Abstract
The p21 RAS subfamily of small GTPases, including KRAS, HRAS, and NRAS, regulates cell proliferation, cytoskeletal organization, and other signaling networks, and is the most frequent target of activating mutations in cancer. Activating germline mutations of KRAS and HRAS cause severe developmental abnormalities leading to Noonan, cardio-facial-cutaneous, and Costello syndrome, but activating germline mutations of NRAS have not been reported. Autoimmune lymphoproliferative syndrome (ALPS) is the most common genetic disease of lymphocyte apoptosis and causes autoimmunity as well as excessive lymphocyte accumulation, particularly of CD4−, CD8− αβ T cells. Mutations in ALPS typically affect CD95 (Fas/APO-1)-mediated apoptosis, one of the extrinsic death pathways involving TNF receptor superfamily proteins, but certain ALPS individuals have no such mutations. We show here that the salient features of ALPS as well as a predisposition to hematological malignancies can be caused by a heterozygous germline Gly13Asp activating mutation of the NRAS oncogene that does not impair CD95-mediated apoptosis. The increase in active, GTP-bound NRAS augments RAF/MEK/ERK signaling, which markedly decreases the proapoptotic protein BIM and attenuates intrinsic, nonreceptor-mediated mitochondrial apoptosis. Thus, germline activating mutations in NRAS differ from other p21 Ras oncoproteins by causing selective immune abnormalities without general developmental defects. Our observations on the effects of NRAS activation indicate that RAS-inactivating drugs, such as farnesyltransferase inhibitors should be examined in human autoimmune and lymphocyte homeostasis disorders.
Keywords: autoimmunity, B cell lymphoma 2-interacting mediator of cell death, intrinsic apoptosis, lymphoma, lymphoproliferation
The RAS genes (NRAS, KRAS, and HRAS) encode 21-kDa proteins that are members of the superfamily of small GTP-binding proteins, which have diverse intracellular signaling functions including control of cell proliferation, growth, and apoptosis (1). Somatic activating mutations in RAS are present in up to 30% of all human cancers (2). Germline RAS pathway mutations have only recently been described as causing the related Costello (HRAS), Noonan (PTPN11, KRAS, SOS1), and cardiofaciocutaneous syndromes (KRAS, BRAF, MEK1, and MEK) (3–7). Individuals with these syndromes typically present with severe developmental anomalies in various combinations of facial abnormalities, heart defects, short stature, skin and genital abnormalities, and mental retardation (8). Defects in the immune system have not been reported. Patients with Costello and Noonan syndromes have an increased propensity to solid and hematopoietic tumors, respectively (3, 8). Germline mutations in NRAS have not yet been described.
The autoimmune lymphoproliferative syndrome (ALPS) (OMIM 601859/603909) is the most common genetic disorder of lymphocyte apoptosis and is characterized by chronic accumulation of nonmalignant lymphocytes, defective lymphocyte apoptosis, and an increased risk for the development of hematological malignancies (9). A signature of the disease is the accumulation of αβ T cells lacking the CD4 and CD8 coreceptors that are termed “double-negative” T cells (DNTs: CD4−, CD8− TCRαβ+ cells). These cells bear no known relationship to thymic DNTs, a stage that occurs before αβ TCR gene rearrangements in ontogeny (10). According to genotype, ALPS can be classified as types Ia, Ib, and II, which are due to germline mutations in CD95 (TNFRSF6), CD95 ligand (TNFSF6), and caspase 10 (CASP10), respectively (10–13). Additionally, somatic mutations of CD95 in αβ DNTs can also cause ALPS of type Im (mosaic) (14). All of these mutations impair extrinsic, Fas receptor-mediated apoptosis (10). An enigma has been the ALPS individuals who have no defects in CD95 pathway apoptosis (some ALPS type III patients) (9, 10). This group encompasses a large number of individuals and is probably genetically heterogeneous. In an attempt to unveil new genetic defects, we investigated alternative apoptosis pathways in ALPS type III and identified one ALPS patient with a unique defect in cytokine withdrawal-induced apoptosis due to an activating NRAS mutation.
