Abstract
NAD(P) has long been known as an essential energy-carrying molecule in cells. Recent data, however, indicate that NAD(P) also plays critical signaling roles in regulating cellular functions. The crystal structure of a human protein, HSCARG, with functions previously unknown, has been determined to 2.4-Å resolution. The structure reveals that HSCARG can form an asymmetrical dimer with one subunit occupied by one NADP molecule and the other empty. Restructuring of its NAD(P)-binding Rossmann fold upon NADP binding changes an extended loop to an α-helix to restore the integrity of the Rossmann fold. The previously unobserved restructuring suggests that HSCARG may assume a resting state when the level of NADP(H) is normal within the cell. When the NADP(H) level passes a threshold, an extensive restructuring of HSCARG would result in the activation of its regulatory functions. Immunofluorescent imaging shows that HSCARG redistributes from being associated with intermediate filaments in the resting state to being dispersed in the nucleus and the cytoplasm. The structural change of HSCARG upon NADP(H) binding could be a new regulatory mechanism that responds only to a significant change of NADP(H) levels. One of the functions regulated by HSCARG may be argininosuccinate synthetase that is involved in NO synthesis.
Keywords: Rossmann fold, signal transduction
NAD and its phosphorylated form, NADP, is a universal cofactor in a large number of redox reactions carried out by a variety of enzymes, especially dehydrogenases. It is widely recognized that NAD(P) is an essential molecule in energy metabolism in all organisms. More recently, mounting evidence suggests that, in addition to its central role in energy metabolism, NAD(P) is also involved in signaling pathways that regulate fundamental processes of cellular functions, including gene transcription and apoptosis (1, 2). This realization generated a link between the energy metabolism and the regulated networks of biological processes.
One of the NAD signaling mechanisms is through gene silencer proteins known as sirtuins, such as Sir2 in yeast, a NAD-dependent histone deacetylase. Increased activities of Sir2 induced by an increase in NAD production extended lifespan (3). It was shown that a product of NAD-dependent deacetylation by Sir2, nicotinamide, strongly inhibits the activity of Sir2 (4, 5). Similar mechanisms are present in other eukaryotes, including humans (6). NAD is also directly related to transcription regulation. PolyADP-ribose polymerases (PARPs) can modify the acceptor proteins by synthesizing a polyADP-ribose molecule using NAD+ as the substrate. The transcription factors that can be modified by PARPs include p53, YY1, NF-κB, and TATA-binding protein (1). In other cases, transcription regulation is not carried out by any enzymatic reaction. For instance, the C-terminal-binding protein CtBP is a corepressor that has an increased affinity to its partners, such as adenovirus E1A or cellular repressor ZEB, when NADH binds to CtBP (7). Crystal structures showed that NADH binding to CtBP induced a conformational switch that stabilizes the dimerization of CtBP, which in turn promotes its binding to the repressors (8). In the case of the negative transcriptional regulator NmrA, NAD+ binding to NmrA controls the rate of nuclear entry of the GATA transcription-activating protein AreA (9, 10). NmrA has a similar structure to short-chain dehydrogenase/reductase (SDR) family proteins but no enzyme activities, because of the lack of conserved active-site residues (9).
NAD(P) exerts its functions by association with proteins. The protein fold that binds NAD(P) was discovered by Rossmann when the crystal structure of lactate dehydrogenase was determined (11). The Rossmann fold is the most common fold, based on its predicted occurrence from the genes known today (12). This motif consists of six β-strands connected by α-helices with the NAD(P) molecule bound at the top [supporting information (SI) Fig. 5]. The Rossmann fold has always been present as a rigid-body domain in all other known crystal structures until the structure of HSCARG was determined. We found that the common αE that connects β5 to β6 in the Rossmann fold is deformed as an extended loop when NADP is not bound with HSCARG. This allows the formation of an asymmetric dimer between a subunit with NADP bound and an empty subunit. The NADP(H)-free HSCARG form with the deformed Rossmann fold could stay as a resting state when the level of NADP(H) is normal in the cell and be restructured only in response to a higher intracellular level of NADP(H). Responses could also be in the reverse direction by restructuring to the deformed Rossmann fold if the cofactor is released at lower NADP(H) levels. The immunofluorescent imaging study showed that HSCARG switched from being associated with intermediate filaments to being dispersed in the nucleus and the cytoplasm when the intracellular NADPH/NADP level was changed by dehydroepiandrosterone (DHEA). Interactions of HSCARG with argininosuccinate synthetase (ASS), a possible limiting step in NO synthesis (13), were detected by coimmunoprecipitation, which suggests that HSCARG may regulate the activity of ASS in response to an increased level of NADP, thus potentially correlating NADP signaling with that of NO. The restructuring of HSCARG represents a mechanism to mount a rapid response to high levels of intracellular NADP while staying dormant under normal NADP levels.
Results
Overall Structure of Human HSCARG.
