Abstract
The use of molecular hydrogen as electron donor for energy generation is a defining characteristic of the hydrogenotrophic methanogens, an ancient group that dominates the phylum Eury archaeota. We present here a global study of changes in mRNA abundance in response to hydrogen availability for a hydrogenotrophic methanogen. Cells of Methanococcus maripaludis were grown by using continuous culture to deconvolute the effects of hydrogen limitation and growth rate, and microarray analyses were conducted. Hydrogen limitation markedly increased mRNA levels for genes encoding enzymes of the methanogenic pathway that reduce or oxidize the electron-carrying deazaflavin, coenzyme F420. F420-dependent redox functions in energy-generating metabolism are characteristic of the methanogenic Archaea, and the results show that their regulation is distinct from other redox processes in the cell. Rapid growth increased mRNA levels of the gene for an unusual hydrogenase, the hydrogen-dependent methylenetetrahydromethanopterin dehydrogenase.
Keywords: coenzyme F420, microarrays, Methanococcus maripaludis
Methanogenic Archaea may have been among earth's earliest organisms and today play essential roles in a variety of anaerobic habitats (1, 2). The methanogens are phylogenetically widespread, comprising three of the seven classes in the phylum Euryarchaeota (3). Most species are hydrogenotrophic, obtaining energy by using H2 to reduce carbon dioxide to methane. H2 is a major electron donor for methanogenesis in a variety of anaerobic habitats and a key intermediate in the anaerobic food web. During interspecies H2 transfer, the concentration of the H2 that is produced by heterotrophic bacteria and eukaryotes during the fermentation of complex organic compounds is maintained at very low levels by the hydrogenotrophic methanogens. These low H2 concentrations are essential for the complete degradation of complex organic compounds in anaerobic environments. In addition, on early earth geochemical H2 may have supported methanogenesis (1). Because methane is a potent greenhouse gas, this process may have contributed to an early global warming effect.
Because of the central role of H2 in interspecies H2 transfer and as electron donor for hydrogenotrophic methanogens, H2 is expected to play an important role in the global regulation of gene expression. Previous studies (e.g., ref. 4) have observed specific changes in gene expression with changes in H2 availability but they could not distinguish the effects of H2 availability from the effects of growth rate because nutrient limitation slows growth in batch culture. A critical feature of our analysis was the use of continuous culture. This approach was necessary not only for superior reproducibility, but also for determination of the effects of hydrogen and growth rate independently. Methanococcus maripaludis, a mesophilic hydrogenotrophic methanogen from marine sediments, served as our representative species. As a laboratory model, M. maripaludis is distinguished by relatively rapid growth, a well developed genetic system (5, 6), a complete genome sequence (7), established systems for transcriptome arrays and quantitative proteomics (8), and continuous culture under defined nutrient conditions (9).
Results and Discussion
Transcriptome microarrays were used to compare mRNA abundance in M. maripaludis in chemostat cultures where growth was limited by H2 or two other nutrients, leucine or phosphate. During the comparisons of H2- to phosphate-limited growth and H2- to leucine-limited growth, growth rate and cell density were held constant. Thus, the effects of the nutritional limitations were independent of growth rate and cell density. By using three limiting nutrients, it was also possible to identify the causative factor for the regulatory responses. For example, a mRNA that is increased by H2 limitation would show an increase under H2 limitation relative to leucine limitation as well as under H2 limitation relative to phosphate limitation. In addition, cultures were compared at 4.5- to 4.8-fold different growth rates while cell density was held constant, and the limiting nutrient was either H2 or phosphate. Thus, it was possible to clearly identify genes affected by growth rate.
All mRNA ratios determined in this study and their P values are tabulated in supporting information (SI) Table 3. Differential levels were generally regarded as significant if P < 10−5. Consistent trends across operons were considered significant at higher P values.
Distinct Effect of H2 Limitation on Genes Encoding F420-Dependent Processes.
