Abstract
In contrast to rapid progress in the molecular biology of olfaction, there are few physiological data to characterize the odor response properties of different populations of olfactory receptor neurons (ORNs) and their spatial distributions across the epithelium, which is essential for understanding the coding mechanisms underlying odor discrimination and recognition. We have tested the hypothesis that the ORNs are arranged in a functional mosaic, using an intact epithelial preparation from the mouse, in which odor responses of many ORNs in situ can be monitored simultaneously with calcium imaging techniques. ORNs responding to a given odor were widely distributed across epithelium and intermingled with ORNs responding to other odors. Tight clusters of ORNs responding to the same odor were observed. For a given odor, more ORNs were recruited when the concentration was increased. ORNs were able to distinguish between pairs of enantiomers by showing distinct but somewhat overlapping patterns. The results provide evidence regarding the response spectra of ORNs in situ, supporting the combinatorial coding of odor quality and intensity by different ORN subsets.
The mammalian olfactory system has the remarkable ability to discriminate among thousands of odor molecules. In contrast to other sensory systems, such as visual and somatosensory, in which the spatial information of the sensory input is encoded and processed by neural space, the olfactory system processes a basically nonspatial modality. We want to test how the neural space in the olfactory epithelium, with subsequent projection to the olfactory bulb and cortex, is used to encode the nonspatial olfactory input.
The molecular basis for the spatial organization of the olfactory system has emerged recently from a series of studies, starting with the cloning of olfactory receptor genes (1). It is generally believed that a single olfactory receptor neuron (ORN) expresses only one (or at most a few) of a repertoire of 1,000 receptor genes (2–6). The ORNs that express a given receptor gene send their axons to a pair of glomeruli to form a highly organized topographic map in the olfactory bulb (7–9). In the epithelium, the ORNs expressing a particular receptor gene are selectively distributed in one among four spatially distinct zones, where they are widely dispersed and intermingled with other subtypes of ORNs within each zone (2, 3, 10, 11).
In parallel with the finding of the zonal expression of olfactory receptor proteins, broad topographic patterns of responsiveness to odor molecules in rodent olfactory epithelium have been demonstrated by both eletroolfactogram (EOG) recordings (12–15) and voltage-sensitive dye imaging (16, 17). These studies have resolution at the regional level, by recording the summated activities of many ORNs. There is a critical need to obtain resolution at the cellular level, by relating the physiological responses of individual ORNs to their molecular properties.
To begin to obtain such information, we have applied calcium imaging techniques to a recently developed intact preparation of mouse olfactory epithelium (18). This has allowed us to record simultaneously the odor responses from up to several hundred dendritic knobs of ORNs at the resolution of individual cells with their spatial relations intact. The results provide evidence for a functional mosaic of ORNs responding to different odors. We have characterized the properties of ORNs responding to odors with distinct chemical structures, different intensities, and different enantiomers and discussed their implications in the peripheral coding of odor quality and intensity.
Materials and Methods
Intact Epithelial Preparation.
The method has been described in detail (18). Briefly, female adult C57BL/6 mice (4–16 weeks) were anesthetized by injection of ketamine (500 mg/kg of body weight) and decapitated. Swatches of olfactory epithelia were peeled off the underlying bone in the nasal cavity and kept in oxygenated saline, which contained 124 mM NaCl, 3 mM KCl, 1.3 mM MgSO4, 2 mM CaCl2, 26 mM NaHCO3, 1.25 mM NaH2PO4, 15 mM glucose, pH 7.6 and 305 mOsm. A small sheet of epithelium was transferred to a recording chamber with mucus layer facing up. The dendritic knobs of ORNs were visualized through an upright infrared differential interference contrast microscope (Olympus BX50WI) equipped with a ×40 water-immersion objective and a cooled CCD-300-RC camera (Dage–MTI, Michigan City, IN) for patch clamping or a SensiCam camera for calcium imaging. Oxygenated saline was continuously perfused into the recording chamber at a rate of about 3 ml/min throughout the experiments. All experiments were carried out at 25 ± 2°C.
Perforated Patch-Clamp Recording of Odor Responses.
Perforated patch clamp was performed on the dendritic knobs of mouse ORNs under voltage clamp mode, using 260 μM nystatin in the pipette to permeablize the membrane of a cell-attached patch. The recording pipettes were filled with the following solution: 17.7 mM KCl, 105.3 mM KOH, 82.3 mM methanesulfonic acid, 5.0 mM EGTA, 10 mM Hepes, 70 mM sucrose, pH 7.2 (KOH), and 310 mOsm. Voltage-clamp recordings were controlled by an EPC-9 patch clamp amplifier combined with the pulse/pulsefit software package (HEKA Electronics, Lambrecht/Pfalz, Germany) running on a Macintosh computer. The odor stimuli were delivered by pressure ejection using Picospritzer II (General Valve, Fairfield, NJ) through multibarrel pipettes, which were placed 5–10 μm downstream from the dendritic knobs. All odors [Aldrich, except (+)/(−) carvone and (+)/(−) limonene from Fluka] were prepared as 0.5 M stock solutions in DMSO and diluted to the final concentrations by adding saline.