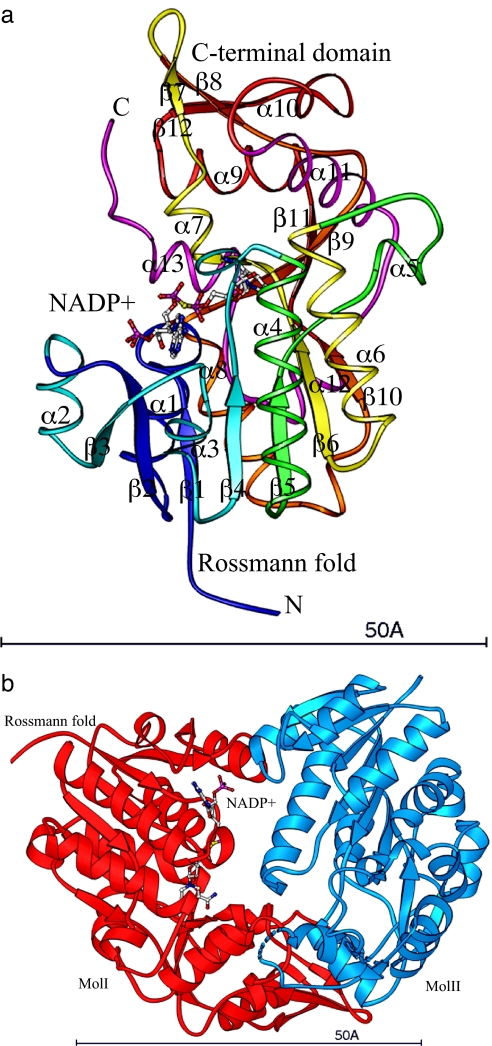
The topology of HSCARG is nearly identical to that of NmrA (Fig. 1a). The protein can essentially be divided into two domains, the N-terminal domain composed of residues 1–219 and the C-terminal domain composed of residues 220–299. The N-terminal domain contains a typical Rossmann fold with a central β sheet (strands β1–β6 and α1–α6). α5 is a short helix that proceeds the helix α6 (α6 is equivalent to αE in a conventional Rossmann fold). Residues 238–280 in the C-terminal region actually extend to the N-terminal domain, forming an additional β strand (β10) parallel to strand β6. As a result, residues 238–280 should be structurally considered as part of the N-terminal domain. The C-terminal domain is composed of two small β sheets and seven α-helices. β sheet I is formed by three short β strands, β7, β8, and β12. β7 and β8 are antiparallel to each other, whereas β12 is parallel to β8. β sheet II is also formed by three short β strands, the C-terminal half of β8, β9, and β11. All three are antiparallel to each other. Helix α7 is between strands β6 and β7; α8, between strands β9 and β10; α9, between strands β10 and β11. Helices α10 and α11 follow strand β11 and are antiparallel to each other. Helix α12 proceeds strand β12, and helix α13 is the last helix at the C terminus. The NADP+-binding site is located between the two domains. No dehydrogenase activity could be associated with HSCARG, because the catalytic tyrosine in the active site triad of Ser-Tyr-Lys is replaced by a histidine.
Fig. 1.
Crystal structure of HSCARG. (a) The tracing of HSCARG in complex with the NADP+ molecule. The protein is shown as ribbons, and the NADP+ molecule is shown as the stick model (35). The color scheme of the protein is from the N terminus (dark blue) to the C terminus (magenta). Secondary structure elements are labeled according to their positions in the protein sequence. (b) The asymmetric dimer of HSCARG with molecules I (red) binding one NADP+ molecule (stick model) and molecule II empty (blue).
NADP-Binding Site.
The asymmetric unit of the HSCARG crystal contains a dimer with a NADP+ molecule bound only in one (molecule I) of the two monomers (Fig. 1b). No additional NADP+ was added during purification or crystallization. Our data showed that NADPH was bound in HSCARG when first purified but oxidized later to NADP+. From the absorbance of freshly purified HSCARG examined (10 h after cells were lysed), bound NADPH was calculated to be 57 μM in 300 μM HSCARG by comparison with the absorbance of A340 nm = 0.0042 by 1 μM NADPH, suggesting ≈20% of the protein is bound with NADPH when expressed in Escherichia coli. The cofactor must be bound during expression of human HSCARG in E. coli and retained through purification and crystallization. Therefore, the association of the cofactor appears to be very tight, because it survived two steps of chromatographic purification without the addition of any cofactor in the buffer.
The NADP+ molecule binds to a similar region in the Rossmann fold as the NAD molecule in NmrA. However, there are nine hydrogen bonds between the nicotinamide-pyrophosphate and the protein and 11 between the AMP moiety and the protein. The details of these interactions are tabulated in Table 1. The AMP moiety is lifted up from the protein in comparison with that of NAD in NmrA.
Table 1.
Hydrogen bonds between NADP+ and HSCARG
| NADP+ | HSCARG | |
|---|---|---|
| 2′-P-AMP | Adenine–NH2 | Asp-58–Oδ1of molecule I |
| Adenine–N1 | Gln-59–N of molecule I | |
| 2′-phosphate-O2 | Arg-37–Nδof molecule I | |
| Arg-37–N of molecule I | ||
| 2′-phosphate-O3 | Thr-13–Oγof molecule I | |
| Lys-41–Nζof molecule I | ||
| 2′-phosphate-O4 | Lys-41–Nζof molecule I | |
| Gln-62–Nε2of molecule II | ||
| Arg-37–NH2ζ2 of molecule I | ||
| Ribose-OH3' | Gly-11–N of molecule I | |
| Gly-11–O of molecule I | ||
| 5′-phosphate-O2 | Lys-92–Nζ of molecule II | |
| 5′-phosphate-O3 | Ala-15–N of molecule I | |
| NMP | Nicotinamide-O | Asn-158–NH2δ2 of molecule I |
| Tyr-155–N of molecule I | ||
| Ribose-OH2' | Lys-133–NH2ζof molecule I | |
| Ribose-OH3 | Lys-133–NH2ζof molecule I | |
| Tyr-81–N of molecule I | ||
| Thr-79–O of molecule I | ||
| Ribose-O1' | Gln-16–Oε1 of molecule I | |
| 5′-phosphate-O2 | Tyr-155–Oηof molecule I | |
| 5′-phosphate-O3 | Gln-16–N of molecule I |
The hydrogen bonds were calculated by using Xfit.
