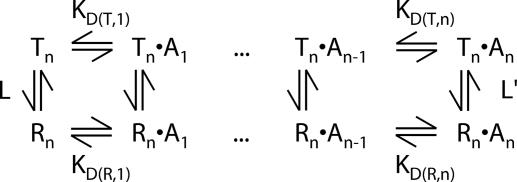Abstract
The 306-kDa aspartate transcarbamoylase is a well studied regulatory enzyme, and it has emerged as a paradigm for understanding allostery and cooperative binding processes. Although there is a consensus that the cooperative binding of active site ligands follows the Monod–Wyman–Changeux (MWC) model of allostery, there is some debate about the binding of effectors such as ATP and CTP and how they influence the allosteric equilibrium between R and T states of the enzyme. In this article, the binding of substrates, substrate analogues, and nucleotides is studied, along with their effect on the R–T equilibrium by using highly deuterated, 1H,13C-methyl-labeled protein in concert with methyl-transverse relaxation optimized spectroscopy (TROSY) NMR. Although only the T state of the enzyme can be observed in spectra of wild-type unliganded aspartate transcarbamoylase, binding of active-site substrates shift the equilibrium so that correlations from the R state become visible, allowing the equilibrium constant (L′) between ligand-saturated R and T forms of the enzyme to be measured quantitatively. The equilibrium constant between unliganded R and T forms (L) also is obtained, despite the fact that the R state is “invisible” in spectra, by means of an indirect process that makes use of relations that emerge from the fact that ligand binding and the R–T equilibrium are linked. Titrations with MgATP unequivocally establish that its binding directly perturbs the R–T equilibrium, consistent with the Monod–Wyman–Changeux model. This study emphasizes the utility of modern solution NMR spectroscopy in understanding protein function, even for systems with aggregate molecular masses in the hundreds of kilodaltons.
Keywords: allostery, Monod–Wyman–Changeux (MWC) model, NMR spectroscopy, methyl-transverse relaxation optimized spectroscopy, ligand binding
Quantitative, site-specific studies of proteins by NMR spectroscopy for the most part have been restricted to systems with molecular masses on the order of 50 kDa or less. With the development of new labeling schemes along with experiments that optimally preserve NMR signals, it now has become possible to investigate much larger complexes (1). For example, Wüthrich, Horwich, and coworkers have used 1H-15N cross-correlated relaxation-induced polarization transfer (CRIPT) spectroscopy to establish which residues of highly deuterated GroES interact with GroEL in a GroES–GroEL complex that is 900 kDa (2) and to study interactions between substrates and GroEL (3). Sprangers and coworkers have exploited methyl-transverse relaxation optimized spectroscopy (TROSY) of 1H,13C-methyl-labeled probes in the 300-kDa highly deuterated protease ClpP to quantify dynamics and relate it to function (4). In a second study, this methyl-based approach has been used to measure site-specific dynamics in the 20S proteasome (670 kDa) along with binding to target molecules (5).
The methodology that now is available opens the possibility for the study of a wide range of molecular machines in a site-specific and quantitative manner. One such fascinating molecule is the enzyme aspartate transcarbamoylase (ATCase), which catalyzes the first step in pyrimidine biosynthesis, the reaction of aspartic acid and carbamoyl phosphate (CbmP) to form carbamoyl aspartate. It was observed many years ago that the plot of reaction velocity versus [aspartate] is not hyperbolic (i.e., does not follow standard Michaelis–Menten kinetics), but rather it is sigmoidal, indicating that binding of substrate is cooperative (homotropic effect) (6). Sedimentation studies showed that the 300-kDa enzyme is composed of two kinds of polypeptides (7): regulatory chains (r), of which there are six, where effectors such as ATP and CTP bind as well as six copies of catalytic chains (c) that form the substrate binding sites. X-ray results establish that the structure is made up of a dimer of catalytic trimers (c3) along with three copies of regulatory domain dimers (r2) (8). A large body of data on the enzyme has been obtained over the past 40 years that supports the idea that the homotropic effect can be explained by the Monod–Wyman–Changeux (MWC) model of allostery (9, 10). For an oligomeric protein with n binding sites, this model postulates the existence of two conformations in equilibrium, T and R, with the T form having a lower affinity for substrates (11). All of the chains within an oligomer are identical (i.e., all are either T or R), and all microscopic equilibrium dissociation constants for ligands are assumed to be the same. As more sites in the enzyme become bound with substrate, the equilibrium is shifted from the T state in the unliganded form toward R, producing an apparent increase in affinity and sigmoidal binding.
There is not a consensus, however, regarding binding of effectors (heterotropic effect). It has been argued that the MWC model also applies for binding of ATP and CTP, with preferential binding to the R and T states, respectively (9). In contrast, in a competing model, the binding of nucleotides to the regulatory chains is assumed to modify the relative affinities of R and T states for substrate without changing the R–T equilibrium per se (i.e., there are structural changes with R and T converted to R′ and T′, respectively) (12). Small-angle x-ray scattering (SAXS) studies (13) have been interpreted in terms of the latter model, whereas ultracentrifuge experiments and binding data support the MWC model (9, 14).