Results
Defective IL-2 Withdrawal-Induced Apoptosis in a Patient with Clinical and Laboratory Hallmarks of ALPS.
The intrinsic mitochondrial pathway of apoptosis can be triggered by developmental cues in the thymus or bone marrow, cytokine deprivation, DNA damage, or treatment with cytotoxic drugs (15, 16). To screen for defects in this pathway, we exposed activated lymphocytes from individuals with salient features of ALPS (lymphadenopathy and increased αβ DNTs) but normal CD95-mediated apoptosis, to inducers of intrinsic apoptosis including staurosporine, γ-radiation, and cytokine withdrawal. We identified an individual whose lymphocytes clearly resisted death induced by IL-2 withdrawal (Fig. 1A).
Fig. 1.
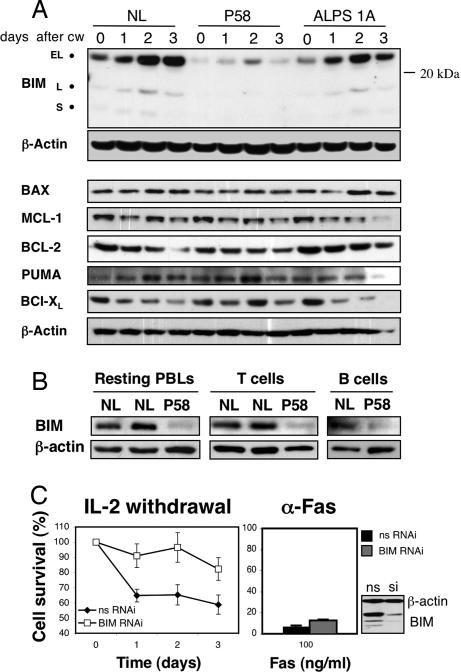
Defective cytokine withdrawal-induced apoptosis in P58 lymphocytes. (A–D) Activated peripheral blood mononuclear cells (PBLs) from normal volunteers (NL A and B), from a patient with an inactivating Fas mutation (ALPS 1A), and from P58 were cultured in media without IL-2 for the indicated periods of time (A); or treated for 18 h with anti-Fas (Apo1.3) antibody (B), staurosporine (C), or γ-irradiation (D) at the indicated doses. (E) (a–d) Merged views of active BAX (green) and Hoechst nuclear staining (blue) of cells from P58 and a normal control (NL) at 0 h (a and b) and 72 h after cytokine withdrawal (c and d). (e–h) Merged views of staining with the mitochondrial marker Hsp60 (green), cytochrome c (red), and Hoechst staining (blue) at 0 h (e and f) or 72 h after IL-2 withdrawal (g and h). (F) Quantitation using fluorescence microscopy of the relative proportion of cells in the experiment in E showing active BAX expression (activ. BAX), diffuse cytochrome c (diff. cyt. c), or apoptotic (apop.) nuclei. Data shown are the representative of two or three independent experiments. Shown is mean ± SD.
The affected individual [National Institutes of Health (NIH) cohort patient 58, P58] is a 49-year-old male with lifelong overexpansion of lymphocytes, and an unusual history of two malignancies: childhood leukemia and early adulthood lymphoma, both successfully treated [supporting information (SI) Table 1]. Peripheral blood immunophenotyping revealed a sustained elevation in αβ DNT cells over several years and other findings frequently seen in ALPS, including an elevated percentage of CD5+ B cells and low numbers of CD27+ B cells (17). However, other features such as low CD25/HLA-DR ratio and high numbers of CD3+CD57+ were not seen (SI Table 1). Lymph node biopsy performed elsewhere and reviewed at the NIH revealed reactive follicular hyperplasia and sinus histiocytosis, but DNT cells were not prominent. Several serum autoantibodies were detected and elevations of several T helper 2 cytokines including IL-5, -6, -8, -10, and -13 were observed (SI Tables 1 and 2). Based on the published NIH ALPS diagnostic criteria of elevated DNT on peripheral blood, chronic lifelong nonmalignant hyperplasia and defective lymphocyte apoptosis, P58 received a provisional clinical diagnosis of ALPS with recognition that this is not a typical clinical presentation of this disease.