It is unusually high for a typical SDR family protein to have a total of 20 hydrogen bond interactions between a bound NADP+ molecule and the protein. This may explain the tight association of the NADP+ molecule with one monomer in the HSCARG dimer. Moreover, the unoccupied monomer (molecule II) also interacts directly with the NADP+ molecule, which could further stabilize the NADP+ molecule in molecule I. The side chain of Lys-92 in molecule II bonds to the phosphate in the dinucleotide, and the side chain of Gln-62 bonds to the phosphate in the AMP moiety.
Restructuring upon NADP Binding.
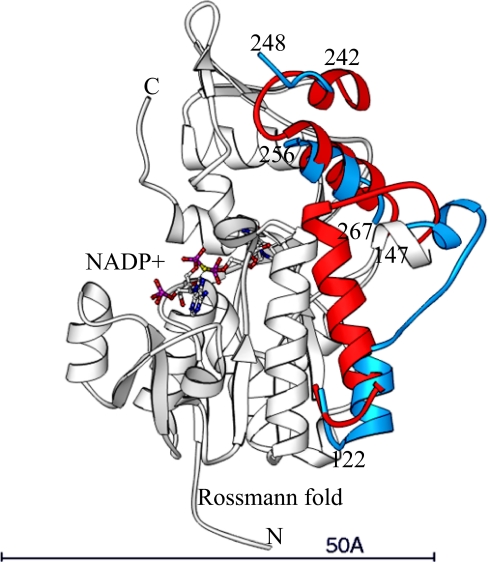
SDR family proteins usually form dimers or tetramers, which is required for their enzyme activities. Binding of the coenzyme causes only subtle changes around the binding site (14, 15). HSCARG is also found as a dimer in the crystallographic asymmetric unit. However, it is completely unexpected that the two monomers form an asymmetrical dimer, with one monomer (molecule I) bound with a NADP+ molecule and the other (molecule II) unoccupied, and the two protein molecules have dramatically different conformation (Fig. 2). The most visible changes are from residue 123 to residue 133. In the NADP+ free form (molecule II), residues 123–126 are part of a short α-helix (α5), followed by residues 127–133 that form an extended stretch. Residues 134–143 form an α-helix that is similar as the α5 connecting β5 and β6 in a conventional Rossmann fold but only half of the normal size near strand β6. Upon NADP+ binding, residue 123 makes a 180° rotation around its original α-helix. Residues 127–133 now change to α-helical to complete the α-helix (αE) continuous to residue 143. Residues 123–126 are in a new loop connecting the two helices. This structural change corresponds to three residues coming out of an α-helix and seven residues moving into a new α-helix. This large structural change may be caused by the reorientation of Tyr-81 that has to be moved out of the way to allow NADP+ to bind to the Rossmann fold. As a result, residue 129 is permitted to get closer to the bound NADP+ molecule and the long helix is allowed to be completed. Helix α4 also moves closer to the NADP+ binding site. In addition, helix α10 and α11 in the C-terminal domain are translated away from the NADP+-binding site by one turn of an α-helix. Residues 249–252 are disordered in the NADP+ free form and become an ordered loop in the NADP+-bound form. Another region, residues 174–182, also becomes ordered only in the NADP+-bound form. All of the structural changes caused by NADP+ binding occur in one continuous area near the NADP+-binding site (Fig. 2).
Fig. 2.
Structural changes of HSCARG upon NADP+ binding. Restructuring of HSCARG NADP+ upon binding. The blue regions represent the conformation of HSCARG before NADP+ binding and the red after NADP+ binding. The full structure of HSCARG and the bound NADP+ molecule is shown as white ribbons and the stick model, respectively. The residues that define the restructured regions are labeled by their residue numbers.
This unusual restructuring suggests that after a HSCARG protein molecule binds NADP(H), a second HSCARG protein molecule that is free of cofactor can associate with the occupied HSCARG to form this asymmetrical dimer. The presence of the dimeric form of HSCARG in solution was confirmed by dynamic light-scattering studies (SI Text and SI Table 3). Binding of the second protein molecule further stabilizes the NADP molecule bound in the first protein molecule and prevents the second protein molecule from binding a cofactor, as shown by the fact that addition of NADP or NADPH did not increase the presence of monomers in the dynamic light scattering studies (SI Text). The asymmetrical dimer presents unique surface characteristics that are different from a completely cofactor free protein or a fully cofactor occupied protein (Fig. 2). HSCARG may therefore exist in possibly three different forms in the cell: a cofactor free form, an asymmetrical dimer, and a cofactor occupied form (SI Fig. 6). Based on this observation, we hypothesized that when the concentration of NADP(H) increases, HSCARG becomes occupied by NADP(H) and assumes a new conformation. The cofactor occupied HSCARG can form an asymmetrical dimer with another cofactor-free HSCARG molecule. If the concentration of NADP(H) decreases, on the other hand, HSCARG can be changed to having the deformed Rossmann fold by releasing the cofactor. The restructuring of HSCARG upon binding to the cofactor prompted us to study whether HSCARG will change its subcellular distribution when the concentration of NADP changes in the cytoplasm, because the homologous protein NmrA appears to change the nucleus nuclear entry of AreA when bound with NAD+ (9, 10).