In principle, solution NMR spectroscopy is a powerful monitor of binding, especially for micromolar to millimolar interactions, a range that is relevant for many of the ligand–ATCase complexes. Indeed, NMR studies of ATCase span several decades, with initial work monitoring relaxation rates and chemical shifts of substrates (or their analogues) and allosteric effectors to study ligand–enzyme interactions and ionization equilibria (15, 16). In addition, the enzyme was probed with 13C spectroscopy of 13Cγ-labeled aromatic residues (17, 18), 19F NMR of incorporated 3-fluorotyrosines (19) and 1H NMR focusing on 1Hδ Tyr probes in a highly deuterated aromatic background (20). These studies concentrated on ligand binding and associated conformational changes and attempted to better define the catalytic mechanism of the enzyme. All of this work was performed with one-dimensional spectroscopy and often involved interpretation of low-intensity signals that suffered from overlap. Here, we significantly extend these studies by using two-dimensional 1H,13C NMR spectroscopy of methyl groups that are sensitive and well resolved probes of molecular structure and ligand binding (21). We show that, despite the fact that only the T state of unliganded wild-type (WT) enzyme is observed in spectra, it nevertheless is possible to quantify the R–T equilibrium indirectly through a study of ligand binding. Results of binding of substrates, substrate analogues, and nucleotides are presented, with the observed shifts in R–T equilibria completely explained in terms of the MWC model. A model in which nucleotide binding does not affect equilibrium R–T ratios can be ruled out unequivocally.
Results and Discussion
Model of Ligand Binding and Conformational Changes.
Fig. 1 shows the classic MWC model for ligand (A) binding to a protein that can exist in either R or T states (11). In the case of ATCase, binding may be to either c (substrate) or r (effector) sites, and six substrates/effectors can bind to each molecule (i.e., n = 6). Because each of the six c chains (or r chains) is assumed equivalent in this model, all microscopic dissociation constants for a given state are the same (KD, RMicro and KD, TMicro for states R and T, respectively) and KD(T, j) = j/(n − j + 1)KD, TMicro. The linked binding equilibria of Fig. 1 form a thermodynamic cycle so that L′ = cnL, where L = [Tn]/[Rn], L′ = [Tn·An]/[Rn·An], and c = KD(R, j)/KD(T, j) = KD, RMicro/KD, TMicro. Because of its inherent simplicity, the MWC model has been used to explain ligand binding in many cooperative systems (11), and in the case of ATCase, a large body of accumulated evidence over an extensive period supports this mode of binding (9). In principle, NMR spectroscopy is an extremely powerful tool for testing the model further and for obtaining definitive answers that are less forthcoming from other techniques. For example, with other biophysical approaches that do not probe specific sites independently but only report average properties, local effects attributable to substrate/effector binding cannot be separated from the accompanying global shifts in the R–T equilibrium. By contrast, we show below that changes in chemical shifts in the vicinity of binding sites in response to ligand binding provide sensitive reporters of ligand–protein binding equilibria, whereas the concomitant appearance of separate resonances corresponding to both R and T states leads to definitive conclusions about the effects of ligand binding on the R–T equilibrium. This ability to “dissect” the complex equilibria of Fig. 1 has been exploited, for example, to establish conclusively that the nucleotide binding shifts the R–T ratio, a point that has remained controversial (13, 14).
Fig. 1.
MWC model of binding allostery. The protein is composed of n chains, all of which must be in the same state, T or R. Molecules in state T or R with j equivalents of bound ligand (A) are denoted by Tn·Aj or Rn·Aj, respectively. The vertical equilibria represent T–R isomerization steps with L = [Tn]/[Rn] and L′ = [Tn·An]/[Rn·An], whereas the horizontal equilibria correspond to binding or dissociation of ligand A (macroscopic dissociation constants KD(T, j) or KD(R, j). See Results and Discussion for further explanation.
Methyl-TROSY of ATCase.
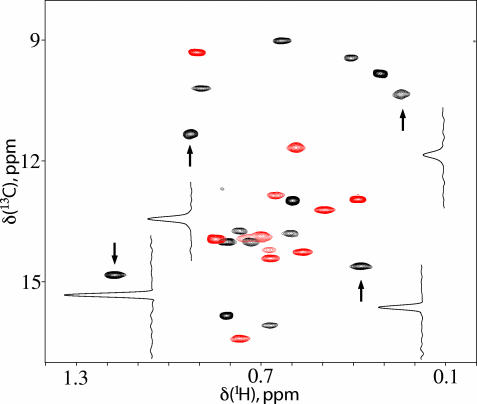
Fig. 2 shows a 1H-13C methyl-TROSY heteronuclear multiple quantum correlation (HMQC) spectrum of U-[2H]Ile-[δ113CH3]-labeled unliganded WT-ATCase recorded at 800 MHz (1H frequency) in 40 min on a spectrometer equipped with a room-temperature probe head at 37°C (0.1 mM in complex, 0.6 mM in monomer). The high signal-to-noise ratio of the data set and the short measuring time required suggest that methyl groups will be valuable probes of molecular conformation and dynamics in this system, allowing the effects of a wide range of ligands to be studied in relatively short order. Further, because holoenzyme molecules can be assembled from r and c chains (with different NMR labeling patterns if desired) (19), it is possible to readily assign correlations to chain type and, where necessary, to study intact enzyme that is labeled in only one of the two chains. As Fig. 2 illustrates, the Ile region of the 1H,13C correlation map of ATCase is well resolved, and by preparing samples with label confined to either r or c chains, it is possible to separate the two or three overlapping cross-peaks and to count all 27 correlations that originate from the 27 Ile residues in the molecule. Because in the absence of ligands the R–T equilibrium is shifted highly to T (9), the spectrum of Fig. 2 thus corresponds to that of the T state. Furthermore, because only a single correlation is observed for each Ile, each c (r) chain must be structurally identical or, alternatively, any differences in structure between chains must be averaged rapidly on the NMR chemical-shift time scale (i.e., fast exchange). Although it likely is possible that individual peaks could be assigned to specific sites in the protein by using approaches similar to those described in the context of our work on the proteasome (5), we have not done so here. Instead, we show that quantitative inferences about binding and allostery can be made simply by analysis of how spectra change as a function of ligand, without time-consuming steps that are involved in resonance assignment.