Calcium Imaging of Odor Responses.
Calcium-sensitive dye calcium green-1/AM or fluo-4/AM (Molecular Probes) was prepared as a 5 mM stock solution in DMSO with 20% Pluronic F-127 (Molecular Probes) and then diluted to a loading solution of 10–20 μM by adding saline. Olfactory epithelia attached to the septal wall were kept in the loading solution for 20–40 min, followed by a washing-out period of at least 20 min, before they were transferred to a recording chamber and stabilized by a customized net. The calcium imaging system consisted of a SensiCam super VGA Camera (PCO, Kelheim, Germany) mounted on an upright microscope (BX50WI) equipped with fluorescence attachment (Olympus BX-FLA), a green light filter set (XF22, Omega Optical, Brattleboro, VT), a Xenon short arc lamp (USHIO, Tokyo), and imaging workbench 2.1 (Axon Instruments, Foster City, CA) as software running on a PC (Dell). Odor stimulation (4–5 sec) was applied through bath perfusion by using a custom-designed gravity perfusion system. The interstimulus interval was 3–5 min. The time courses of the onset (τ = 1.4 s) and decay (τ = 3.3 s) of the odor delivery system were estimated by measuring the changes of light transmission when saline containing 0.5% of food color was perfused. This test also revealed the relatively even application of ligands within the viewing area.
Efforts were made to find a positive control for the mouse ORNs in the intact epithelial preparations during calcium imaging recordings. High K+ solution (100 mM K+ instead of Na+ in saline) failed to serve as an indicator of cell viability in this preparation as in the study of dissociated ORNs (19), because it introduced movement artifacts during perfusion. IBMX (3-isobutyl-1-methyl-xanthine), a potent inhibitor of phosphodiesterase, did not serve as an effective positive control for two reasons. First, only half of rodent ORNs responded to IBMX in patch clamp studies (18, 20). It has been reported that signal transduction pathways other than cAMP pathway may exist in rodent ORNs (21, 22). Second, in the patch clamp studies, IBMX could be rapidly applied and then quickly removed, because it was directly puffed onto the knob and cilia through a pipette (18). In contrast, slow bath perfusion in the imaging studies led to dilution of ligands at the site of action. Therefore, much higher concentrations of IBMX in the reservoir were needed to achieve 300 μM in the bath, which tended to produce movement artifacts during perfusion.
To make sure that the cells were alive and the responses we recorded were real, we performed other control experiments. When 0.04% trypan blue solution was used to test the viability of neurons after the preparation was loaded with calcium green-1, no double-labeled cells were found (data not shown). This indicated that cells loaded with calcium-sensitive dyes should be alive (23). Applying saline did not induce fluorescence changes and removal of Ca2+ by perfusing low Ca2+ saline (0 Ca and 10 mM EGTA) eliminated the fluorescence changes during odor stimulation. Virtually the same patterns of responsive cells were recorded when two identical stimuli were sequentially delivered to one preparation. Cells responded to a given odor at lower concentrations almost always responded to the same odor at higher concentrations (see Results). We may have underestimated the percentage of responsive cells because of the lack of a suitable positive control in the preparation. This is unlikely to affect the main conclusions drawn from the current study.
Data Analysis.
Calcium imaging data were acquired at 0.5–1 Hz with five frames as baseline before the onset of odor delivery. The fluorescence changes were calculated for individual dendritic knobs of mouse ORNs as follows: ΔF/F = [(F1 − B1) − (F0 − B0)]/(F0 − B0), where F1 and B1 were fluorescence in the knob and background, respectively, and F0 and B0 were fluorescence at the beginning of the experiment, which were averaged from the first five control frames. Each knob corresponded to ≈20 pixels. An ORN was considered to respond to odor stimulation when ΔF/F was more than 4%, which gave the confidence level above 95% (or P < 0.05), because the maximum noise level was less than 2%. The relative size of ΔF/F was not used as a criterion to define the strength of a response; in other words, the response of an ORN was considered to be all or none. This simplified the analysis by excluding any possible effects caused by saturation of calcium-sensitive dyes in such small compartments as the dendritic knobs or by adaptation during repetitive stimulations.
Results
Odor Responses of Single ORNs Recorded by Patch Clamp.
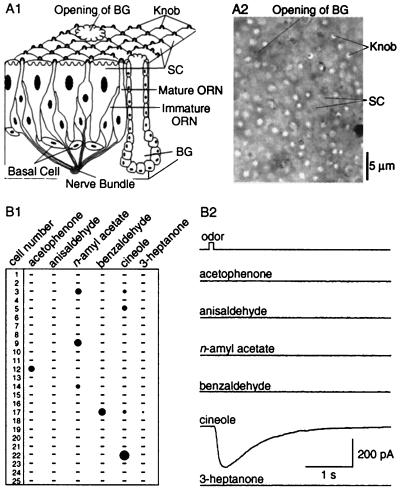
As described (18), the intact olfactory epithelial preparation permitted visualization of the dendritic knobs of mouse ORNs under infrared differential interference contrast microscopy. As seen in an en face view (Fig. 1A), the olfactory epithelium was characterized by the numerous dendritic knobs of ORNs filling the areas between large supporting cells; the evenly distributed openings of Bowman's glands were also visible.