Subcellular Localization of HSCARG.
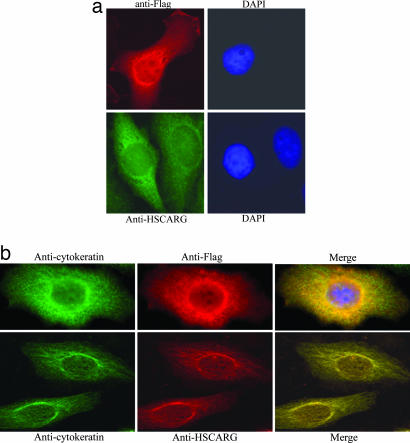
To investigate its subcellular localization, human HSCARG was overexpressed as a fusion protein with an N-terminal Flag tag by transfection of HeLa cells. The intracellular distribution of HSCARG was visualized by fluorescence microscopy. HSCARG is predominantly localized in the cytoplasm surrounding the nucleus (Fig. 3a). The localization of endogenously expressed HSCARG in the cytoplasm was further confirmed by an immunofluorescence assay. HeLa cells were stained with a mouse antiserum raised against human HSCARG. The nucleus was stained with a DNA dye, DAPI. There is no overlap between the fluorescent stain of HSCARG (green) and the nucleus (blue). Inspired by the similar distribution pattern of HSCARG and intermediate filaments, possible association of overexpressed Flag-tagged HSCARG with cytokeratin was first investigated in HeLa cells. Cells were transfected with Flag-HSCARG plasmid, cultured for 24 h, and then incubated with mouse anti-Flag antibody and rabbit anticytokeratin antibody simultaneously. There was a significant overlap between overexpressed HSCARG (red) and cytokeratin (green) in the cytoplasm, especially around the nucleus (Fig. 3b). Colocalization of endogenously expressed HSCARG with cytokeratin was also observed by immunofluorescence microscopy (Fig. 3b). These results indicate that HSCARG is associated with the intermediate filaments in the cytoplasm under normal growth conditions.
Fig. 3.
Subcellular localization of HSCARG in HeLa cells. (a) Overexpressed HSCARG was detected by using monoclonal anti-flag antibody (Upper). Endogenously expressed HSCARG was detected by using polyclonal anti-HSCARG generated from rat (Lower). (b) Colocalization of HSCARG with cytokeratin in HeLa cells. In Upper, HeLa cells were transfected with an expression plasmid for Flag-tagged HSCARG and double immunofluorescent stained with polyclonal anticytokeratin and anti-Flag. In Lower, untransfected HeLa cells were double-stained with polyclonal anticytokeratin and polyclonal anti-HSCARG. (Right) The overlay image of cytokeratin and HSCARG.
DHEA Treatment-Induced Redistribution of HSCARG Within Cells.
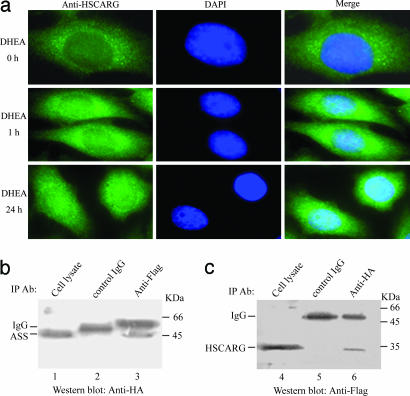
The Kd values determined for NADPH and NADP+ from the fluorescence measurements were 4.86 × 10−7 M and 1.35 × 10−5 M, respectively (SI Fig. 7). The stoichiometry n values determined for NADPH and NADP+ were 0.88 and 0.79, respectively, which is consistent with one cofactor molecule binding in one HSCARG molecule. Under normal conditions, intracellular HSCARG that is cofactor occupied is likely to be associated with NADPH because of the 360-fold higher affinity than that for NADP+. NADPH/NADP+ ratio is a critical modulator of intracellular redox states and is mainly determined by the pentose phosphate pathway through the action of glucose-6-phosphate dehydrogenase (G6PD) (16, 17). DHEA is an endogenous steroid (18, 19), which is known to be an uncompetitive inhibitor of G6PD, among other functions (20). Inhibition of G6PD could lead to a 30–40% decrease in the NADPH/NADP+ ratio (21). To detect whether the change of NADPH/NADP+ concentration will induce redistribution of HSCARG within cells, we treated HeLa cells with DHEA and examined the subcellular location of HSCARG by immunofluorescence microscopy. The results showed that in cells treated with DHEA, HSCARG was observed both in the cytoplasm and the nucleus (Fig. 4a). This suggests that changes of NADPH/NADP+ levels induce relocalization of HSCARG from being associated with intermediate filaments to being distributed in the cytoplasm and the nucleus, which may be associated with the regulatory functions of HSCARG. Immunoprecipitation experiments followed by mass spectrometry analysis were performed to identify what target proteins that HSCARG may act on. When protein samples precipitated from HeLa cell lysates with the anti-HSCARG sera were analyzed, several specific bands were found in SDS/PAGE, and mass spectrometry analysis revealed that ASS was one of the proteins that are associated with HSCARG (SI Text). Coimmunoprecipitation using lysates of human 293T cells transfected with expression plasmids for Flag-tagged HSCARG and HA-tagged ASS showed that HSCARG and ASS were able to coprecipitate (Fig. 4 b and c), suggesting that ASS, an enzyme in NO synthesis, may be a target that HSCARG regulates. HSCARG targeted proteins in the nucleus remain to be identified.