Despite defective IL-2 withdrawal death, we found no abnormalities in apoptosis induced by an agonistic anti-APO-1 (CD95) antibody (Apo1.3), staurosporine, or γ-radiation (Fig. 1 B–D). Moreover, activated T cells did not spontaneously proliferate or secrete cytokines, but did show persistent proliferation after IL-2 withdrawal compared with normal cells (SI Fig. 6 and data not shown). Thus, lymphocytes from P58 demonstrated a specific defect in the intrinsic pathway of apoptosis.
During cytokine withdrawal, the proapoptotic B cell lymphoma 2 (BCL-2) family members BAX and BAK are activated and oligomerize at mitochondrial surfaces, causing permeabilization of the outer membrane, which releases cytochrome c and triggers apoptosis (15, 16). After IL-2 withdrawal, we found that P58 cells were markedly defective in BAX activation and mitochondrial cytochrome c release, correlating with a decreased percentage of apoptotic nuclei compared with control cells after cytokine withdrawal, but not staurosporine treatment, clearly indicating a selective mitochondrial apoptosis defect (Fig. 1 E and F, and SI Fig. 7 A and B).
Diminished BCL-2-Interacting Mediator of Cell Death (BIM) Levels in P58.
Critical initiators of intrinsic apoptosis that act upstream of BAX and BAK are the BCL-2 homology 3 (BH3) subclass of the BCL-2 family (15). Knockout mice lacking BIM, a BH3-only protein, manifest immune abnormalities including lymphocyte accumulation, autoimmunity, and various apoptosis defects, specially cytokine withdrawal-induced cell death (18). We therefore measured BIM levels and found markedly reduced basal and induced levels after IL-2 withdrawal from activated T cells in P58 compared with normal or ALPS type 1A controls (Fig. 2A). This defect was specific to BIM because other proapoptotic (PUMA, BAX) and antiapoptotic (BCL-2, BCL-XL, MCL-1) BCL-2 family members were unaffected (Fig. 2A). We also observed a constitutive BIM deficiency in resting peripheral blood lymphocytes (PBLs) as well as purified T and B cells from P58 (Fig. 2B).
Fig. 2.
BIM down-regulation in P58 lymphocytes. (A) Analysis by immunoblotting of BIM and other BCL-2 members expression after IL-2 withdrawal (CW) in PBLs from a NL, P58, and an ALPS 1A patient. The isoforms extra-long (EL), long (L), and short (S) are indicated. β-Actin is a loading control. (B) Resting ex vivo PBLs and purified T and B cells from normals (NL) and P58 were lysed and assessed for BIM expression by immunoblotting. (C) Activated human lymphocytes were transfected with either nonsilencing RNAi (nsRNAi) or with a small interfering oligonucleotide directed at BIM (BIM RNAi) for 3 days, and then deprived of IL-2 or treated with anti-Fas antibody. Silencing efficiency was assessed by immunoblotting (Lower Right) for the nonspecific (ns) and the silencing (si) transfections. Data shown are the representative of three independent experiments. Shown is mean ± SD.
In an additional effort to demonstrate the importance of BIM for mitochondrial apoptosis in human cells, we used small interfering RNA (siRNA) to silence BIM expression in otherwise normal human lymphocytes. BIM suppression caused resistance to IL-2 withdrawal-, but not Fas- or staurosporine-induced apoptosis, as seen in P58 (Fig. 2C and data not shown).
Taken together, these data demonstrate that cells from P58 have a selective decrease of the proapoptotic protein BIM, which seems critical for cytokine withdrawal-induced apoptosis.
P58 Has a Gain-of-Function NRAS Mutation.