Figure 1.
(A) Visualization of dendritic knobs of mouse ORNs. (A1) Schematic drawing of olfactory epithelium. BG, Bowman's gland. SC, supporting cell. The 10–15 cilia emanating from each knob are not drawn. (A2) The dendritic knobs of mouse ORNs were seen as bright spots in an en face view under infrared differential interference contrast microscope. (B) Responses to different odors by single ORNs, revealed by perforated patch-clamp recordings. (B1) The table summarizes the responses of 25 cells to six different odors. Identical 100-ms pulses of six different odors at 300 μM were sequentially delivered to a cell. The interval between two consecutive pulses was >30 sec. If a cell did not respond to a mixture of the six odors, it was considered not to respond to any individual odor. The area of a filled circle represents the relative amplitude of the odor-induced current; a dash indicates no detectable current. The holding potential was −60 mV. The cells were chosen randomly from all areas of olfactory epithelium. (B2) Raw data from cell 22. Of the six odors, the cell responded only to cineole.
We first tested the capabilities of individual ORNs in the intact epithelial preparations to respond to different odors by patch clamping on in situ knobs (Fig. 1B). Six different odors at 300 μM were sequentially puffed onto the epithelial surface. Of 25 cells, five responded to only one odor, one responded to two odors, and one responded to three odors with different amplitudes (Fig. 1B1). The raw data from cell no. 22 is shown; the cell responded only to cineole (Fig. 1B2). Even with the limited numbers of cells and odors tested, one can see that a single ORN can respond to different odors with distinct chemical structures (nos. 3 and 17); and a given odor (cineole) can be recognized by different subsets of ORNs. For instance, nos. 5 and 22 had different response spectra from both no. 3 and 17, despite the fact that they all responded to cineole.
Although this approach, involving patch clamping the dendritic knobs of ORNs, can provide information about how single ORNs respond to different odors, it is technically difficult and cannot efficiently provide information about spatial relations between ORNs responding to different odors. This led us to apply imaging methods to record many ORNs simultaneously.
Mosaic Organization of ORN Subsets Revealed by Calcium Imaging.
Basic response properties.
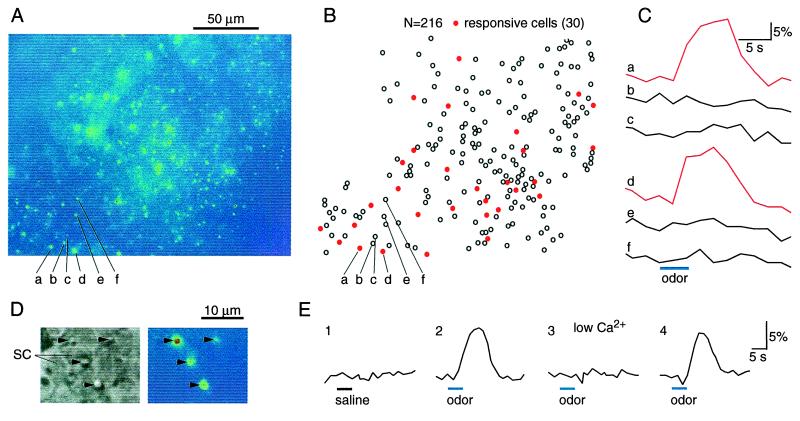
The intact epithelial preparation allowed us to visualize large populations of dendritic knobs simultaneously, which made it possible to monitor odor responses from a group of ORNs with their spatial relations intact by using calcium imaging. After a swatch of epithelium was loaded with calcium-sensitive dye, up to 400 dendritic knobs could be seen as bright spots under fluorescence illumination in a viewing area of 215 μm by 172 μm (Fig. 2A). No supporting cells were labeled. The fluorescence intensity changes (ΔF/F) of individual dendritic knobs were measured in response to a 4-sec odor stimulus containing 10 μM of each of the six odors listed in Fig. 1B. Thirty of 216 cells responded by showing increased fluorescence intensity (Fig. 2 B and C).
Figure 2.
Imaging odor responses of mouse ORNs in the intact epithelial preparation by calcium imaging techniques. (A) The preparation was viewed under fluorescence illumination, after being loaded with calcium green. (B) A total of 216 dendritic knobs are mapped for further analysis. Each circle represents a cell. Red circles (30 of 216) stand for the responsive cells to a 4-sec odor stimulus (a mixture containing 10 μM of each of the six odors listed in Fig. 1B). (C) The fluorescence intensity changes (ΔF/F) induced by the odor stimulus are measured and calculated for individual dendritic knobs; the traces corresponding to cells a–f are shown. (D) Comparison between transmitted (Left) and fluorescent (Right) images confirmed that the bright spots were dendritic knobs. The arrows mark the dendritic knobs. SC, supporting cell. (E) The fluorescence changes (ΔF/F) induced by odor stimulation was caused by calcium entry. Perfusion of saline (trace 1) or removal of Ca2+ (trace 3) eliminated odor-induced fluorescence changes. The stimulus was saline (trace 1) or an odor mixture as in Fig. 1B (traces 2–4). The bath was normal (traces 1, 2, and 4) or low Ca2+ saline (trace 3).