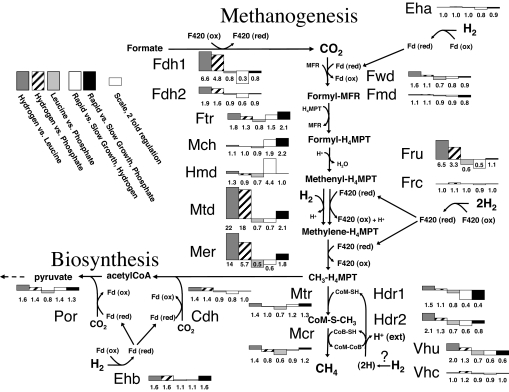
A distinct set of genes displayed mRNA levels that were 4- to 22-fold higher under H2 limitation than under phosphate or leucine limitation (Table 1 and Fig. 1). All of these genes encoded enzymes of the methanogenic pathway (10) that use the electron-carrying deazaflavin, coenzyme F420. The markedly regulated genes included those encoding the selenocysteine-containing coenzyme F420-reducing hydrogenase (Fru; Mmp1382–1385), the primary reducer of F420 during growth on H2 when selenium is present (11), as is the case in our medium. Also included was a cluster that contains genes for one of two formate dehydrogenases (Fdh1; Mmp1297–1302). A second Fdh (Fdh2; Mmp0138–0139) was similarly regulated although not as markedly. The Fdhs reduce F420 when formate instead of H2 is the electron donor (12). The remaining genes that were markedly regulated by H2 were those encoding F420-dependent methylenetetrahydromethanopterin dehydrogenase (Mtd; Mmp0372) and F420-dependent methylenetetrahydromethanopterin reductase (Mmp0058), central steps in methanogenesis. In addition to increased mRNA abundance with H2 limitation, the fru and fdh genes had significantly decreased mRNA abundance with increasing growth rate when H2 was the limiting nutrient. However, no growth rate effect was observed when phosphate was the limiting nutrient. In continuous culture the limiting nutrient concentration typically increases with growth rate, so the decreased mRNA levels with growth rate could actually be an effect of H2.
Table 1.
Expression ratios of selected genes
| ORF number | Gene name | H2/leu |
H2/Pi |
Rapid/slow (H2-lim) |
Rapid/slow (Pi-lim) |
||||
|---|---|---|---|---|---|---|---|---|---|
| Ratio | P value | Ratio | P value | Ratio | P value | Ratio | P value | ||
| F420-interacting proteins | |||||||||
| Mmp0058 | mer | 3.81 (14.0) | 1.8E-15 | 2.51 (5.7) | 4.8E-11 | −0.65 (0.64) | 3.4E-04 | 0.87 (1.8) | 8.4E-04 |
| Mmp0372 | mtd | 4.49 (22.5) | 5.6E-16 | 4.14 (17.6) | 2.3E-10 | −0.50 (0.71) | 2.2E-02 | 1.09 (2.1) | 1.0E-01 |
| Mmp1298 | fdhA1* | 2.73 (6.6) | 2.3E-05 | 2.26 (4.8) | 1.5E-04 | −1.77 (0.29) | 9.0E-03 | −0.25 (0.84) | 1.7E-02 |
| Mmp1385 | fruB* | 2.73 (6.6) | 3.7E-12 | 1.83 (3.6) | 3.0E-09 | −1.12 (0.46) | 4.1E-08 | 0.14 (1.1) | 3.9E-01 |
| Other methanogenesis and carbon assimilation proteins | |||||||||
| Mmp0127 | hmd | 0.34 (1.3) | 7.9E-02 | −0.20 (0.87) | 2.6E-01 | 2.12 (4.3) | 1.1E-08 | 0.03 (1.0) | 5.9E-01 |
| Mmp1155 | hdrB1* | 0.66 (1.6) | 9.8E-05 | 0.22 (1.2) | 1.5E-02 | −1.38 (0.38) | 1.1E-10 | −1.41 (0.38) | 6.1E-10 |
| Mmp1559 | mcrA* | 0.48 (1.4) | 7.2E-03 | −0.30 (0.81) | 6.7E-03 | −0.18 (0.88) | 4.6E-02 | 0.26 (1.2) | 3.0E-04 |
| Mmp1609 | ftr | 0.81 (1.8) | 6.3E-13 | 0.42 (1.3) | 6.9E-07 | 0.62 (1.5) | 5.1E-11 | 1.04 (2.1) | 4.0E-11 |
| Mmp1622 | ehbM* | 0.62 (1.5) | 4.4E-06 | 0.66 (1.6) | 5.0E-08 | 0.10 (1.1) | 4.8E-03 | 0.65 (1.6) | 9.2E-04 |
Ratios are log2. Nonlogged ratios are given in parentheses.
