conformation that might prevent the lone pair of electrons from the further reaction with a second AdoMet molecule (9). A molecular dynamics study (21) supported this suggestion and further showed that the active-site conformation in a model of SET7/9 complexed with AdoMet and H3-K4me is indeed not favorable. It has been observed that the Tyr-305→Phe mutation in SET7/9 not only led to an increase of the enzyme's activity for monomethylation, but also changed SET7/9 into a dimethyltransferase (11). The relatively high activity of Y305F on H3-K4 and its ability to efficiently produce dimethylated lysine product suggest that the active-site structures for the methyl transfers from AdoMet to H3-K4 as well as from AdoMet to H3-K4me may become more favorable as a result of the Y305→F mutation. Nevertheless, the x-ray structures for the Y305F complexes are not available and a previous MD study (21) was unable to reproduce the experimental findings.
In this study, the free-energy simulations showed that the free-energy barrier for the second methyl transfer from AdoMet to H3-K4me is ≈5 kcal/mol higher than the barrier for monomethylation in wild type (Fig. 2A). Therefore, the addition of the second methyl group becomes considerably more difficult. The analysis of the trajectories of the QM/MM MD simulations on the reactant complexes supported the earlier suggestions that the decrease of the activity for the second methyl transfer is related to the inability of the active site to adopt a favorable conformation (6, 9, 21). For Y305F, the free-energy barriers for the both first and second methyl transfers are lower than or similar to the barrier of monomethylation in wild type (Fig. 2B), and the mutant is therefore a dimethyltransferase. The examination of the active-site structures showed that, unlike in wild type, the lone pair of electrons is well aligned with the methyl group of AdoMet in the both reactant complexes, leading to the efficient methyl transfers from AdoMet to H3-K4 as well as from AdoMet to H3-K4me.
One of the outstanding questions for understanding the action of PKMTs is how the positively charged lysine and methyl lysine are deprotonated before their methylation by the enzymes. Multiple rounds of methylation of the substrates by PKMTs presumably include the following: binding of the substrate and AdoMet, methylation of the target lysine, dissociation of AdoHcy from the enzyme, binding of the second AdoMet molecule and methylation of methyl lysine, and so on. The enzymes (or bulk solvent) must perform multiple rounds of deprotonation to prepare for each of the methylation steps, and the mechanism for deprotonation of positively charged lysine and methyl lysine is still unknown. The experimental observations (11, 16) that certain mutants of monomethyltransferases (e.g., Y305F of SET7/9) could efficiently produce the dimethylated lysine products indicate that, even for monomethyltransferases, the ability of the enzymes and/or bulk solvent to perform multiple rounds of deprotonation may already be in place, even though the further methylation cannot proceed.
Y335 is one of the most important residues in SET7/9 and is absolutely conserved in the SET domain PKMTs. One surprising observation from our simulations is that the free-energy barrier for the methyl transfer in Y335F is not affected at all by the mutation, indicating that its role may not be for the promotion of the methyl transfer. The x-ray structures of the PKMT product complexes (8–16) showed that this residue donates a conserved hydrogen bond to the backbone carboxyl oxygen of Ala-295 and is too far away from the target lysine or methyl lysine to perform the deprotonation function. However, the deprotonation may occur after dissociation of AdoHcy from the enzyme and before binding of the next AdoMet. To explore this possibility, the QM/MM MD simulations were performed on the Y305F product complex from monomethylation with AdoHcy removed. It was found that the hydroxyl group of Tyr-335 moved to donate a hydrogen bond to bulk water molecules. This result seems to be consistent with an apo structure containing the core SET domain of SET7/9 and the N-flanking region (18) that showed that Tyr-335 was at a different location without AdoHcy bound. However, the lack of the complete C-flanking domain (which is required for the activity) in the apo structure leads to some uncertainty as to whether the different location of Tyr-335 would be due to the lack of the C-flanking domain or AdoHcy.
The exposure of Tyr-335 to bulk solvent as observed from the simulations provides the opportunity for Y335 to be deprotonated by bulk solvent and then act as the general base in the deprotonation of positively charged methyl lysine. Indeed, our simulations showed that the deprotonated Y335 underwent conformational transitions and was able to remove the proton from the positively charged H3-K4me very efficiently. The results therefore suggest that Y335 may be the general base for the deprotonation after dissociation of AdoHcy and before binding of AdoMet (Fig. 4D). The similar results were obtained for the wild-type reactant complex with positively charged H3-K4 and without AdoMet bound (see SI Fig. 7).