Comparisons between fluorescent and transmitted images confirmed that the bright spots were identical with dendritic knobs (Fig. 2D). Furthermore, they could be easily distinguished from other nonknob fluorescent objects, such as the openings of Bowman's glands. Control experiments established that the fluorescence changes during odor stimuli were likely caused by Ca2+ entry and not mechanical responses to the odor application, because applying saline did not generate such responses and removal of Ca2+ eliminated the responses (Fig. 2E).
Spatial patterns evoked by different odors.
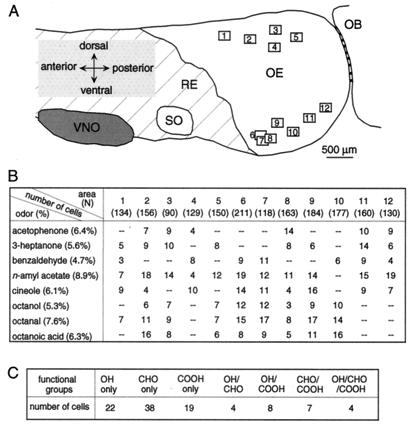
A mosaic arrangement of rodent ORNs expressing a given receptor gene subfamily within one zone has been shown by in situ hybridization (2, 3, 10). However, the spatial distribution of ORNs responding to a given odor has not been reported. For this purpose, we recorded simultaneously the responses of ORN populations induced by different odors. The epithelium attached to the septal wall was used because it is relatively easy to map the physical position of each recorded area and relate them to the gene expression zones. For instance, areas in the dorsal aspect are likely from the most dorsal zone (zone 1, ref. 2; Fig. 3A).
Figure 3.
Summary of the data obtained from the septal epithelium. (A) The positions of the recorded areas (rectangles of 215 μm by 172 μm) are numbered from 1 to 12. The standard contour of the epithelium was obtained from a 6-week-old female mouse. RE, respiratory epithelium. OE, olfactory epithelium. VNO, vomeronasal organ. SO, septal organ. OB, olfactory bulb. (B) Summary of the number of ORNs in response to different odors. The viewing areas nos. 1–12 shown in A were from 12 different preparations. N (in parentheses) stands for the total number of cells tested in each area. The percentage after each odor is the averaged percentage of cells that responded to the odor from the 12 preparations. A dash indicates the fact that an odor was not tested. All odors were at 50 μM. (C) Summary of the number of ORNs in response to odors with different functional groups. A total of 102 responsive cells were from 1,846 tested. The odors used were alcohols (OH), aldehydes (CHO), and carboxylic acids (COOH) with straight seven or eight carbon chains at a concentration of 50 μM.
The data obtained in 12 viewing areas from 12 different preparations are summarized (Fig. 3 A and B). Two examples, areas no. 8 and no. 12, are shown in Figs. 4 and 5, respectively. ORNs responding to a given odor were widely distributed and intermingled with other subtypes of ORNs (Figs. 4B and 5B), suggesting a functional mosaic similar to the molecular mosaic of ORNs expressing the same receptor gene. However, closer inspection revealed several aspects of the functional mosaic that were of particular interest.
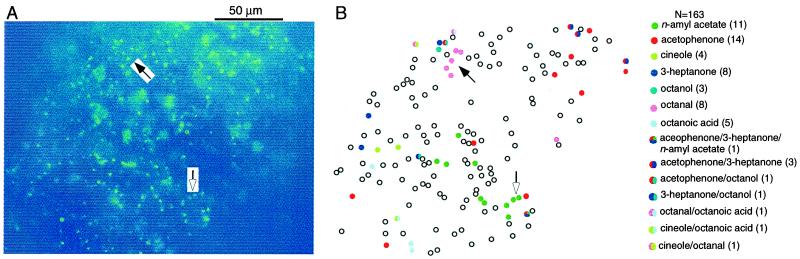
Figure 4.
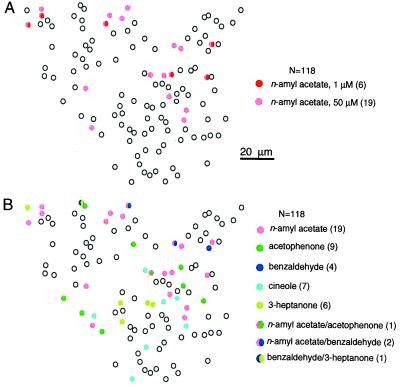
Mosaic organization of mouse ORNs in responding to different odors. (A) The preparation was viewed under fluorescence illumination, after being loaded with calcium green. (B) ORNs responded to different odors with distinct patterns. A total of 163 cells are mapped. Responsive ORNs to different odors are represented by different colors. Cells responding to more than one odor are drawn in multiple colors. The numbers in parentheses are the numbers of responsive cells to the given odor(s). Filled and hollow arrows mark the tight clusters of octanal and n-amyl acetate responsive cells, respectively. All odors were at 50 μM.