Fig. 4.
Effects of NADPH/NADP+ levels. (a) Changes in NADPH/NADP+ concentrations induced redistribution of HSCARG within cells. HeLa cells were treated with 25 μM DHEA for 1 and 24 h, respectively. Immunofluorescence analysis was performed as described in Materials and Methods. Images shown represent three independent experiments. (b and c) In vivo interaction of HSCARG with ASS. 293T cells were cotransfected with expression plasmids for Flag-tagged HSCARG and HA-tagged ASS. Cell extracts were prepared after 48 h and immunoprecipitated with an anti-Flag monoclonal antibody (lane 3) in b or an anti-HA monoclonal antibody (lane 6) in c or control IgG (lanes 2 and 5). Western blot analysis was performed with anti-HA antibody (b) or anti-Flag antibody (c) followed by an anti-mouse IgG secondary step that also recognizes the IgG heavy chain. Expression of HSCARG and ASS was confirmed by Western blot analyses of the cell lysates with anti-Flag antibody (c, lane 4) and anti-HA antibody (b, lane 1), respectively. IP, immunoprecipitation.
Discussion
The absence of the active tyrosine that is highly conserved among SDR enzymes suggests that HSCARG, like NmrA, most likely plays a regulatory role in a cellular process, rather than a metabolic pathway. However, the presence of the Rossmann fold that can tightly bind a NADP molecule indicates that HSCARG may be sensitive to the redox potential when carrying out its possible regulatory functions. This functional feature defines a group of NAD/NADP-dependent proteins that play a regulatory role by sensing the cellular level of NAD/NADP. This group of proteins includes NmrA, HSCARG, CC3, and CtBP, which seem to regulate the activities related to gene transcription factors. It may also be possible that such proteins are involved in regulation of the functions of other proteins.
The most remarkable discovery from the crystal structure of HSCARG is the restructuring of the Rossmann fold induced by association with the cofactor. An asymmetrical dimer can be formed by the association of one protein molecule that contains a NADP molecule tightly bound in its Rossmann fold with an empty subunit. The structure switch and the formation of the unique dimer can be considered as a unique response mechanism to changes of intracellular NADPH or NADP levels. In the normal cellular redox state, it is possible that HSCARG is expressed and associated with intermediate filaments. Depending on the intracellular concentration of NADPH/NADP, HSCARG may exist in one of the three possible forms; a cofactor-free form, an asymmetrical dimer, and a cofactor-occupied form. When the concentration of intracellular NADPH/NADP changes, restructuring of HSCARG will be triggered, and HSCARG is activated to act on other proteins, such as ASS. This hypothesis describes a mechanism of a regulatory role by a NADP(H)-binding protein, which senses the fine concentration of intracellular NADP(H). HSCARG swings into action only when the intracellular NADPH/NADP concentration changes. The preexisting reservoir of the HSCARG forms allows a rapid response as soon as a certain cofactor level is reached, because no new expression of HSCARG genes needs to be turned on. The identification of ASS as a potential target protein regulated by HSCARG suggests that the redox state may be correlated the NO signaling pathway, because ASS is one of the limiting enzymes in NO synthesis (13, 22). This link between the two processes may be significant, because changes of NADP+ levels and NO signaling are involved in apoptosis (23, 24).
Materials and Methods
Protein Expression and Purification.
Molecular cloning of HSCARG and purification of recombinant protein from E. coli were carried out as described (25) by using the Gateway cloning system. The Selenium-labeled protein was overexpressed in E. coli strain B834 (Novagen, Madison, WI). A two-step purification strategy by using a Ni chelating column followed by a Superdex-75 size-exclusion column was performed to obtain near homogenous protein. Approximately 3 mg of HSCARG could be achieved from 1L culture with purity >95%. The amount of NADPH in freshly purified HSCARG protein was evaluated by absorbance at 340 nm by using a spectrophotometer (Ultrospec 2000, GE Healthcare, Piscataway, NJ) and the protocol reported (26), because only NADPH has significant absorbance at 340 nm. Oxidation of bound NADPH in HSCARG was measured in cell-free solution containing 300 μM HSCARG in 20 mM Tris·HCl/150 mM NaCl (pH 7.5) buffer, by following the decrease of NADPH absorbance at different time points: 0, 3, 9, 17, and 72 h. As a control, a known amount of NADPH (50 μM) was evaluated alone in the same buffer following the same time course.
Fluorescence Measurements of Kd.
Fluorescence measurements were conducted in a spectrofluorimeter (Synergy HT, BioTek, Winooski, VT) to determine the binding constant Kd and stoichiometry n values for the binding of NADPH and NADP+ to HSCARG, following the procedure described by Venard et al. (27). The fluorescence of NADPH was measured with excitation at 340 and emission at 450 nm. The experiments were carried out in 200 mM Tris·HCl (pH 8.0) buffer by mixing 9.7 μM HSCARG (≈80% cofactor free) with various concentrations of NADPH (2–16 μM) in a final volume of 500 μl. Fluorescence of NADPH at 450 nm was enhanced when HSCARG was added, which allowed us to follow NADPH binding to HSCARG to determine its Kd.