*Proteins encoded by multiple genes; the values given are for the gene with the median H2/leu value.
Fig. 1.
Pathways of methanogenesis and CO2 assimilation in M. maripaludis. Enzymes are listed next to each reaction. Bar graphs show the log2 expression ratios for the gene(s) coding for each enzyme. For enzymes with multiple genes, the bar graph shows the average log2 expression ratio. Numbers below bars indicate corresponding nonlogged ratios. Fmd, formylmethanofuran dehydrogenase (molybdenum-containing); Ftr, formylmethanofuran:tetrahydromethanopterin formyltransferase; Mch, methenyltetrahydromethanopterin cyclohydrolase; Mer, methylenetetrahydromethanopterin reductase; Mtr, methyltetrahydromethanopterin:coenzyme M methyltransferase; Cdh, carbon monoxide dehydrogenase/acetyl CoA synthase. Log2 ratios for leucine limitation vs. phosphate limitation are from unpublished work.
F420 is unusually abundant in methanogenic Archaea, where it is coupled to many of the redox reactions in methanogenesis (13, 14). The distinct effect of H2 limitation on genes encoding F420-dependent processes suggests that cells may use F420 to compartmentalize electron flow. Recent evidence demonstrates that the intracellular pool of reduced F420 is in equilibrium with the H2 concentration in the medium (15). Therefore, F420-linked processes would be directly coupled to and most sensitive to H2 levels. In this light, the up-regulation of F420-dependent processes appears to be a strategy to maintain electron flow for the methanogenic pathway by increasing the levels of enzymes when substrate (H2) is low. In contrast, processes linked to ferredoxin or NAD(P), which were not markedly affected by H2 limitation (below), may be buffered from rapid changes in H2 concentrations, allowing the cells to perform biosynthetic reactions that depend on reduced ferredoxin or NAD(P)H (16–18) when H2 levels are low.
Genes Moderately Regulated by Hydrogen Limitation.
Genes for redox functions that do not involve F420 were at most moderately affected by H2 limitation. Genes for the biosynthetic energy-conserving hydrogenase B (Ehb; Mmp1621–1629), carbon monoxide dehydrogenase/acetyl CoA synthase (Mmp0979–0985), and pyruvate oxidoreductase (Por; Mmp1502–1507), which use ferredoxins as electron carriers, had only moderately increased mRNA levels under H2 limitation. We previously observed a similar increase in mRNA for these genes when Ehb was mutationally inactivated (8, 19), suggesting a response to maintain flux through this pathway when the electron donor is limiting. Genes for the step of the methanogenic pathway that uses ferredoxin [the energy-conserving hydrogenase A (Eha)] were not significantly affected by H2 limitation, nor could an effect of H2 limitation be discerned for the methylreductase (Mcr), the non-F420-reducing hydrogenases (Vhu or Vhc), or the heterodisulfide reductases (Hdrs). Similarly, numerous biosynthetic steps depend on NAD or NADP; none was found to be significantly affected by H2 limitation.
Genes Regulated by Growth Rate.
The gene for the H2-dependent methylenetetrahydromethanopterin dehydrogenase (Hmd; Mmp0127) was increased 4-fold in mRNA abundance with rapid growth under hydrogen-limited conditions. Although not reflected in the arrays, real-time RT-PCR indicated moderate (1.8-fold) increased mRNA with increased growth rate under phosphate limitation as well (Table 2). The more moderate effect of growth rate when phosphate was the limiting nutrient indicates complex regulation that depends on the limiting nutrient as well as growth rate. It is notable that Hmd was the enzyme of methanogenesis whose mRNA was most increased with growth rate. In contrast, the genes for the F420-associated proteins, including Mtd, which functions in the same step with Hmd, were not significantly affected by growth rate. These results suggest that Hmd, which is an unusual iron–sulfur cluster-free hydrogenase (20) with low affinity for hydrogen (21), takes on increasing importance as growth rate increases. Genes for two steps in methanogenesis, formylmethanofuran:tetrahydromethanopterin formyltransferase (Mmp1609) and methenyltetrahydromethanopterin cyclohydrolase (Mmp1191), had ≈2-fold increased mRNA levels with increased growth rate (Fig. 1 and Table 1). These steps are consecutive in the pathway and are the only steps that do not directly involve electron transfer or energy conservation.