Figure 5.
Mouse olfactory epithelia respond to stronger stimuli by recruiting more ORNs. (A) More cells responded to 50 μM (pink) n-amyl acetate than to 1 μM (red). Of 118 cells, 19 responded to 50 μM and only six responded to 1 μM n-amyl acetate. (B) Cells responding to the other four odors are mapped in the same preparation as shown in A. Responsive ORNs to different odors are represented by different colors. Cells responding to more than one odor are drawn in multiple colors. The numbers in parentheses are the numbers of responsive cells to the given odor(s). All odors were at 50 μM.
First, the distribution of the responsive ORNs appeared not to be restricted within one zone. ORNs responding to all of the odors tested here were found in both the dorsal and ventral aspect of the septal epithelium, across different gene expression zones (Fig. 3 A and B). This contrasts with the restriction of ORNs expressing a given receptor within a given zone.
Second, compared with the low percentage (0.1%) of ORNs expressing a given receptor gene, the percentage of ORNs responding to a given odor was much higher. Although the numbers varied among different preparations, the averaged percentage was 4–9% (Fig. 3B). The n-amyl acetate responsive cells in viewing area no. 12 reached 16% for higher odor concentrations (Figs. 3 and 5). In all of the preparations tested, different odors induced distinct but somewhat overlapping patterns. Even with the limited set of odors tested, cells responding to more than one odor were regularly observed. In the experiment shown in Fig. 4, nine of 43 responsive cells responded to multiple odors. The implications of these results for odor encoding by ORNs are discussed later.
Third, in contrast to the random distribution reported for ORNs expressing a given receptor in the rat and mouse (2, 3), the functional mosaic of ORNs responding to a given odor in the mouse often showed some degree of clustering. Examples of two adjacent cells responding to a common stimulus were found in almost every preparation and with all of the odors tested. Tight clusters of up to five ORNs were observed in some cases. For instance, there appeared to be clustering of cells responding to n-amyl acetate and octanal in the experiment shown in Fig. 4. In the cluster responding to n-amyl acetate (green), these cells were arranged in a semicircle around the perimeter of a supporting cell when examined under the microscope (cf. Fig. 1A2). A simple hypothesis would be that these clustered cells come from a common precursor cell and express the same or similar receptor genes to give similar response spectra.
Selections for functional groups.
It has been hypothesized that functional groups in chemical compounds are essential in determining their binding properties to receptors, supported by both physiological recordings (5, 19) and computational modeling (24). Here we tested how ORNs in the intact epithelium responded to odors with different functional groups but similar carbon chain length, specifically, heptanol (7-OH), octanol (8-OH), heptanal (7-CHO), octanal (8-CHO), heptanoic acid (7-COOH), and octanoic acid (8-COOH). The odors were presented individually or as mixtures of two with the same functional group, such as 7-OH and 8-OH, etc. The data are summarized according to the tuning specificity of ORNs to different functional groups (Fig. 3C). In total, 102 of 1,846 cells from 14 preparations responded to any one of the six odors mentioned above. Of the 102 responsive cells, 79 responded specifically to one of the three functional groups, i.e., 22 to alcohol, 38 to aldehyde, and 19 to carboxylic acid only. Nineteen ORNs responded to any combination of two functional groups, i.e., four to alcohol/aldehyde, eight to alcohol/acid, and seven to aldehyde/acid. Four cells responded to all chemicals with three different functional groups. The fact that cells responding to any combination of all three functional groups are present suggests that there are a variety of ORN subtypes contributing to the recognition of odors containing these functional groups.
Effects of odor concentrations.
Individual ORNs respond to stronger odor stimuli with increased currents as seen in dose–response curves obtained by patch-clamp recordings (18, 25, 26); comparable results have been obtained by using calcium imaging of dissociated cells (19). We tested the hypothesis that olfactory epithelia respond to stronger stimuli by recruiting more ORNs. This is demonstrated clearly in Fig. 5A. Six cells responded to 1 μM n-amyl acetate, whereas 13 more cells responded to 50 μM. Mouse ORNs respond to stronger stimuli by increasing responses from individual cells and recruiting more cells, presumably because of the broad tuning and different affinities of these receptors in binding odor molecules.
Responses to pairs of enantiomers.