Because the binding of NADP+ to HSCARG cannot be measured by fluorescence, we determined the K′d of NADP+ binding to HSCARG by an indirect method. When the fluorescence enhancement of NADPH by HSCARG binding was measured in the presence of 15 μM NADP+, the K′d of NADP+ could be derived from the Kd of NADPH and the n factor according to the following modified Klotz equation:
where X = fraction of molecules occupied by NADP+ for a total NADP+ concentration [LNADP], whereas Y and LNADPH refer to those corresponding to NADPH. Kd and K′d are the binding constants of NADPH and NADP+, respectively. E indicates the concentration of HSCARG monomer, whereas n is the n factor.
Crystallization and Data Collection.
The purified protein was concentrated to ≈13 mg/ml, and crystallization was performed at 277 K by using hanging-drop method of vapor diffusion. Hampton Research Crystal Screen kit I, kit II and Index kit (Hampton Research, Aliso Viejo, CA) were used for initial screening. Two microliters of protein solution mixed with 1 μl of reservoir solution was equilibrated against 400 μl of reservoir solution. Crystals appeared in many initial conditions. Large crystals were obtained from the solution contains 1.2 M sodium potassium phosphate pH 7.3 after optimizing the pH and the concentration of precipitants.
X-ray diffraction data were collected on Mar225 CCD Detector at SER-CAT 22BM synchrotron source at a wavelength of 0.97928 Å.
During data collection, the crystal was maintained at 100 K by using nitrogen gas, with 20% glycerol as the cryoprotectant. The data were collected and processed with in-house software at SER-CAT and HKL2000 (28), respectively. The crystallographic parameters and data collection statistics are given in Table 2.
Table 2.
Refinement statistics for HSCARG structure
| Space group | F23 |
| Unit cell a, b, and c, Å | a = b = c = 223.30 |
| Resolution | 50–2.4 (2.49–2.4) |
| Reflections, unique/total observed | 34,021/365,081 |
| Completeness (50–2.40 Å) | 95.3 (100)* |
| I/σ (50–2.40 Å) | 20.87 (7.19) |
| Rmerge(50–2.40 Å)† | 0.123 (0.435) |
| Rcryst‡ | 0.228 |
| Rfree§ | 0.264 |
| Model | |
| Number of atoms | 4,736 |
| Number of reflections | 33,998 |
| rmsd | |
| Bonds, Å | 0.008 |
| Angles, ° | 1.720 |
*Values in parentheses are for the last resolution shell.
†Rmerge = ΣI − 〈I〉 /Σ 〈I〉.
‡Rcryst= Σ∥Fo − Fc∥/Σ Fo.
§Rfree is the Rcryst for the selected subset (10%) of the reflections not included in prior refinement calculations.
Structure Determination and Refinement.
The structure of HSCARG was solved by Se-SAD method (Protein Data Bank ID code 2EXX). The sites of Se were determined with program of SHELXD (29) by using data from 20 Å to 3.5 Å. Refinement of heavy atom parameters and phasing work was carried out with the program SHARP (30). The electron-density map for automatic model tracing was calculated by using the program Resolve (31). Approximately 50% of proportions of the amino acids were autotraced, and the remainder of HSCARG structure was built manually by using the program O (32).
Structure refinement was conducted to 2.4 Å by using the program CNS (33), which contained simulated annealing, energy minimization, and individual B factor refinement. Reference and model modulation were made with 2Fo−Fc and Fo−Fc by using the program O. The quality of the model was checked with the program PROCHECK. The final statistics are tabulated in Table 2, and the coordinates have been deposited in the Protein Data Bank (ID code 2EXX).
Transfection and Immunofluorescence Assays.
To determine the subcellular localization of the HSCARG protein in human cells, the cDNA of HSCARG was inserted downstream of flag cDNA into the SalI-NotI sites of the pRK-Flag vector (a gift from H. Shu, Peking University, Beijing, China) by using the following primers, 5′-ACGCGTCGACTATGGTGGACAAGAAACTG and 5′-ATAAGAATGCGGCCGCTCACAGCAGGTTGAAGTC, and transfected into HeLa cells by the standard calcium phosphate precipitation (34). The cells were incubated with monoclonal anti-flag generated from mouse (Sigma, St. Louis, MO) followed by a fluorescent-labeled secondary antibody (1:100 in PBS) (goat anti-mouse IgG conjugated with FITC or TRITC), and visualized under Zeiss (Thornwood, NY) Axioskop fluorescence microscope (Axiovert 2000M). Nuclear DNA was stained by using 1 μg/ml DAPI, a fluorescent DNA-intercalating dye.
Immunofluorescence assays with the mouse antiserum raised against HSCARG were carried out following similar procedures to those described but without the step of transfection.
Colocalization of HSCARG with cytokeratin was performed in HeLa cells by using mouse anti-HSCARG and monoclonal anticytokeratin antibody, respectively. A radiance confocal microscope system (Leica, Wetzlar, Germany; Tcs-spII) was used to visualize the distribution of cytokeratin and HSCARG.
Measurements of Redistribution of HSCARG.