The SET domain PKMTs are often active at the basic condition (e.g., Dim-5 has optimum pH of ≈10). It has been hypothesized (15) that bulk solvent might play a role in the direct deprotonation of lysine or methyl lysine (typical pKa values of ≈10). Our simulations cannot rule out this possibility, because the lysine and methyl lysine seem to have some limited exposure to bulk solvent as well (see Fig. 4A). However, it seems that the mechanism proposed here from the simulations is reasonable and able to explain the role of Y335 as well (which would presumably have a similar or lower pKa than lysine and methyl lysine).
Conclusions
The results of the QM/MM MD and free-energy simulations suggest that Tyr-335 may play the role as the general base for the deprotonation of lysine and methyl lysine after dissociation of AdoHcy and before binding of AdoMet (Fig. 4D). The conformational changes could bring Y335 to the target for the proton abstraction, as suggested from the simulations. The deprotonation of lysine or methyl lysine along with binding of AdoMet then prepares the system for methyl addition. The free-energy profiles for methyl transfers were reported and analyzed for wild type and certain mutants and the active-site interactions that are of importance for the enzyme's function were discussed. The results of this study led to a better understanding of experimental observations and provided important insights into catalytic process and origin of product specificity for PKMTs.
Methods
QM/MM free-energy (potential of mean force) simulations were applied for the study of the mono- and dimethylation processes catalyzed by SET7/9 and its mutants. The CHARMM (version c32b2) program (22) was used for the simulations. The self-consistent charge density functional tight-binding method (23) was used for the QM description in the present study. The results of the self-consistent charge density functional tight-binding and B3LYP/6–31G** (MP2/6–31G**) methods in the description of methyl transfer in a small model system were first compared, and corrections were introduced and used for improving the free-energy profiles of methyl transfers in SET7/9 and its mutants. Only the free-energy profiles based on the B3LYP corrections were reported here, because the same conclusions were obtained with the MP2 correction. AdoMet, AdoHcy, Tyr-335 (involved in deprotonation), and the target lysine/methylated lysine side chains were treated by QM and the rest of the system by MM. The all-hydrogen potential function (PARAM22) (24) was used for the MM atoms. The link-atom approach (25) available in the CHARMM program was used to separate the QM and MM boundaries. A modified TIP3P water model (26, 27) was used for the solvent. The stochastic boundary molecular dynamics method (28) was used for the QM/MM MD and free-energy simulations. The reference center for partitioning the system was chosen to be the Cβ atom of the target K-4 or the methylated K-4. The resulting systems contain 5,500≈5,700 atoms, including ≈800–900 water molecules (200 crystal water molecules).
The initial coordinates were based on the crystallographic ternary complex (PDB ID code 1O9S) containing SET7/9, AdoHcy, and a histone H3-K4me peptide (9). The reactant complex containing SET7/9, AdoMet, and the H3-K4 peptide was generated from the structure of the product complex by using the free-energy simulations. For the study of the second methyl transfer from AdoMet to H3-K4me, a methyl group was manually added to AdoHcy to change it into AdoMet. This leads to the reactant complex for the second methyl transfer. The Y305F (Y335F) mutant was generated simply by changing Tyr-305 (Tyr-335) in wild type to Phe. In addition, AdoHcy in the Y305F product complex resulting from monomethylation and AdoMet in the wild-type reactant complex for monomethylation (with a proton added to H3-K4) were removed to produce the models without the cofactor bound and to generate the states before deprotonation of H3-K4me and H3-K4, respectively.
QM/MM MD simulations (1.5 ns) were carried out for each of the reactant and product complexes and the distributions of r(CM ζ) and θ were monitored. The free energies for the formation of the active reactant complexes were estimated based on the probability densities ρ[r(CM
ζ) and θ were monitored. The free energies for the formation of the active reactant complexes were estimated based on the probability densities ρ[r(CM ζ)] and ρ(θ) (29). To test the hypothesis whether the negatively charged Y335 could act as the general base to deprotonate the positively charged methyl lysine and lysine, Y335 was manually deprotonated in each of the corresponding models and MD simulations were performed. A similar approach has been used previously in the study of chorismate mutase (30).