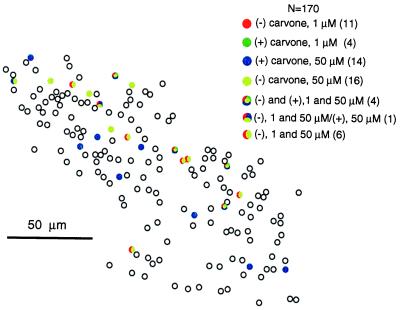
We tested the ability of mouse ORNs to distinguish between two pairs of enantiomers. Fig. 6 shows the result from an experiment where (+) and (−) carvone at 1 or 50 μM were presented. Eleven ORNs responded to (−) carvone at 1 μM, and four of the 11 cells responded to (+) carvone, too. When the concentration was increased to 50 μM, these two isomers still evoked quite different patterns. Although a common glomerulus is reported to be activated by both (+) and (−) carvone in the rat olfactory bulb (27), the different patterns of responsive cells to the two isomers in the mouse epithelium (Fig. 6) lead us to believe that there must be some glomeruli activated by only one of the two isomers. Similar to carvone, (+) and (−) limonene elicited distinct but somewhat overlapping patterns of responsive ORNs (data not shown). These data indicate that different enantiomers are also distinguished by combinations of ORN subsets in the epithelium.
Figure 6.
Mouse ORNs can distinguish between pairs of enantiomers. ORNs responded to (+) and (−) carvone at different concentrations with distinct patterns. A total of 170 cells are mapped. Responsive ORNs to the two isomers at different concentrations are represented by different colors. The numbers in parentheses are the numbers of responsive cells to the given odor(s).
Discussion
Imaging Odor Responses in the Intact Epithelium and its Potential Applications.
There are several advantages of the recording method used in this study. The ORNs in the intact epithelial preparation are closer to in vivo physiological conditions than dissociated ORNs (18). Presumably only mature ORNs are examined and recorded here because the preparation is viewed from the surface (ref. 28; cf. Fig. 1A). The recordings are made from the dendritic knobs, which are close to the cilia where olfactory signal transduction takes place. The calcium changes in the dendritic knobs have been recorded in dissociated vertebrate ORNs (29–32). Most importantly, the activities of hundreds of ORNs can be monitored simultaneously at the resolution of individual cells.
Physiological studies can potentially be carried out on these preparations with known locations in different olfactory receptor expression zones, with maintained spatial relations among neighboring ORNs. By preventing loss of cells during dissociation, the preparation is especially useful in studies carried out in gene-targeted mice in which molecular markers are introduced into the olfactory epithelium. This method has the potential to be a valuable tool to study peripheral coding of olfactory information by providing physiological data with greater efficiency than would be possible by other methods.
Implications of the Functional Mosaic Organization of ORNs for Peripheral Coding.
In the present study, we have demonstrated the mosaic organization of ORNs responding to different odors by applying calcium imaging to the intact olfactory epithelial preparation. ORNs responding to a given odor are widely distributed in the epithelium and intermingled with ORNs responding to other odors. Different odors evoke distinct but somewhat overlapping patterns in the epithelium. These results strongly support the hypothesis that odor quality and intensity are encoded by multiple combinations of ORN subsets.
How is odor quality encoded in the epithelium?
A key issue in olfactory coding is how broadly the receptors are tuned. This is not an easy question to address because of the tremendous numbers we have to deal with, i.e., millions of ORNs and thousands of odors. In this study, we have tried to answer this question from another point of view: for a given odor, how many receptors can respond?
Approximately 0.1% neurons express a single receptor gene, given the assumptions that an individual neuron only expresses a single receptor gene (4–6), that there are about 1,000 receptor genes (1) and that the total number of neurons expressing a given gene is similar among different genes. This is supported by results from in situ hybridization (2, 3, 10, 33). However, in our study the percentage of neurons responding to a given odor ranged from 4% to 9%, i.e., much higher than 0.1%, and these neurons were widely distributed, across different gene expression zones (Fig. 3). Even though these ORNs can respond to a common stimulus, it is likely that they express different receptor genes, and they share some overlapping in their response spectra because of the broad tuning of the expressed receptor proteins. We therefore estimate that approximately 40–90 different receptors are able to respond to a given odor under the conditions in the current study. This implies clearly that the quality of an odor is determined by the combinations of a variety of ORN subsets in the epithelium. This broad tuning of receptor neurons is supported by calcium imaging studies on dissociated mouse ORNs (5, 19, 34) and single-unit recordings from in vivo ORNs in rat (35).
In contrast, recent evidence from functional expression of olfactory receptor genes has pointed to relatively narrowly tuned receptors (6, 36–39). The apparent difference in the degree of olfactory receptor specificity obtained by physiological recordings and functional expression could be caused by several factors. The two approaches may be dealing with different and limited sets of odors and receptors under different experimental conditions. It is possible that broadly tuned receptors have not been found in the functional expression studies, and narrowly tuned ORNs are difficult to identify in the physiological studies. Alternatively, ORNs may indeed have broader response spectra than individual expressed receptor proteins, because of expression of multiple receptor genes in ORNs (40), or some unknown modifications of receptor proteins under physiological conditions. These questions will require further investigation.
How is odor intensity encoded in the epithelium?