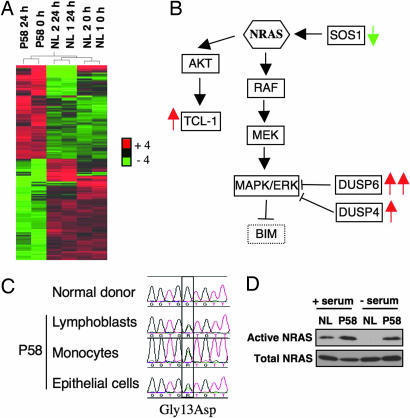
To uncover the genetic basis of the BIM defect in P58, we sequenced the genomic locus of BIM and some of its regulators, including FOXO3a, FOXO1, FOXO4, ERK1/2, and JNK1/2, but found no mutations (15, 19). We then carried out an unbiased screen by interrogating mRNA expression patterns in healthy controls and P58 lymphocytes at 0 and 24 h after IL-2 withdrawal by using microarrays. Two hundred and five probe sets were differentially expressed in P58 compared with controls (Fig. 3A; a complete list is available in SI Dataset, and the microarray dataset is deposited in the GEO public database). Surprisingly, the gene expression pattern in P58 indicated the possible constitutive activation of the small GTPase, neuroblastoma RAS (NRAS) (Fig. 3B). This included up-regulation of two dual-specificity phosphatases (DUSP4 and -6) that are inhibitors of ERK signaling and were previously shown to be overexpressed in tumor cell lines containing somatic activating mutations in NRAS (20).
Fig. 3.
Identification of a de novo gain-of-function NRAS mutation in P58. (A) Heat map and relational dendrogram results of a microarray study demonstrating the 205 probe sets differentially expressed by cells from P58 compared with cells from two normal subjects, NL1 and NL2. Activated lymphocytes were lysed at 0 and 24 h after IL-2 withdrawal, and mRNA expression was analyzed by using oligonucleotide microarrays (Affymetrix U133Plus2.0). (B) Simplified diagram demonstrating differential expression in P58 cells of genes in the NRAS/RAF/ERK pathway. Red and green arrows indicate the genes up- or down-regulated in P58, respectively, compared with controls. (C) Sequencing of NRAS by using genomic DNA from P58 lymphoblasts, monocytes, and buccal epithelial cells, all demonstrating a heterozygous G-to-A substitution, which changes codon 13 from glycine to aspartic acid. (D) Active GTP-bound NRAS was immunoprecipitated before and after serum withdrawal in a NL and P58, by using a Raf-1 (RBD)-GST fusion protein as bait. The total quantity of NRAS in cell lysates before immunoprecipitation is also shown.
We examined the nucleotide sequence of NRAS in P58 and found a single heterozygous G-to-A transition causing a nonconservative aspartic acid substitution for glycine at codon 13 (G13D) (Fig. 3C). To exclude the possibility that this mutation was a somatic event, we analyzed genomic DNA from various cells and tissues including lymphoblasts, unseparated resting peripheral blood mononuclear cells, purified monocytes, EBV-transformed B cells generated years before the diagnosis, as well as buccal epithelial cells (Fig. 3C and data not shown). The mutation was found in all samples suggesting it is germline in origin. Because neither of the patient's parents, his sibling, or his two children harbored the mutant allele (data not shown), the activating mutation is probably a de novo event, as characteristically observed with KRAS and HRAS or, less likely, a very early embryonic mutation causing mosaicism (3–6). Importantly, the very same amino acid change in NRAS arises somatically in human pediatric and adult myeloid and lymphoid malignancies, but was never before documented in nonmalignant cells or tissues (21, 22).
Mutations in codons 12, 13, and 61 are known to stabilize RAS proteins in an active, GTP-bound state by reducing intrinsic GTPase activity and causing resistance to GTPase-activating proteins (23, 24). Consistent with the genetic alteration, we found that active NRAS was increased in P58 lymphocytes in serum-rich and, especially, in serum-poor conditions (Fig. 3D). We therefore hypothesized that the “gain-of-function” NRAS mutation may result in hyperactivation of the RAS/RAF/ERK pathway, which can negatively regulate BIM expression (25–27).
Hyperactive NRAS Induces BIM Down-Regulation Through ERK in Human Lymphocytes.