Table 2.
Comparison of real-time RT-PCR and array results
| ORF number | Gene name | H2/leu |
H2/Pi |
Rapid/slow (H2-lim) |
Rapid/slow (Pi-lim) |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| RT-PCR | Array | P value | RT-PCR | Array | P value | RT-PCR | Array | P value | RT-PCR | Array | P value | ||
| Mmp0009 | 0.11 (1.1) | −0.03 (0.98) | 6.5E-01 | 0.08 (1.1) | 0.05 (1.0) | 5.4E-01 | 0.06 (1.04) | −0.02 (0.99) | 6.8E-01 | 0.12 (1.1) | 0.13 (1.1) | 4.9E-02 | |
| Mmp0127 | hmd | 0.00 (1.0) | 0.34 (1.3) | 7.9E-02 | 0.09 (1.1) | −0.20 (0.87) | 2.6E-01 | 1.80 (3.5) | 2.12 (4.3) | 1.1E-08 | 0.82 (1.8) | 0.03 (1.0) | 5.9E-01 |
| Mmp0372 | mtd | 3.13 (8.8) | 4.49 (22.5) | 5.6E-16 | 2.61 (6.1) | 4.14 (17.6) | 2.3E-10 | 0.32 (1.2) | −0.50 (0.71) | 2.2E-02 | 1.79 (3.5) | 1.09 (2.1) | 1.0E-01 |
| Mmp1100 | 2.40 (5.3) | 1.42 (2.7) | 5.4E-06 | 0.10 (1.1) | −0.12 (0.92) | 3.9E-01 | −1.01 (0.50) | −2.39 (0.19) | 4.0E-10 | −0.36 (0.78) | −1.37 (0.39) | 6.2E-07 | |
| Mmp1503 | porE | 0.57 (1.5) | 0.61 (1.5) | 1.1E-05 | 0.07 (1.0) | 0.40 (1.3) | 2.3E-07 | 1.55 (2.9) | 0.64 (1.6) | 2.6E-07 | 1.81 (3.5) | 0.34 (1.3) | 1.9E-01 |
Ratios are log2. Nonlogged ratios are given in parentheses. PCR values are average log2 expression ratios from two biological replicates.
Some genes of methanogenesis had moderately decreased mRNA with increased growth rate. M. maripaludis contains genes for two Hdrs and two non-F420-reducing hydrogenases. An operon containing genes for Hdr1 B and C subunits followed by genes of unknown function (Mmp1154–1165) was significantly down-regulated 2.5-fold with increasing growth rate (Fig. 1 and Table 1). Genes for the Hdr2 B and C subunits (Mmp1053–1054) were also moderately but significantly down-regulated with rapid growth (Fig. 1). An apparent operon (Mmp1691–1697) containing genes for Vhu along with a Hdr A-subunit as well as the B-subunit of tungsten-containing formylmethanofuran dehydrogenase (FwdB) showed similar trends, as did an operon containing genes for the remaining subunits of Fwd (Mmp1244–1249). The similar trend observed for the Hdr and Vhu genes is consistent with the hypothesis that these enzymes function in the pathway of electron flow from H2 to the heterodisulfide intermediate that forms between coenzyme M and coenzyme B during methanogenesis. Genes for Eha (Mmp1447–1465) were also slightly, but consistently, down-regulated with increasing growth rate.
Finally, many genes of the methanogenic pathway and early biosynthetic steps had decreased mRNA levels with leucine limitation. The effects of leucine and phosphate limitation, as well as additional effects of growth rate, will be reported (unpublished work).
Real-Time RT-PCR.
To check specific results and assess the overall quality of the array results, real-time RT-PCR was conducted for selected genes. The analysis included two genes, mtd and hmd (Mmp0372 and Mmp0127) that encode alternative enzymes for the second reduction step in methanogenesis (Fig. 1) and were markedly regulated in certain comparisons, and an important biosynthetic gene porE (Mmp1503), that was moderately regulated. Also included were a putative transcriptional regulator gene (Mmp1100) and Mmp0009, a putative DNA primase gene that was chosen for its apparent lack of regulation. Most genes showed similar trends with the two measurement techniques (Table 2). In only one case did real-time RT-PCR fail to detect regulation where the arrays had indicated moderate regulation, porE in the hydrogen vs. phosphate comparison.