ζ)] and ρ(θ) (29). To test the hypothesis whether the negatively charged Y335 could act as the general base to deprotonate the positively charged methyl lysine and lysine, Y335 was manually deprotonated in each of the corresponding models and MD simulations were performed. A similar approach has been used previously in the study of chorismate mutase (30).
The umbrella sampling method (31) implemented in the CHARMM program along with the weighted histogram analysis method (32) was applied to determine the change of the free energy (potential of mean force) as a function of the reaction coordinate for the methyl transfer from AdoMet to H3-K4 (H3-K4me) in each case. The reaction coordinate is defined as a linear combination of r(CM ζ) and r(CM
ζ) and r(CM δ) [i.e., R = r(CM
δ) [i.e., R = r(CM δ) − r(CM
δ) − r(CM ζ)]. Twenty-one windows were used for each methylation process. Within each window, 100-ps productive runs were performed after the equilibration. The force constants of the harmonic biasing potentials used in the potential of mean force simulations were 100–500 kcal·mol−1·Å−2. See SI Methods for a more detailed description of methods.
ζ)]. Twenty-one windows were used for each methylation process. Within each window, 100-ps productive runs were performed after the equilibration. The force constants of the harmonic biasing potentials used in the potential of mean force simulations were 100–500 kcal·mol−1·Å−2. See SI Methods for a more detailed description of methods.
Supplementary Material
Acknowledgments
We thank Prof. Martin Karplus (Harvard University, Cambridge, MA) for a gift of the CHARMM program and Qin Xu for useful discussions. This work was supported in part by the Center of Excellence for Structural Biology and University of Tennessee–Oak Ridge National Laboratory Science Alliance, University of Tennessee, and the Petroleum Research Fund.
Abbreviations
- PKMT
protein lysine methyltransferase
- AdoHcy
S-adenosyl-l-homocysteine
- AdoMet
S-adenosyl-l-methionine
- QM/MM
quantum mechanical/molecular mechanical
- MD
molecular dynamics.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0702981104/DC1.
References
- 1.Strahl BD, Allis CD. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 2.Martin C, Zhang Y. Nat Rev Mol Cell Biol. 2005;6:838–849. doi: 10.1038/nrm1761. [DOI] [PubMed] [Google Scholar]
- 3.Marmorstein R. Trends Biochem Sci. 2003;28:59–62. doi: 10.1016/S0968-0004(03)00007-0. [DOI] [PubMed] [Google Scholar]
- 4.Ruthenburg AJ, Allis CD, Wysocka J. Mol Cell. 2007;25:15–30. doi: 10.1016/j.molcel.2006.12.014. [DOI] [PubMed] [Google Scholar]
- 5.Cheng X, Collins RE, Zhang X. Annu Rev Biophys Biomol Struct. 2005;34:267–294. doi: 10.1146/annurev.biophys.34.040204.144452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xiao B, Wilson JR, Gamblin SJ. Curr Opin Struct Biol. 2003;13:699–705. doi: 10.1016/j.sbi.2003.10.003. [DOI] [PubMed] [Google Scholar]
- 7.Dillon SC, Zhang X, Trievel RC, Cheng X. Genome Biol. 2005;6:227. doi: 10.1186/gb-2005-6-8-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wilson JR, Jing C, Walker PA, Martin SR, Howell SA, Blackburn GM, Gamblin SJ, Xiao B. Cell. 2002;111:105–115. doi: 10.1016/s0092-8674(02)00964-9. [DOI] [PubMed] [Google Scholar]
- 9.Xiao B, Jing C, Wilson JR, Walker PA, Vasisht N, Kelly G, Howell S, Taylor IA, Blackburn GM, Gamblin SJ. Nature. 2003;421:652–656. doi: 10.1038/nature01378. [DOI] [PubMed] [Google Scholar]