To investigate the relationship between hyperactive NRAS and BIM suppression, we transfected human lymphocytes with wild-type (NRASWT) or mutant NRAS (NRASG13D). Overexpression of NRASWT and especially NRASG13D reduced BIM in H-9 cells and primary normal human lymphocytes (Fig. 4 A and B), reminiscent of HRAS overexpression in epithelial cells (28). We consistently observed a 3- to 4-fold reduction in BIM, although this could be an underestimate of the effect because the transfection efficiency did not reach 100% in all cases.
Fig. 4.
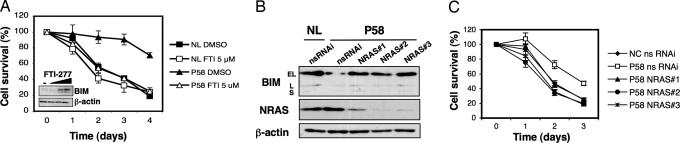
BIM down-regulation by NRAS through ERK. (A and B) Expression constructs for hemagglutinin (HA)-tagged wild type (WT) or active mutant (G13D) NRAS were transfected into the human lymphoid cell line H-9 (A) or primary human activated lymphocytes (B), followed 48 h later by quantification of BIM expression by immunoblotting. The numbers beneath each lane are the relative intensities of the bands in arbitrary units compared to the corresponding control lane. (C) Activated lymphocytes from two NL and from P58 were deprived of IL-2 and treated with either the DMSO vehicle or MEK1 inhibitors PD98059 (PD) (20 μM) or U0126 (10 μM) for 18 h, after which expression of BIM and BAX were analyzed by immunoblotting. (D) Lymphocytes from P58 and a normal control (NL) were deprived of IL-2 and treated daily with DMSO or PD98059 (20 μM). Apoptosis was measured daily by flow cytometry. Data shown are representative of three or more independent experiments. Shown is mean ± SD. EV, empty vector.
The most widely studied RAS effector proteins are either in the RAF/MEK/ERK pathway or the phosphatidylinositol 3-kinase pathway (1, 2). We found that chemical inhibition of MEK1, the MAP kinase kinase immediately upstream of ERK1/2, by using the drugs PD98059 or U0126 rescued BIM protein expression in P58 lymphocytes and NRAS-overexpressing H9 cells (Fig. 4C and data not shown). Remarkably, ERK inhibition also restored apoptosis after IL-2 withdrawal in P58 lymphocytes (Fig. 4D). Despite the strong increase in BIM protein levels after MEK inhibition, BIM mRNA varied little (SI Fig. 8A), consistent with a posttranscriptional, rather than transcriptional, down-regulation of BIM by ERK. This was also true when resting lymphocytes from P58 were treated with MEK inhibitors in IL-2-rich conditions, and BIM protein and mRNA levels were measured (SI Fig. 8 B and C). By contrast, the inhibitors LY294002 and wortmannin had no effect on BIM levels in P58 lymphocytes indicating that the phosphatidylinositol 3-kinase pathway was not involved (SI Fig. 9 and data not shown). Thus, active NRAS down-regulates BIM through the induction of the RAF/MEK/ERK pathway.
Rescue of the Apoptotic Defect by Farnesylation Inhibition and NRAS Silencing.
To further demonstrate the causative role of active NRAS in decreased BIM and defective lymphokine withdrawal apoptosis, we used two approaches. First, we used farnesyltransferase inhibitors (FTIs) that block the membrane localization and function of RAS proteins and are currently under clinical investigation for the treatment of human cancers (29). Treatment with one such pharmaceutical, FTI-277, corrected the apoptotic defect and increased BIM levels in P58 lymphocytes (Fig. 5A). This drug had a negligible effect on apoptosis and BIM levels in normal cells (Fig. 5A).
Fig. 5.
Correction of the apoptotic defect in P58 by inhibition of farnesylation and RNA silencing. (A) Activated lymphocytes from P58 and a normal control (NL) were deprived of IL-2 and treated daily with DMSO or FTI-277 (5 μM). (Inset) Resting lymphocytes from P58 were treated with DMSO (−) or 2, 5, or 10 μM FTI-277, and BIM expression was analyzed by immunoblotting. (B) Activated lymphocytes from a NL and P58 were transfected with nonsilencing (nsRNAi) or three different NRAS-targeted siRNA oligonucleotides (NRAS#1/2/3), and BIM expression was measured 3 days later by immunoblotting. (C) Activated lymphocytes from a NL and P58 were transfected as described in B and subjected to IL-2 withdrawal, and apoptosis was measured daily for the indicated period of time. Data shown are representative of three or more independent experiments. Shown is mean ± SD.