Comparison with Previous Studies.
Several previous studies have reported the effects of different hydrogen delivery rates, or of different degrees of mixing, on the expression of methanogenesis genes. However, because of the use of batch culture, the effects of hydrogen availability could not be distinguished from the effects of growth rate. Comparison with our results suggests that both effects may have occurred, and that similar patterns of gene expression may occur in diverse species of methanogens. Among those genes of methanogenesis that we found to be markedly up-regulated by hydrogen limitation in M. maripaludis, studies in Methanothermobacter thermautotrophicus (4, 22) led to a similar conclusion for homologs of Mtd, methylenetetrahydromethanopterin reductase, F420-reducing hydrogenases, and Fdh (23). Similarly, an analysis at the protein level in Methanocaldococcus jannaschii found an elevated level of Mtd at lower partial pressure of H2 (24). Genes for Fdhs in M. maripaludis were expressed at higher levels when formate replaced hydrogen as the electron donor (12), consistent with our findings here under hydrogen limitation. It is notable that in those experiments, the differential expression of the Fdh genes was observed with promoter-lacZ fusions, indicating regulation at the level of transcription initiation. On the other hand, our findings suggest that other effects may actually have been caused by growth rate. hmd in M. thermautotrophicus had decreased mRNA under a low hydrogen regime, and a proteomic analysis in M. jannaschii found lower Hmd with lower partial pressure of H2 (24). In batch culture low hydrogen coincides with low growth rate, and the effects may have been a result of growth rate differences, consistent with our observation in M. maripaludis of decreased hmd mRNA levels with lower growth rate. Alternatively, regulation by hydrogen of Hmd at a post-mRNA level could occur. In other cases, differences between our results and previous ones could be caused by species differences. The M. thermautotrophicus study reported increased mcr mRNA levels under low hydrogen. In contrast, we observed no significant regulation of mcr in M. maripaludis with hydrogen limitation or growth rate. Among the differences between the two species is the presence of a second Mcr in M. thermautotrophicus, Mrt, which is lacking in M. maripaludis. In M. thermautotrophicus mrt was affected in the opposite direction to mcr and regulation might be associated with the presence of the two alternative enzymes.
Conclusion.
A microarray comparison of M. maripaludis chemostat cultures revealed distinct sets of genes whose mRNA levels vary with hydrogen limitation or growth rate. Genes for F420-dependent functions were distinguished by marked increases of mRNA levels with hydrogen limitation, whereas mRNA abundance for Hmd was positively affected by rapid growth. More modest effects of hydrogen limitation and growth rate were also observed among the genes of methanogenesis and early biosynthetic steps. The mechanisms for these effects on mRNA abundance remain to be investigated.
Methods
Bacterial Strains, Media, and Culture Conditions.
M. maripaludis strain S52, a leucine auxotroph, has been described (9). Cultures (1-liter volumes) of S52 were grown at 37°C in chemostats as described (9). The standard medium pumped into the chemostat vessels was McA, a defined complex medium. The standard gassing regime was 110 ml/min H2, 40 ml/min CO2, 35 ml/min N2, and 15.5 ml/min H2S/N2 mixture (1:99). Cultures were stirred at 1,000 rpm with two 52-mm, six-bladed, Rushton-type impellers. For nutrient limitation comparisons, growth rate was 0.125 h−l and conditions were modified as follows. For hydrogen-limited cultures H2 was lowered to 20 ml/min and N2 was increased to 125 ml/min. For phosphate limitation phosphate was lowered from the standard 0.8 mM to 0.15 mM. For leucine limitation, leucine was lowered from the standard 0.5 mM to 0.15 mM. For hydrogen-limited growth rate comparisons, cultures were grown at 0.042 or 0.2 h−l. For phosphate-limited growth rate comparisons, cultures were grown at 0.042 or 0.19 h−l. For hydrogen-limited cultures at 0.042 h−l, H2 was delivered at 8 ml/min and N2 was delivered at 135 ml/min. For hydrogen-limited cultures at 0.2 h−l, H2 was delivered at 29 ml/min and N2 was delivered at 115 ml/min. For phosphate-limited cultures at both growth rates, phosphate was 0.15 mM. All cultures were allowed to stabilize and remain at a constant cell density, OD660 = 0.6, for at least three retention times before harvesting. For comparison, when the standard medium and gassing regime were used (no limiting conditions), steady-state OD660 was >1.0.