- 10.Zhang X, Tamaru H, Khan SI, Horton JR, Keefe LJ, Selker EU, Cheng X. Cell. 2002;111:117–127. doi: 10.1016/s0092-8674(02)00999-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang X, Yang Z, Khan SI, Horton JR, Tamaru H, Selker EU, Cheng X. Mol Cell. 2003;12:177–185. doi: 10.1016/s1097-2765(03)00224-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kwon T, Chang JH, Kwak E, Lee CW, Joachimiak A, Kim YC, Lee JW, Cho Y. EMBO J. 2003;22:292–303. doi: 10.1093/emboj/cdg025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Trievel RC, Beach BM, Dirk LMA, Houtz RL, Hurley JH. Cell. 2002;111:91–103. doi: 10.1016/s0092-8674(02)01000-0. [DOI] [PubMed] [Google Scholar]
- 14.Trievel RC, Flynn EM, Houtz RL, Hurley JH. Nat Struct Biol. 2003;10:545–552. doi: 10.1038/nsb946. [DOI] [PubMed] [Google Scholar]
- 15.Xiao B, Jing C, Kelly G, Walker PA, Muskett FW, Frenkiel TA, Martin SR, Sarma K, Reinberg D, Gamblin SJ, et al. Genes Dev. 2005;19:1444–1454. doi: 10.1101/gad.1315905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Couture J-F, Collazo E, Brunzelle JS, Trievel RC. Genes Dev. 2005;19:1455–1465. doi: 10.1101/gad.1318405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Couture J-F, Hauk G, Thompson MJ, Blackburn GM, Trievel RC. J Biol Chem. 2006;281:19280–19287. doi: 10.1074/jbc.M602257200. [DOI] [PubMed] [Google Scholar]
- 18.Jacobs SA, Harp JM, Devarakonda S, Kim Y, Rastinejad F, Khorasanizadeh S. Nat Struct Biol. 2002;9:833–838. doi: 10.1038/nsb861. [DOI] [PubMed] [Google Scholar]
- 19.Chuikov S, Kurash JK, Wilson JR, Xiao B, Justin N, Ivanov GS, McKinney K, Tempst P, Prives C, Gamblin SJ, et al. Nature. 2004;432:353–360. doi: 10.1038/nature03117. [DOI] [PubMed] [Google Scholar]
- 20.Couture J-F, Collazo E, Hauk G, Trievel RC. Nat Struct Mol Biol. 2006;13:140–146. doi: 10.1038/nsmb1045. [DOI] [PubMed] [Google Scholar]
- 21.Hu P, Zhang Y. J Am Chem Soc. 2006;128:1272–1278. doi: 10.1021/ja056153+. [DOI] [PubMed] [Google Scholar]
- 22.Brooks BR, Bruccoleri RE, Olafson BD, States DJ, Swaminathan S, Karplus M. J Comput Chem. 1983;4:187–217. [Google Scholar]
- 23.Cui Q, Elstner M, Kaxiras E, Frauenheim T, Karplus M. J Phys Chem B. 2001;105:569–585. [Google Scholar]
- 24.Mackerell AD, Jr, Bashford D, Bellott M, Dunbrack RL, Jr, Evanseck JD, Field MJ, Fischer S, Gao J, Guo H, Ha S, et al. J Phys Chem B. 1998;102:3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 25.Field MJ, Bash PA, Karplus M. J Comput Chem. 1990;11:700–733. [Google Scholar]
- 26.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. J Chem Phys. 1983;79:926–935. [Google Scholar]
- 27.Neria E, Fisher S, Karplus M. J Chem Phys. 1996;105:1902–1921. [Google Scholar]
- 28.Brooks CL, III, Brunger A, Karplus M. Biopolymers. 1985;24:843–865. doi: 10.1002/bip.360240509. [DOI] [PubMed] [Google Scholar]
- 29.Brooks CL, III, Karplus M, Pettitt BM. Proteins: A Theoretical Perspective of Dynamics, Structure, and Thermodynamics. New York: Wiley; 1988. [Google Scholar]
- 30.Ma JP, Zheng XF, Schnappauf G, Braus G, Karplus M, Lipscomb WN. Proc Natl Acad Sci USA. 1998;95:14640–14645. doi: 10.1073/pnas.95.25.14640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Torrie GM, Valleau JP. Chem Phys Lett. 1974;28:578–581. [Google Scholar]
- 32.Kumar S, Bouzida D, Swendsen RH, Kollman PA, Rosenberg JM. J Comput Chem. 1992;13:1011–1021. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.