Fig. 2.
Two-dimensional 1H-13C correlation spectrum of ATCase. 1H-13C methyl-TROSY correlation map of U-[2H] Ile-[δ113CH3]-labeled ATCase (0.6 mM in monomer, 37°C, 800 MHz, 40-min total acquisition time). Traces through a number of cross-peaks are indicated to illustrate the sensitivity of the spectrum. Peaks from r and c chains are colored red and black, respectively. The “central part” of the spectrum is shown as an overlay of a pair of data sets with labeling confined to either r or c chains.
Binding of Active Site Ligands and Analogues.
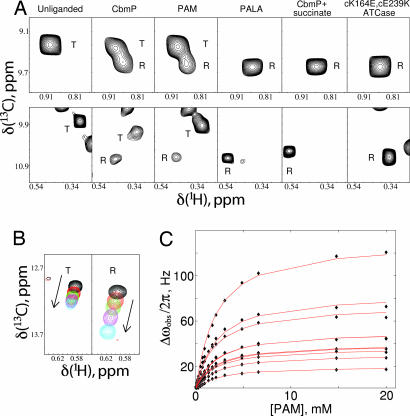
Upon titration of U-[2H] Ile-[δ113CH3]-labeled WT-ATCase with substrate CbmP or its nonhydrolyzable analogue phosphonoacetamide (PAM), a number of the correlations derived from the c chain shift linearly, consistent with weak binding (millimolar affinity) and exchange rates that are fast on the NMR chemical shift time scale [see below and supporting information (SI) Fig. 6]. In contrast, cross-peaks from residues in the r chain (that are all distant from the binding site) do not shift during the titration. During the course of the titration, a second set of correlations emerges, consistent with a second conformation in slow exchange with the T state. This finding is illustrated in Fig. 3A, which focuses on an Ile correlation from the r chain with chemical shifts (ω13C,ω1H) = (9.3 ppm, 0.90 ppm) in the unliganded WT state (Fig. 3A Upper, first column) and a second Ile correlation (9.8 ppm, 0.31 ppm) derived from the c chain (Fig. 3A Lower). Addition of saturating amounts of CbmP or PAM produces a second set of peaks for both correlations, as indicated in the second and third columns of Fig. 3A. To establish the identity of this second state, we have recorded spectra of ATCase with different substrates or analogues that are known to shift the equilibrium to R and of a double mutant (cK164E, cE239K) that has been shown to be exclusively in the R form even when unliganded (22). Columns 4 and 5 of Fig. 3A show how spectra change with saturating amounts of the high-affinity, bisubstrate analogue phosphonoacetyl-l-aspartate (PALA) ([PALA]/[ATCase]monomer = 1.5, where [ATCase]monomer is the concentration of the c chains that form the PALA binding site) and saturating quantities of CbmP and succinate, an analogue of the reactant aspartate ([CbmP]/[ATCase]monomer = 30, [succinate]/[ATCase]monomer = 75), respectively. The corresponding region of the Ile correlation map of unliganded cK164E, cE239K–ATCase is shown in column 6. The excellent correspondence between chemical shifts of the second conformer in spectra of CbmP or PAM-loaded WT-ATCase and the correlations in the subspectra of columns 4–6 that are known to derive from the R state establishes that the observed second state in CbmP- or PAM-saturated ATCase is indeed the R form of the enzyme. Note that the correspondence in shifts is better for residues in the r chain (Fig. 3A Upper) than in the c chain (Fig. 3A Lower) because residues proximal to substrate binding sites formed by the c chains sense both the binding event and the T to R conformational transition. Further, the similarity in shifts between spectra of unliganded cK164E, cE239K–ATCase and R states of the enzyme generated through the addition of ligands argues strongly that the double mutant form of the enzyme is a good model of the R state (see SI Fig. 7). The titration data also establish unequivocally that both CbmP and PAM shift the equilibrium toward R and thus bind preferentially to this state, in agreement with the MWC model. This finding is in contrast to results from earlier literature in which it was argued that CbmP binding would not shift the equilibrium (23). Finally, the absence of exchange cross-peaks connecting correlations between R and T states in magnetization exchange experiments recorded on a PAM-saturated sample allows an upper limit of ≈1/s to be placed on the exchange between the two conformers.
Fig. 3.