There is evidence for at least two mechanisms in coding odor concentrations at the epithelial level. For a single ORN, a stronger response is elicited by a stronger stimulus within a certain concentration range (18, 19, 25, 26). For an area of epithelium, more ORNs are recruited in responding to stronger odor stimuli, presumably because these ORNs can respond to the same odor but with higher thresholds (Figs. 5A and 6A). If these ORNs express the same receptor gene but have different thresholds to a given odor because of some intrinsic mechanisms, such as different maturity stage, one may predict stronger odor stimuli will cause stronger activity at the glomeruli in the olfactory bulb, but with a similar map as weaker stimuli. On the other hand, if the different thresholds of these ORNs in responding to a given odor are because of the different receptor genes they express, one may predict that stronger odor stimuli will both induce stronger activity and recruit more glomeruli than weaker stimuli. The second possibility is supported by both 2-deoxyglucose mapping and optical imaging from the olfactory bulb (27, 41). With increased odor concentration, stronger activity for a given glomerulus and activities from more glomeruli are recorded. As with odor quality, odor intensity is encoded by combinations of ORN subsets.
Acknowledgments
We thank Drs. Wei R. Chen, Charles A. Greer, and Michael S. Singer for their insightful discussions and comments on an earlier draft of the manuscript. This work was supported by grants to G.M.S. from the National Institute on Deafness and Other Communication Disorders (R01), National Institute on Aging, National Aeronautics and Space Administration, and National Institute of Mental Health (Human Brain Project); Army Research Office (Multidisciplinary University Research Initiative); and a Public Health Service training grant to M.M.
Abbreviation
- ORN
olfactory receptor neuron
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.220301797.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.220301797
References
- 1.Buck L, Axel R. Cell. 1991;65:175–187. doi: 10.1016/0092-8674(91)90418-x. [DOI] [PubMed] [Google Scholar]
- 2.Ressler K J, Sullivan S L, Buck L B. Cell. 1993;73:597–609. doi: 10.1016/0092-8674(93)90145-g. [DOI] [PubMed] [Google Scholar]
- 3.Vassar R, Ngai J, Axel R. Cell. 1993;74:309–318. doi: 10.1016/0092-8674(93)90422-m. [DOI] [PubMed] [Google Scholar]
- 4.Chess A, Simon I, Cedar H, Axel R. Cell. 1994;78:823–834. doi: 10.1016/s0092-8674(94)90562-2. [DOI] [PubMed] [Google Scholar]
- 5.Malnic B, Hirono J, Sato T, Buck L B. Cell. 1999;96:713–723. doi: 10.1016/s0092-8674(00)80581-4. [DOI] [PubMed] [Google Scholar]
- 6.Touhara K, Sengoku S, Inaki K, Tsuboi A, Hirono J, Sato T, Sakano H, Haga T. Proc Natl Acad Sci USA. 1999;96:4040–4045. doi: 10.1073/pnas.96.7.4040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ressler K J, Sullivan S L, Buck L B. Cell. 1994;79:1245–1255. doi: 10.1016/0092-8674(94)90015-9. [DOI] [PubMed] [Google Scholar]
- 8.Vassar R, Chao S K, Sitcheran R, Nunez J M, Vosshall L B, Axel R. Cell. 1994;79:981–991. doi: 10.1016/0092-8674(94)90029-9. [DOI] [PubMed] [Google Scholar]
- 9.Mombaerts P, Wang F, Dulac C, Chao S K, Nemes A, Mendelsohn M, Edmondson J, Axel R. Cell. 1996;87:675–686. doi: 10.1016/s0092-8674(00)81387-2. [DOI] [PubMed] [Google Scholar]
- 10.Strotmann J, Wanner I, Helfrich T, Beck A, Meinken C, Kubick S, Breer H. Cell Tissue Res. 1994;276:429–438. doi: 10.1007/BF00343941. [DOI] [PubMed] [Google Scholar]
- 11.Qasba P, Reed R R. J Neurosci. 1998;18:227–236. doi: 10.1523/JNEUROSCI.18-01-00227.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mackay-Sim A, Kesteven S. J Neurophysiol. 1994;71:150–160. doi: 10.1152/jn.1994.71.1.150. [DOI] [PubMed] [Google Scholar]
- 13.Scott J W, Davis L M, Shannon D, Kaplan C. J Neurophysiol. 1996;75:2036–2049. doi: 10.1152/jn.1996.75.5.2036. [DOI] [PubMed] [Google Scholar]
- 14.Scott J W, Shannon D E, Charpentier J, Davis L M, Kaplan C. J Neurophysiol. 1997;77:1950–1962. doi: 10.1152/jn.1997.77.4.1950. [DOI] [PubMed] [Google Scholar]