HeLa cells were seeded on 13-mm coverslips with 105 cells/cm2 density and incubated for 16 h at 37°C 5% CO2 with 10% calf serum in DMEM, followed by further 1- or 24-h incubation in the absence or presence of 25 μM G6PD inhibitor DHEA. Cells were washed three times with PBS, fixed with 3.7% paraformaldehyde in PBS for 20 min at room temperature, and then permeabilized for 10 min with 0.3% Triton X-100 in an ambient environment. After blocking with 3% BSA in PBS for 20 min at room temperature, the cells on coverslips were incubated with mouse polyclonal antibody against human HSCARG (dilution 1:50) overnight at 4°C followed by FITC-conjugated goat anti-mouse IgG (1:50) (Zhongshan Biotech, Beijing, China) for 1 h at 37°C in dark. Cells were counterstained with DAPI and visualized by using fluorescence microscopy.
Immunoprecipitation Experiment.
To look for the partner of HSCARG protein, the immunoprecipitation experiment was carried out as follows. HeLa cells (≈109) were lysed in 20 ml of lysis buffer (1% Nonidet P-40/1 mM EDTA/150 mM NaCl/50 mM Tris·HCl, pH 8.0/1 mg/ml leupeptin/1 mM PMSF) for 2 h on ice and were centrifugated at 15,000 × g for 30 min at 4°C. The cell lysates were precleared with protein G Sepharose (50% slurry) (GE Healthcare) for 3 h at 4°C, and the supernatants were divided into two parts and incubated with 10 μg of polyclonal antibody against HSCARG and 10 μg of rabbit IgG (Zhongshan), respectively, for 12 h at 4°C. Then 800 μl of protein G Sepharose was added to precipitate the protein complexes followed by washing three times with cell lysis buffer. The proteins bound to protein G were eluted with SDS loading buffer and examined on 10% SDS-polyacrylamide gels. The identified specific bands were analyzed in an Ultraflex MALDI TOF mass spectrometer (Bruker Daltonik, Bremen, Germany).
Coimmunoprecipitation and Western Blot Analysis.
To examine the interaction between the HSCARG and ASS, the hscarg and ass genes were inserted in the expression vectors pRK-Flag and pRK-HA, respectively. Ten micrograms of the Flag-tagged HSCARG and 10 μg of HA-tagged ASS plasmids were cotransfected in HEK-293T cells by the standard calcium phosphate precipitation. Forty-eight hours later, cells were collected and lysed in 1 ml of lysis buffer. The cell lysates were precleared with protein G Sepharose for 1 h, and the supernatant was incubated with anti-Flag or -HA monoclonal antibodies or control IgG (Sigma) overnight at 4°C. The following day, 80 μl of protein G Sepharose was added and incubated for 3 more hours. The Sepharose beads were washed three times with lysis buffer. The precipitated proteins were fractionated on 15% SDS/PAGE and transferred to nitrocellulose membrane (Hybond Extra, GE Healthcare). Subsequent Western blotting analyses were performed by using anti-Flag or -HA monoclonal antibodies, respectively, followed by a HRP-conjugated goat anti-mouse secondary antibody (Huamei, Beijing, China).
Supplementary Material
Acknowledgments
We thank Professor Yicheng Dong and Professor Haifu Fan for help with structure determination. We thank Xu Zhou and Chan Li for help with dynamic light-scattering experiments and Yiyu Li for assistance with protein purification. This work was supported by National Science Foundation of China Grant 30670416 and International Centre for Genetic Engineering and Biotechnology Project CRP/CHN05-01.
Abbreviations
- SDR
short-chain dehydrogenase/reductase
- DHEA
dehydroepiandrosterone
- ASS
argininosuccinate synthetase
- G6PD
glucose-6-phosphate dehydrogenase.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates reported in this paper have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 2EXX).
This article contains supporting information online at www.pnas.org/cgi/content/full/0700480104/DC1.
References
- 1.Berger F, Ramirez-Hernandez MH, Ziegler M. Trends Biochem Sci. 2004;29:111–118. doi: 10.1016/j.tibs.2004.01.007. [DOI] [PubMed] [Google Scholar]
- 2.Ying W. Front Biosci. 2006;11:3129–3148. doi: 10.2741/2038. [DOI] [PubMed] [Google Scholar]
- 3.Hekimi S, Guarente L. Science. 2003;299:1351–1354. doi: 10.1126/science.1082358. [DOI] [PubMed] [Google Scholar]
- 4.Anderson RM, Bitterman KJ, Wood JG, Medvedik O, Sinclair DA. Nature. 2003;423:181–185. doi: 10.1038/nature01578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lin SJ, Kaeberlein M, Andalis AA, Sturtz LA, Defossez PA, Culotta VC, Fink GR, Guarente L. Nature. 2002;418:344–348. doi: 10.1038/nature00829. [DOI] [PubMed] [Google Scholar]
- 6.Longo VD, Kennedy BK. Cell. 2006;126:257–268. doi: 10.1016/j.cell.2006.07.002. [DOI] [PubMed] [Google Scholar]