Effects of binding of substrate and analogues on the spectrum of ATCase. (A) Observation of R conformation on ligand binding. Portions of methyl-TROSY spectra of U-[2H] Ile-[δ113C1H3]-labeled ATCase (800 MHz, 37°C) are shown that include a correlation derived from an Ile residue from the r chain (Upper) and from the c chain (Lower). Column 1, unliganded WT-ATCase; column 2, WT-ATCase saturated with 15 equiv. of CbmP (i.e., [CbmP]/[ATCase]monomer = 15); column 3, WT-ATCase saturated with 58 equiv. of PAM; column 4, WT-ATCase saturated with 1.5 equiv. of PALA; column 5, WT-ATCase saturated with 30 equiv. of CbmP and 75 equiv. of succinate; column 6, unliganded cK164E, cE239K–ATCase. The letters “T” and “R” label peaks derived from ATCase in either the T or R conformation, respectively. (B) Titration of a correlation derived from the T state of WT-ATCase (Left, labeled with “T”) and from the R state (Right, labeled with “R”) with PAM. Overlay of spectra with PAM concentrations ranging from 0 (black) to saturating (blue). The direction of peak shift is indicated by an arrow in each case. Only three intermediate PAM concentrations are shown for clarity. (C) Global fitting of all shifting peaks from the titration of cK164E, cE239K–ATCase (R state) to a binding model with a single KD.
Because the exchange between R and T conformers is slow on the NMR chemical-shift time scale (i.e., separate correlations are observed for each state), it is possible to measure the R–T equilibrium under saturating conditions of PAM or CbmP (see below), where correlations derived from both states are visible. In the absence of chemical-shift assignments of correlations in R and T states, care must be taken to ensure that equilibrium constants are derived from intensities of cross-peaks that belong to the same residue. Two residues have been identified for which the R–T pairs can be assigned with certainty, corresponding to the correlations illustrated in Fig. 3A (SI Fig. 8), and values of L′ can be estimated from them. Differential relaxation of magnetization from R and T states during the course of the NMR experiment that could perturb the relative intensities of correlations, and hence L′ values, also must be accounted for, as described in Materials and Methods. A value of L′ = 3.4 ± 1.2 has been calculated with PAM saturation, corresponding to ΔGT−R = −0.76 ± 0.22 kcal/mol. Because of the lability of CbmP, only an approximate value of L′ = 1.5 (ΔGT−R ∼ −0.24 kcal/mol) could be obtained for this compound. Values of L′ for CbmP and PAM are in reasonable agreement with the previously reported value of 7 for CbmP, considering that different pH values were used in the two sets of measurements (1 pH unit different) (10), the known sensitivity of ATCase function to pH (24), the different solvents used (H2O versus D2O), and the fact that ATCase used in the present set of experiments is highly deuterated.
Measuring the R–T Equilibrium Constant.
The ability to monitor R and T conformations independently provides NMR with a unique advantage over many other spectroscopic techniques. It can be shown that if ligand binding is in the fast exchange limit and separate signals are observed from the R and T states (i.e., slow exchange), then chemical shift changes of cross-peaks belonging to either R or T states that accompany ligand binding can be described by a simple hyperbolic isotherm with the extracted dissociation constant KD, RMicro or KD, TMicro (see SI Text). In contrast, titration curves derived from techniques that monitor contributions from both R and T states simultaneously must be analyzed by taking the complete reaction scheme into account (Fig. 1), and the resulting binding curves will be sigmoidal. Fig. 3B shows the titration of a correlation from WT-ATCase (Left) as a function of added PAM along with the corresponding titration for the same residue from cK164E, cE239K–ATCase (Right). All titration curves that derive from binding to either T (WT-ATCase) or R (cK164E, cE239K–ATCase; Fig. 3C) states of the enzyme were fit simultaneously to a simple PA ⇌ P + A model. Excellent fits were obtained with this hyperbolic binding model, as expected. Values of KD, TMicro = 3.8 ± 0.3 mM and KD, RMicro = 1.8 ± 0.1 mM were extracted from fits of 8 and 10 titration curves, respectively, and by using these KDMicro values along with L′ = 3.4 ± 1.2 and the relation L′ = c6L (see above), a value of 300 ± 190 is calculated for L = [T6]/[R6] (Fig. 1), corresponding to ΔGT−R = −3.5 ± 0.4 kcal/mol, which agrees well with a previously published value of L = 250 based on analytical ultracentrifugation (10) and less well with L = 70 obtained from SAXS (25). Thus, even though correlations from the R state cannot be observed in spectra of unliganded WT-ATCase, the relative populations of R and T still can be established, albeit indirectly, by using the linked binding equilibria of Fig. 1 and data obtained from titration of R and T states with ligand.
Effects of Ligand Binding to the Regulatory Chains.