- 15.Scott J W, Brierley T, Schmidt F H. J Neurosci. 2000;20:4721–4731. doi: 10.1523/JNEUROSCI.20-12-04721.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Youngentob S L, Kent P F, Sheehe P R, Schwob J E, Tzoumaka E. J Neurophysiol. 1995;73:387–398. doi: 10.1152/jn.1995.73.1.387. [DOI] [PubMed] [Google Scholar]
- 17.Kent P F, Mozell M M, Murphy S J, Hornung D E. J Neurosci. 1996;16:345–353. doi: 10.1523/JNEUROSCI.16-01-00345.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ma M, Chen W R, Shepherd G M. J Neurosci Methods. 1999;92:31–40. doi: 10.1016/s0165-0270(99)00089-8. [DOI] [PubMed] [Google Scholar]
- 19.Sato T, Hirono J, Tonoike M, Takebayashi M. J Neurophysiol. 1994;72:2980–2989. doi: 10.1152/jn.1994.72.6.2980. [DOI] [PubMed] [Google Scholar]
- 20.Lowe G, Gold G H. Nature (London) 1993;366:283–286. doi: 10.1038/366283a0. [DOI] [PubMed] [Google Scholar]
- 21.Ronnett G V, Cho H, Hester L D, Wood S F, Snyder S H. J Neurosci. 1993;13:1751–1758. doi: 10.1523/JNEUROSCI.13-04-01751.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Okada Y, Teeter J H, Restrepo D. J Neurophysiol. 1994;71:595–602. doi: 10.1152/jn.1994.71.2.595. [DOI] [PubMed] [Google Scholar]
- 23.Maue R A, Dionne V E. Pflügers Arch. 1987;409:244–250. doi: 10.1007/BF00583472. [DOI] [PubMed] [Google Scholar]
- 24.Singer M S. Chem Senses. 2000;25:155–165. doi: 10.1093/chemse/25.2.155. [DOI] [PubMed] [Google Scholar]
- 25.Firestein S, Picco C, Menini A. J Physiol (London) 1993;468:1–10. doi: 10.1113/jphysiol.1993.sp019756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kurahashi T, Menini A. Nature (London) 1997;385:725–729. doi: 10.1038/385725a0. [DOI] [PubMed] [Google Scholar]
- 27.Rubin B D, Katz L C. Neuron. 1999;23:499–511. doi: 10.1016/s0896-6273(00)80803-x. [DOI] [PubMed] [Google Scholar]
- 28.Farbmann A I. In: Cell Biology of Olfaction. Barlow P W, Bray D, Green P B, Slack J M W, editors. Cambridge, U.K.: Cambridge Univ. Press; 1992. p. 58. [Google Scholar]
- 29.Restrepo D, Okada Y, Teeter J H. J Gen Physiol. 1993;102:907–924. doi: 10.1085/jgp.102.5.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tareilus E, Noe J, Breer H. Biochim Biophys Acta. 1995;1269:129–138. doi: 10.1016/0167-4889(95)00105-2. [DOI] [PubMed] [Google Scholar]
- 31.Leinders-Zufall T, Greer C A, Shepherd G M, Zufall F. J Neurosci. 1998;18:5630–5639. doi: 10.1523/JNEUROSCI.18-15-05630.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Leinders-Zufall T, Rand M N, Shepherd G M, Greer C A, Zufall F. J Neurosci. 1997;17:4136–4148. doi: 10.1523/JNEUROSCI.17-11-04136.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Strotmann J, Wanner I, Helfrich T, Beck A, Breer H. Cell Tissue Res. 1994;278:11–20. doi: 10.1007/BF00305773. [DOI] [PubMed] [Google Scholar]
- 34.Bozza T C, Kauer J S. J Neurosci. 1998;18:4560–4569. doi: 10.1523/JNEUROSCI.18-12-04560.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Duchamp-Viret P, Chaput M A, Duchamp A. Science. 1999;284:2171–2174. doi: 10.1126/science.284.5423.2171. [DOI] [PubMed] [Google Scholar]
- 36.Krautwurst D, Yau K W, Reed R R. Cell. 1998;95:917–926. doi: 10.1016/s0092-8674(00)81716-x. [DOI] [PubMed] [Google Scholar]
- 37.Zhao H, Ivic L, Otaki J M, Hashimoto M, Mikoshiba K, Firestein S. Science. 1998;279:237–242. doi: 10.1126/science.279.5348.237. [DOI] [PubMed] [Google Scholar]
- 38.Murrell J R, Hunter D D. J Neurosci. 1999;19:8260–8270. doi: 10.1523/JNEUROSCI.19-19-08260.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wetzel C H, Oles M, Wellerdieck C, Kuczkowiak M, Gisselmann G, Hatt H. J Neurosci. 1999;19:7426–7433. doi: 10.1523/JNEUROSCI.19-17-07426.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rawson N E, Eberwine J, Dotson R, Jackson J, Ulrich P, Restrepo D. J Neurochem. 2000;75:185–195. doi: 10.1046/j.1471-4159.2000.0750185.x. [DOI] [PubMed] [Google Scholar]
- 41.Stewart W B, Kauer J S, Shepherd G M. J Comp Neurol. 1979;185:715–734. doi: 10.1002/cne.901850407. [DOI] [PubMed] [Google Scholar]