- 7.Zhang Q, Piston DW, Goodman RH. Science. 2002;295:1895–1897. doi: 10.1126/science.1069300. [DOI] [PubMed] [Google Scholar]
- 8.Nardini M, Spano S, Cericola C, Pesce A, Massaro A, Millo E, Luini A, Corda D, Bolognesi M. EMBO J. 2003;22:3122–3130. doi: 10.1093/emboj/cdg283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stammers DK, Ren J, Leslie K, Nichols CE, Lamb HK, Cocklin S, Dodds A, Hawkins AR. EMBO J. 2001;20:6619–6626. doi: 10.1093/emboj/20.23.6619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lamb HK, Ren J, Park A, Johnson C, Leslie K, Cocklin S, Thompson P, Mee C, Cooper A, Stammers DK, Hawkins AR. Protein Sci. 2004;13:3127–3138. doi: 10.1110/ps.04958904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Adams MJ, Ford GC, Koekoek R, Lentz PJ, McPherson A, Jr, Rossmann MG, Smiley IE, Schevitz RW, Wonacott AJ. Nature. 1970;227:1098–1103. doi: 10.1038/2271098a0. [DOI] [PubMed] [Google Scholar]
- 12.Pearl F, Todd A, Sillitoe I, Dibley M, Redfern O, Lewis T, Bennett C, Marsden R, Grant A, Lee D, et al. Nucleic Acids Res. 2005;33:D247–51. doi: 10.1093/nar/gki024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Husson A, Brasse-Lagnel C, Fairand A, Renouf S, Lavoinne A. Eur J Biochem. 2003;270:1887–1899. doi: 10.1046/j.1432-1033.2003.03559.x. [DOI] [PubMed] [Google Scholar]
- 14.Jornvall H, Persson B, Krook M, Atrian S, Gonzalez-Duarte R, Jeffery J, Ghosh D. Biochemistry. 1995;34:6003–6013. doi: 10.1021/bi00018a001. [DOI] [PubMed] [Google Scholar]
- 15.Oppermann U, Filling C, Hult M, Shafqat N, Wu X, Lindh M, Shafqat J, Nordling E, Kallberg Y, Persson B, Jornvall H. Chem Biol Interact 143–. 2003;144:247–53. doi: 10.1016/s0009-2797(02)00164-3. [DOI] [PubMed] [Google Scholar]
- 16.Kletzien RF, Harris PK, Foellmi LA. FASEB J. 1994;8:174–181. doi: 10.1096/fasebj.8.2.8119488. [DOI] [PubMed] [Google Scholar]
- 17.Pandolfi PP, Sonati F, Rivi R, Mason P, Grosveld F, Luzzatto L. EMBO J. 1995;14:5209–5215. doi: 10.1002/j.1460-2075.1995.tb00205.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Farquharson C, Milne J, Loveridge N. Bone Miner. 1993;22:105–115. doi: 10.1016/s0169-6009(08)80222-x. [DOI] [PubMed] [Google Scholar]
- 19.Gordon GB, Shantz LM, Talalay P. Adv Enzyme Regul. 1987;26:355–382. doi: 10.1016/0065-2571(87)90023-9. [DOI] [PubMed] [Google Scholar]
- 20.Gordon G, Mackow MC, Levy HR. Arch Biochem Biophys. 1995;318:25–29. doi: 10.1006/abbi.1995.1199. [DOI] [PubMed] [Google Scholar]
- 21.Tian WN, Braunstein LD, Pang J, Stuhlmeier KM, Xi QC, Tian X, Stanton RC. J Biol Chem. 1998;273:10609–10617. doi: 10.1074/jbc.273.17.10609. [DOI] [PubMed] [Google Scholar]
- 22.Goodwin BL, Solomonson LP, Eichler DC. J Biol Chem. 2004;279:18353–18360. doi: 10.1074/jbc.M308160200. [DOI] [PubMed] [Google Scholar]
- 23.Petit B, Leroy K, Kanavaros P, Boulland ML, Druet-Cabanac M, Haioun C, Bordessoule D, Gaulard P. Hum Pathol. 2001;32:196–204. doi: 10.1053/hupa.2001.21569. [DOI] [PubMed] [Google Scholar]
- 24.Gotoh T, Mori M. Arterioscler Thromb Vasc Biol. 2006;26:1439–1446. doi: 10.1161/01.ATV.0000223900.67024.15. [DOI] [PubMed] [Google Scholar]
- 25.Ding HT, Ren H, Chen Q, Fang G, Li LF, Li R, Wang Z, Jia XY, Liang YH, Hu MH, et al. Acta Crystallogr D. 2002;58:2102–2108. doi: 10.1107/s0907444902016359. [DOI] [PubMed] [Google Scholar]
- 26.Zhang Z, Yu J, Stanton RC. Anal Biochem. 2000;285:163–167. doi: 10.1006/abio.2000.4701. [DOI] [PubMed] [Google Scholar]
- 27.Venard R, Jallon JM, Fourcade A, Iwatsubo M. Eur J Biochem. 1975;57:371–378. doi: 10.1111/j.1432-1033.1975.tb02310.x. [DOI] [PubMed] [Google Scholar]
- 28.Otwinowski Z, Minor W. Processing of X-Ray Diffraction Data Collected in Oscillation Mode. New York: Academic; 1996. [DOI] [PubMed] [Google Scholar]
- 29.Sheldrick GM, Dauter Z, Wilson KS, Hope H, Sieker LC. Acta Crystallogr D. 1993;49:18–23. doi: 10.1107/S0907444992007364. [DOI] [PubMed] [Google Scholar]
- 30.de la Fourtelle E, Bricogne G. Methods Enzymol. 1997;276:472–494. doi: 10.1016/S0076-6879(97)76073-7. [DOI] [PubMed] [Google Scholar]
- 31.Terwilliger TC. Methods Enzymol. 2003;374:22–37. doi: 10.1016/S0076-6879(03)74002-6. [DOI] [PubMed] [Google Scholar]
- 32.Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Acta Crystallogr A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 33.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Acta Crystallogr D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 34.Sambrook J, Fritch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 35.Carson M. Methods Enzymol. 1997;277:493–505. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.