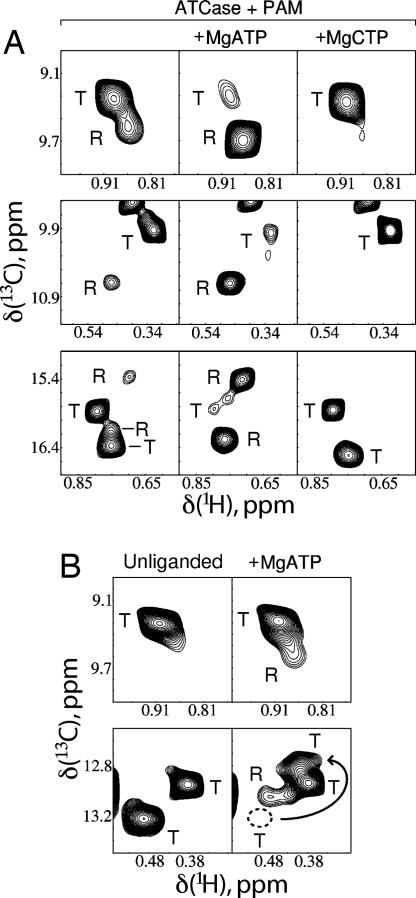
The presence of significant populations of both R and T conformers in the PAM-saturated enzyme (Fig. 3A) allows a straightforward determination of how effectors such as ATP and CTP perturb the R–T equilibrium. Fig. 4A shows examples of how the R–T equilibrium is shifted upon addition of MgATP (Center) or MgCTP (Right) starting from PAM-loaded WT-ATCase (Left). Although it is difficult to quantify precisely the shift upon addition of ATP, because the population of the T state becomes very low, changes in L′ of 15- to 30-fold are estimated from spectra, leading to a decrease in L′ from 3.4 ± 1.2 to 0.1–0.2. Conversely, the effect with CTP is opposite, with correlations from the R conformer disappearing completely from spectra. Such changes are in complete agreement with the MWC model of heterotropic effects, where binding of ATP to the R state is favored and binding of CTP to the T state is preferred.
Fig. 4.
Effect of nucleotides on R–T equilibrium. (A) Portions of spectra of U-[2H] Ile-[δ113CH3]-labeled WT ATCase (800 MHz, 37°C) saturated with PAM ([PAM]/[ATCase]monomer = 58) but in the absence of nucleotide (Left) or with the addition of saturating amounts of either MgATP ([MgATP]/[ATCase]monomer = 46; Center) or MgCTP ([MgCTP]/[ATCase]monomer = 32; Right). Different regions of the 1H-13C methyl-TROSY correlation map are highlighted in three rows. Correlations from R and T conformers are indicated by the letters “R” and “T,” respectively. (B) Portions of spectra of unliganded WT-ATCase without (Left) and with saturating MgATP (Right) ([MgATP]/[ATCase]monomer = 23), illustrating the appearance of the R state conformation upon addition of ATP. Separate regions of the 1H-13C spectrum are shown in each of two rows. Titration of ATP causes some of the T state correlations to move to new locations (Lower) as indicated by the arrow [denoting the displacement from the old position (dotted circle) to the new one]. Each of these spectra (unliganded and +MgATP) were recorded in 17 h, 800 MHz, at 37°C.
The spectra of Fig. 4A, which were obtained with saturating amounts of PAM, could also be explained, however, under the assumption that MgATP binding does not affect the R–T equilibrium directly but rather promotes conformational changes in at least one of the states. These changes would lead to an increase in R state affinity for substrate Asp (12) or for its analogue succinate and by extension also for the CbmP analogue, PAM, and subsequent shifting of the R–T equilibrium only on substrate binding. The latter explanation was put forth to explain the results of SAXS experiments (13, 26). In particular, if this model were operative, we would expect to see significant changes in chemical shifts of probes in the c chain upon ATP binding that reflect the “postulated” changes in structure leading to higher affinity of substrate, which, however, is not what we observed. Of the 22 cross-peaks in 1H,13C correlation spectra of U-[2H] Ile-[δ113CH3] PAM-saturated WT-ATCase that are well resolved (belonging to either of T or R conformers), only 2 change position by at least 0.1 ppm in 1H or 0.4 ppm in 13C when MgATP is added. These two peaks, both from the regulatory chain, are almost certainly rIle12 and rIle86, which contact bound ATP directly (8). The other 20 peaks change positions by <0.025 ppm and 0.2 ppm in 1H and 13C, respectively; peaks obscured by overlap move very little as well. The fact that a set of 20 well dispersed peaks that includes probes in the c chain change position very little in response to the addition of MgATP suggests that even if binding of nucleotide does induce alteration of R and T conformations, these changes must be minor, at least in parts of the molecule remote from the site of nucleotide binding. A similar situation also holds for MgCTP. Results from a second experiment are even more conclusive in favor of the MWC model. From the measured value L = 300 for WT-ATCase and the fact that MgATP shifts the equilibrium to the R state by 15- to 30-fold (see above), L′ is predicted to be in the range of 10–20 in the presence of saturating amounts of ATP. Thus, the ATP-saturated R form of the enzyme is expected to constitute ≈5–10% of the population (effective monomer concentration of 50–100 μM), and with the sensitivity of the NMR methodology used here, such a fraction should be observable, even for a system as large as ATCase. As predicted by our numerical estimates and the MWC model, Fig. 4B shows that the addition of saturating amounts of MgATP to unliganded WT-ATCase does indeed produce measurable amounts of R, estimated to be on the order of 5% on the basis of relative peak heights. Note that in the second set of spectra (Fig. 4B Lower), addition of ATP also causes a shift in one of the correlations from the T state (cross-peak tentatively assigned to rIle12), as indicated by the arrow. The only explanation for the appearance of R state cross-peaks is that MgATP is an allosteric effector that directly alters the R–T equilibrium.