Cell Harvesting.
To harvest cells, the chemostat vessel was pressurized to 10 psi, and culture was allowed to flow out through the sampling tube. Culture was rapidly cooled by either of two methods. In one method, 400 ml of culture were collected in a 1-liter round-bottom flask that had been precooled on ice and rapidly cooled to 4°C by swirling in a dry ice–ethanol bath. Culture was then divided into 50-ml portions, and cells were collected by centrifugation at 4°C at 8,000 × g for 5 min. In the other method, culture was collected directly into centrifuge tubes through stainless-steel tubing bathed in ice water and centrifuged as above. Cell pellets were stored at −80°C.
RNA Extraction, Array Hybridization, and Real-Time RT-PCR.
RNA was extracted, treated with DNase, and checked for quality. Cy3- and Cy5-labeled cDNAs were produced and hybridized to spotted PCR product arrays. Real-time RT-PCR was performed for selected samples. Details are given in ref. 8.
Array Data Collection and Analysis.
Nutrient limitation comparisons were conducted with four biological replicates for each condition. Growth rate comparisons were conducted with three biological replicates. Each comparison involved four technical replicates as described (8). For nutrient limitation comparisons and growth rate comparisons under hydrogen-limited conditions, slides were read at the University of Washington Center for Expression Arrays and data were quantified as described (8). For growth rate comparisons under phosphate-limited conditions, slides were read on a Scan-Array Express reader (PerkinElmer, Wellesley, MA) at the Institute for Systems Biology, Seattle, WA. Average expression ratios and unadjusted P values were calculated for each gene across all replicates by using the S+ array analyzer, using Loess normalization and a t test for significance to calculate P values. Gene expression ratios were viewed by using a TIGR MultiExperiment Viewer (25). Data are available at the Gene Expression Omnibus database under accession no. GSE6747.
Supplementary Material
Acknowledgments
We thank Roger Bumgarner, Murray Hackett, and Fred Taub for assistance with array analysis. This work was supported by National Institutes of Health Grants GM60403 and GM74783, Department of Energy Basic Research for the Hydrogen Fuel Initiative Grant DE-FG02-05ER15709, Department of Energy Microbial Cell Project Grant DE-FG03-01ER15252, and National Aeronautics and Space Administration Astrobiology Institute Grant NCC 2-1273.
Abbreviations
- Eha
energy-conserving hydrogenase A
- Ehb
energy-conserving hydrogenase B
- Fdh
formate dehydrogenase
- Fwd
formylmethanofuran dehydrogenase (tungsten-containing)
- Mtd
coenzyme F420-dependent methylenetetrahydromethanopterin dehydrogenase
- Hmd
H2-dependent methylenetetrahydromethanopterin dehydrogenase
- Fru
F420-reducing hydrogenase
- Frc
F420-reducing hydrogenase (cysteine-containing)
- Vhu
non-F420-reducing hydrogenase (selenocysteine-containing)
- Vhc
non-F420-reducing hydrogenase (cysteine-containing)
- Mcr
methyl coenzyme M reductase
- Hdr
heterodisulfide reductase
- Por
pyruvate oxidoreductase.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE6747).
This article contains supporting information online at www.pnas.org/cgi/content/full/0701157104/DC1.