To study the binding of MgATP to ATCase in more detail, we have recorded an ATP titration series. Unfortunately, the two significantly perturbed Ile correlations in 1H-13C spectra, most likely derived from rIle12 and rIle86 as discussed above, are not particularly useful for quantifying binding because rIle12 titrates into another peak and rIle86 broadens very significantly in response to ATP addition (because of the large 1H chemical-shift difference between free and bound forms, 0.64 ppm). To increase the number of probes available, a U-[2H] Ile-[δ113CH3],Leu,Val-[13CH3,12CD3]-labeled WT-ATCase sample was prepared so that cross-peak positions from Leu and Val residues could be quantified as well. Labeling in the complex was confined to only the r chain, the site of ATP binding. The HMQC correlation map of this Ile-, Leu-, and Val-labeled ATCase sample is provided in SI Fig. 9; there are a total of 27 Leu and Val residues in the r chain, and 50 peaks can be counted in the Leu-Val region of the spectrum. Of interest, one of the Val residues, rVal121, is immediately adjacent to the fully protonated catalytic chain, and its signal would not be expected to be observed because of contributions to transverse relaxation resulting from proximal 1H spins (27); such a situation also occurs for rIle115, which is proximal to the c chain.
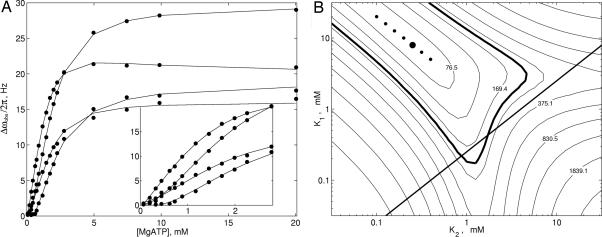
Upon titration of MgATP into the Ile, Leu, and Val methyl-labeled sample of WT-ATCase, the majority of peaks shift in position, although no more than 36 Hz in either 1H or 13C dimensions, with little or no broadening, consistent with fast exchange. A pair of correlations that are assigned tentatively to Leu and Val residues from the amino-terminal end of the r chain that are proximal to the nucleotide binding site shift in position by at least 0.35 ppm in the 1H dimension and broaden beyond detection during the course of the titration. Of note, the trajectories of at least six of the correlations that titrate in the fast exchange limit are not linear, indicating that the binding of MgATP to ATCase is more complex than 1:1, as observed previously (28, 29). To quantify the binding further, we chose 17 titration curves (either 1H or 13C) derived from 13 peaks that were in the fast exchange limit and fitted these data simultaneously to a number of binding models. Several of the titration profiles are shown in Fig. 5A, and the Inset highlighting the response to binding for low [MgATP] makes it clear that the binding cannot be described by a hyperbolic function, which would characterize a 1:1 interaction. Global fits to such a model produced a poor reduced χ2 value (≈5) with noticeable deviations from experiment for low [MgATP]. The data subsequently were fit to a sequential binding model (SI Fig. 10) that assumes a pair of equivalent binding sites where the binding of ligand to the first site alters the affinity at the second (cooperative binding). Although outside the scope of the MWC model, which postulates that the intrinsic affinity of ligand at one site is independent of the binding of ligand to a second site (i.e., same microscopic binding), the sequential model is attractive on the basis of the structure of ATCase in which the regulatory chains that bind nucleotides are arranged as dimers that certainly could “communicate” in response to ligand binding (8).
Fig. 5.
Titration of unliganded WT-ATCase with MgATP. (A) Chemical-shift changes (either 1H or 13C) as a function of added [MgATP] ([ATCase]monomer = 0.69 mM) for 4 of the 17 titration curves used in data fitting. All 17 titration curves (filled circles correspond to experimental points) were fit to a sequential binding model (see SI Text) with the solid lines showing the best fit. (Inset) Enlarged region of the titration curves focusing on low [MgATP]. (B) χ2 surface used to determine optimal K1 and K2 values for the sequential binding model. The global minimum is labeled with a large filled circle, with the six next-best solutions indicated by small filled circles. Contours (spaced on a logarithmic scale) are labeled with χ2 values. The region encompassed by the thick contour corresponds to (K1, K2) values within the 95% confidence interval. The straight line K1 = K2/4 (no cooperativity) separates areas of positive (above) and negative (below) cooperativity.
Fig. 5B shows a χ2 surface of the distribution of K1 and K2 values that are obtained from the fits of the titration curves, where K1 and K2 are the macroscopic dissociation constants associated with the first and second binding events, respectively. The bold line, 4K1 = K2, indicates where microscopic dissociation constants for the first and second binding events are the same so that no binding cooperativity is observed. Points above the line correspond to positive cooperativity of binding (i.e., binding at the first site increases affinity at the second), whereas those below the line indicate negative cooperativity. The seven best solutions obtained from a grid search of (K1, K2) space are indicated by circles in the plot with the best solution corresponding to K1 = 7.9 mM and K2 = 0.25 mM. These solutions lie within a shallow trough on the χ2 surface, where K1× K2 ∼ 2 mM2. Finally, the bold contour line encompasses the range of solutions that lie within the 95% confidence limit (for a model with 221 degrees of freedom). It is worth noting that an alternative model that assumes consecutive binding to two noninteracting sites with different affinities can be rejected via F test statistical criteria. Other studies involving NaATP binding in which data have been interpreted in terms of a consecutive binding model report K1 and K2 values of 0.065 and 1.25 mM, respectively, at 4°C and K1 = 0.14 mM and K2 = 0.67 mM at 24°C, pH 7 (28, 29). Deviations from the values obtained here likely arise from the different experimental conditions used because, for example, it is known that increasing temperature decreases nucleotide affinity (28).
In summary, we have presented an NMR study of how ligand binding affects the allosteric equilibrium in the 306-kDa enzyme ATCase. Despite the size of this system, high-sensitivity 1H,13C correlation maps of Ile, Leu, and Val methyl groups could be obtained in very reasonable measuring times (<1 h) using protein concentrations <1 mM in monomer (<160 μM in complex), so that large numbers of spectra could be recorded as a function of different ligands or ligand concentrations. By using relations that describe linked binding equilibria, a value for L = [T6]/[R6] could be calculated for WT ATCase, despite the fact that the R form of the protein is “invisible.” The effect of binding of a variety of different substrates or substrate analogues and nucleotides on the R–T equilibrium could be well understood within the framework of the MWC model, and the binding of MgATP to ATCase was shown unequivocally to alter this equilibrium, in contrast to observations from a series of other studies (12, 13, 25, 26). This work emphasizes the important role that modern solution NMR spectroscopy can play in providing quantitative information on systems with molecular masses in the hundreds of kilodaltons.
Materials and Methods
NMR Spectroscopy.
All NMR samples were composed of protein (concentrations between 0.28 and 0.93 mM in monomer, corresponding to 0.05 to 0.16 mM in complex) dissolved in 50 mM Na-Hepes, 50 mM KCl, 20 mM 2-mercaptoethanol (pH 7.5, uncorrected), and 100% D2O. Sample preparation details are provided in SI Text.
Methyl-TROSY (HMQC) spectra were recorded at 37°C on an 800-MHz Varian (Palo Alto, CA) Inova spectrometer equipped with a room-temperature pulsed-field gradient triple-resonance probe. The acquisition time in the indirect dimension typically was 26 ms with a spectral window of 9 (19) ppm, and the carbon carrier was placed at 12.5 (18) ppm for experiments recorded on Ile-[δ113CH3] and Ile-[δ113CH3],Leu,Val-[13CH3,12CD3]-labeled samples, respectively. 1H and 13C chemical shifts were referenced against 0.2 mM 2,2-dimethyl-2-silapentane-5-sulfonate (DSS), which was included in each sample. All data were processed with the NMRPipe/NMRDraw suite of programs (30).
Data Processing and Analysis.
Relative populations of molecules in R and T states (PAM-saturated samples) were determined by fitting line-shapes of R–T cross-peaks with line-shape functions that are Lorentzian in each of the 1H and 13C dimensions. To minimize the contributions from differential relaxation in each of the two states, which can skew relative intensities, weighting functions were not used during data processing. 1H transverse relaxation during the magnetization transfer steps in the HMQC scheme (two 3.6-ms intervals) that potentially also could influence relative intensities was accounted for by measuring 1H T2 values in separate experiments and “subtracting out” relaxation effects during these intervals.
Peak positions as a function of titrated ligand were extracted by using the peak picking facility in NMRDraw, with 1H and 13C chemical shifts from each peak treated separately. Titration curves obtained with PAM were fitted to the equation: Δωobs = off + ΔωPAfB, where Δωobs is the chemical-shift displacement at each ligand concentration relative to zero ligand added, ΔωPA is the displacement upon addition of saturating amounts of ligand (A), the fraction of bound ligand, fB, depends on the microscopic binding constant KD because fB = β − (β2 − 4[PT][PAMT])1/2/(2[PT]) with β = [PT] + [PAMT] + KD, [PT] is the total concentration of ATCase c chains (=6 [ATCase]), [PAMT] is the total concentration of added PAM, and “off” is a small offset accounting for possible error in the chemical shift of the unliganded state, which shifts the entire binding curve up or down without altering its KD-dependent profile (offset varies between −2 and 2 Hz). The parameters off and ΔωPA were varied individually for each titration curve, and a single KD was enforced globally.
Titration curves derived from addition of MgATP were fitted to a sequential binding model (see SI Text) where Δωobs = off + ΔωPAfPA + ΔωPA2fPA2. Here, ΔωPA = ωPA − ωo, ΔωPA2 = ωPA2 − ωo, and ωPA, ωPA2 are the chemical shifts when only one ligand is bound (but not the other) or both, respectively, where ωo is the chemical shift in the unliganded state and fPA and fPA2 are the mole fractions of singly and doubly bound regulatory dimers. Values of fPA and fPA2 are obtained by solving a cubic equation that is given in SI Text.
Supplementary Material
Acknowledgments
This work was supported by a grant from the Canadian Institutes of Health Research (CIHR) (to L.E.K.), by a CIHR postdoctoral fellowship (to A.V.), and by National Institute of General Medical Sciences Research Grant GM 12159 (to H.K.S.). L.E.K. holds a Canada Research Chair in Biochemistry.
Abbreviations
- ATCase
aspartate transcarbamoylase
- MWC
Monod–Wyman–Changeux
- TROSY
transverse relaxation optimized spectroscopy
- SAXS
small-angle x-ray scattering
- jPM: HMQC
heteronuclear multiple quantum correlation
- CbmP
carbamoyl phosphate
- PAM
phosphonoacetamide
- PALA
phosphonoacetyl-l-aspartate.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0703347104/DC1.