References
- 1.Kasting JF, Siefert JL. Science. 2002;296:1066–1068. doi: 10.1126/science.1071184. [DOI] [PubMed] [Google Scholar]
- 2.Wolfe RS. ASM News. 1996;62:529–534. [Google Scholar]
- 3.Boone DR, Castenholz RW, editors. The Archaea and the Deeply Branching Phototrophic Bacteria. New York: Springer; 2001. [Google Scholar]
- 4.Reeve JN, Nolling J, Morgan RM, Smith DR. J Bacteriol. 1997;179:5975–5986. doi: 10.1128/jb.179.19.5975-5986.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tumbula DL, Whitman WB. Mol Microbiol. 1999;33:1–7. doi: 10.1046/j.1365-2958.1999.01463.x. [DOI] [PubMed] [Google Scholar]
- 6.Moore BC, Leigh JA. J Bacteriol. 2005;187:972–979. doi: 10.1128/JB.187.3.972-979.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hendrickson EL, Kaul R, Zhou Y, Bovee D, Chapman P, Chung J, Conway de Macario E, Dodsworth JA, Gillett W, Graham DE, et al. J Bacteriol. 2004;186:6956–6969. doi: 10.1128/JB.186.20.6956-6969.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xia Q, Hendrickson EL, Zhang Y, Wang T, Taub F, Moore BC, Porat I, Whitman WB, Hackett M, Leigh JA. Mol Cell Proteomics. 2006;5:868–881. doi: 10.1074/mcp.M500369-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Haydock AK, Porat I, Whitman WB, Leigh JA. FEMS Microbiol Lett. 2004;238:85–91. doi: 10.1016/j.femsle.2004.07.021. [DOI] [PubMed] [Google Scholar]
- 10.Deppenmeier U. Prog Nucleic Acid Res Mol Biol. 2002;71:223–283. doi: 10.1016/s0079-6603(02)71045-3. [DOI] [PubMed] [Google Scholar]
- 11.Noll I, Muller S, Klein A. Genetics. 1999;152:1335–1341. doi: 10.1093/genetics/152.4.1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wood GE, Haydock AK, Leigh JA. J Bacteriol. 2003;185:2548–2554. doi: 10.1128/JB.185.8.2548-2554.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Deppenmeier U. Prog Nucleic Acid Res Mol Biol. 2002;71:223–283. doi: 10.1016/s0079-6603(02)71045-3. [DOI] [PubMed] [Google Scholar]
- 14.Keltjens J, van der Drift C. FEMS Microbiol Rev. 1986;39:259–303. [Google Scholar]
- 15.de Poorter LM, Geerts WJ, Keltjens JT. Microbiology (Reading, UK) 2005;151:1697–1705. doi: 10.1099/mic.0.27679-0. [DOI] [PubMed] [Google Scholar]
- 16.Tersteegen A, Linder D, Thauer RK, Hedderich R. FEBS. 1997;244:862–868. doi: 10.1111/j.1432-1033.1997.00862.x. [DOI] [PubMed] [Google Scholar]
- 17.Lin WC, Yang YL, Whitman WB. Arch Microbiol. 2003;179:444–456. doi: 10.1007/s00203-003-0554-3. [DOI] [PubMed] [Google Scholar]
- 18.Bock AK, Kunow J, Glasemacher J, Schonheit P. FEBS. 1996;237:35–44. doi: 10.1111/j.1432-1033.1996.0035n.x. [DOI] [PubMed] [Google Scholar]
- 19.Porat I, Kim W, Hendrickson EL, Xia Q, Zhang Y, Wang T, Taub F, Moore BC, Anderson IJ, Hackett M, et al. J Bacteriol. 2006;188:1373–1380. doi: 10.1128/JB.188.4.1373-1380.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Korbas M, Vogt S, Meyer-Klaucke W, Bill E, Lyon EJ, Thauer RK, Shima S. J Biol Chem. 2006;281:30804–30813. doi: 10.1074/jbc.M605306200. [DOI] [PubMed] [Google Scholar]
- 21.Thauer RK, Klein AR, Hartmann GC. Chem Rev. 1996;96:3031–3042. doi: 10.1021/cr9500601. [DOI] [PubMed] [Google Scholar]
- 22.Morgan RM, Pihl TD, Nolling J, Reeve JN. J Bacteriol. 1997;179:889–898. doi: 10.1128/jb.179.3.889-898.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nolling J, Reeve JN. J Bacteriol. 1997;179:899–908. doi: 10.1128/jb.179.3.899-908.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mukhopadhyay B, Johnson EF, Wolfe RS. Proc Natl Acad Sci USA. 2000;97:11522–11527. doi: 10.1073/pnas.97.21.11522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Saeed AI, Sharov V, White J, Li J, Liang W, Bhagabati N, Braisted J, Klapa M, Currier T, Thiagarajan M, et al. BioTechniques. 2003;34:374–378. doi: 10.2144/03342mt01. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.