References
- 1.Foster MP, McElroy CA, Amero CD. Biochemistry. 2007;46:331–340. doi: 10.1021/bi0621314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fiaux J, Bertelsen EB, Horwich AL, Wüthrich K. Nature. 2002;418:207–211. doi: 10.1038/nature00860. [DOI] [PubMed] [Google Scholar]
- 3.Horst R, Bertelsen EB, Fiaux J, Wider G, Horwich AL, Wüthrich K. Proc Natl Acad Sci USA. 2005;102:12748–12753. doi: 10.1073/pnas.0505642102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sprangers R, Gribun A, Hwang PM, Houry WA, Kay LE. Proc Natl Acad Sci USA. 2005;102:16678–16683. doi: 10.1073/pnas.0507370102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sprangers R, Kay LE. Nature. 2007;445:618–622. doi: 10.1038/nature05512. [DOI] [PubMed] [Google Scholar]
- 6.Gerhart JC, Pardee AB. J Biol Chem. 1962;237:891–896. [PubMed] [Google Scholar]
- 7.Gerhart JC, Schachman HK. Biochemistry. 1965;4:1054–1062. doi: 10.1021/bi00882a012. [DOI] [PubMed] [Google Scholar]
- 8.Lipscomb WN. Adv Enzymol Relat Areas Mol Biol. 1994;68:67–151. doi: 10.1002/9780470123140.ch3. [DOI] [PubMed] [Google Scholar]
- 9.Schachman HK. J Biol Chem. 1988;263:18583–18586. [PubMed] [Google Scholar]
- 10.Howlett GJ, Blackburn MN, Compton JG, Schachman HK. Biochemistry. 1977;16:5091–5100. doi: 10.1021/bi00642a023. [DOI] [PubMed] [Google Scholar]
- 11.Monod J, Wyman J, Changeux JP. J Mol Biol. 1965;12:88–118. doi: 10.1016/s0022-2836(65)80285-6. [DOI] [PubMed] [Google Scholar]
- 12.Tauc P, Leconte C, Kerbiriou D, Thiry L, Herve G. J Mol Biol. 1982;155:155–168. doi: 10.1016/0022-2836(82)90442-9. [DOI] [PubMed] [Google Scholar]
- 13.Fetler L, Vachette P. J Mol Biol. 2001;309:817–832. doi: 10.1006/jmbi.2001.4681. [DOI] [PubMed] [Google Scholar]
- 14.Eisenstein E, Markby DW, Schachman HK. Biochemistry. 1990;29:3724–3731. doi: 10.1021/bi00467a019. [DOI] [PubMed] [Google Scholar]
- 15.Schmidt PG, Stark GR, Baldeschwieler JD. J Biol Chem. 1969;244:1860–1868. [PubMed] [Google Scholar]
- 16.Roberts MF, Opella SJ, Schaffer MH, Phillips HM, Stark GR. J Biol Chem. 1976;251:5976–5985. [PubMed] [Google Scholar]
- 17.Kleanthous C, Wemmer DE, Schachman HK. J Biol Chem. 1988;263:13062–13067. [PubMed] [Google Scholar]
- 18.Moore AC, Browne DT. Biochemistry. 1980;19:5768–5773. doi: 10.1021/bi00566a016. [DOI] [PubMed] [Google Scholar]
- 19.Wacks DB, Schachman HK. J Biol Chem. 1985;260:11659–11662. [PubMed] [Google Scholar]
- 20.Cohen RE, Takama M, Schachman HK. Proc Natl Acad Sci USA. 1992;89:11881–11885. doi: 10.1073/pnas.89.24.11881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tugarinov V, Hwang PM, Kay LE. Annu Rev Biochem. 2004;73:107–146. doi: 10.1146/annurev.biochem.73.011303.074004. [DOI] [PubMed] [Google Scholar]
- 22.Newell JO, Schachman HK. Biophys Chem. 1990;37:183–196. doi: 10.1016/0301-4622(90)88018-n. [DOI] [PubMed] [Google Scholar]
- 23.England P, Leconte C, Tauc P, Herve G. Eur J Biochem. 1994;222:775–780. doi: 10.1111/j.1432-1033.1994.tb18924.x. [DOI] [PubMed] [Google Scholar]
- 24.Pastra-Landis SC, Evans DR, Lipscomb WN. J Biol Chem. 1978;253:4624–4630. [PubMed] [Google Scholar]
- 25.Fetler L, Tauc P, Herve G, Moody MF, Vachette P. J Mol Biol. 1995;251:243–255. doi: 10.1006/jmbi.1995.0432. [DOI] [PubMed] [Google Scholar]
- 26.Herve G, Moody MF, Tauc P, Vachette P, Jones PT. J Mol Biol. 1985;185:189–199. doi: 10.1016/0022-2836(85)90190-1. [DOI] [PubMed] [Google Scholar]
- 27.Hamel DJ, Dahlquist FW. J Am Chem Soc. 2005;127:9676–9677. doi: 10.1021/ja052517m. [DOI] [PubMed] [Google Scholar]
- 28.England P, Herve G. Biochemistry. 1992;31:9725–9732. doi: 10.1021/bi00155a028. [DOI] [PubMed] [Google Scholar]
- 29.Gray CW, Chamberlin MJ, Gray DM. J Biol Chem. 1973;248:6071–6079. [PubMed] [Google Scholar]
- 30.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.