Second, we decreased NRAS expression in P58 lymphocytes by using siRNA. Each of three different siRNA oligonucleotides reduced NRAS expression and restored BIM levels and sensitivity to apoptosis in P58 cells (Fig. 5 B and C). By contrast, NRAS siRNAs had no significant effect on BIM or sensitivity to IL-2 withdrawal in normal cells (SI Fig. 10). These results verify that the heterozygous mutation causes an abnormal gain-of-function that is reversed by silencing or functional inactivation of NRAS.
Discussion
NRAS was identified as a transforming factor in neuroblastoma and other malignancies, but its principal physiological role in humans has been uncertain (1). Our genetic evidence suggests that an activating germline NRAS mutation causes BIM down-regulation and defective intrinsic mitochondrial apoptosis prominently in lymphocytes, leading to the key features resembling ALPS and hematopoietic malignancies. This study is an example of how gene profiling can facilitate the discovery of the molecular underpinnings of a genetic disorder.
The human phenotype associated with an activating NRAS mutation contrasts with the previously known human genetic disorders due to p21 RAS oncoprotein pathways, including Costello syndrome (HRAS), Noonan syndrome (PTPN11, KRAS, SOS1) and cardiofaciocutaneous syndrome (MEK1, MEK2, B-RAF, and KRAS) that comprise developmental aberrations and neoplastic transformation of various mesodermal and ectodermal tissues (3–7). None of these developmental anomalies is present in P58. One plausible explanation for the different phenotypes is that the RAS isoforms are known to exhibit functional heterogeneity that may be due to their C-terminal variable regions that may influence cellular localization and isoform-specific binding partners (30). Our data suggests that “neuroblastoma” or NRAS is an historical misnomer when considering the physiological function in humans of this gene suggested here, and other data in the literature supports this conclusion. In rodents, an NRAS gene knockout causes subtle immune deficiencies, despite the fact that the mice are overall healthy (31). By contrast, KRAS deficiency causes embryonic lethality, whereas HRAS knockout mice are completely normal (32, 33). Transgenic mice bearing activating NRAS mutations develop several hematological tumors, such as leukemia, lymphoma, mastocytosis, and rare mammary carcinomas (32, 33). The absence of developmental features and the completely different phenotype of a human germline NRAS mutation might explain the fact that no mutation in NRAS was ever detected in any of the developmental syndromes cited above, despite efforts to find one (3–7). Taken together, these data indicate that NRAS has an immune regulatory function and its absence or gain-of-function affects primarily hematopoietic cells.
Our observations reveal a vital role of intrinsic mitochondrial apoptosis in peripheral lymphocyte homeostasis and tolerance in humans. Although gene manipulation in rodents suggested that this pathway was important for peripheral immune homeostasis, our human data validate the hypothesis with some surprising twists. The NRAS mutation caused a clinical phenotype that, in several important aspects, is similar to other ALPS patients, with modestly elevated TCR-αβ+CD4−CD8− T cells, chronic lymphoid accumulation, and a clear propensity to hematological tumors. However, certain other immunophenotypic and histological features of ALPS were not seen such as marked expansions of DNTs in the lymph nodes and elevated HLA-DR+ T cells and CD57+ T cells in the periphery (17). The apoptosis defect is also clearly different from all previous ALPS cases and underscores a confusing point in the literature. Originally, the ALPS phenotype was based on defects in TCR-induced apoptosis indicating a deficiency of the propriocidal regulatory mechanism. Because many cases over the years were identified with mutations in the Fas receptor, it was generally assumed that ALPS could be defined by a defect in Fas-induced apoptosis. If we adopt a broader view of apoptosis defects that could disturb peripheral lymphocyte homeostasis, then P58 reveals another derangement of this regulatory process. Because we favor this view, it is reasonable to consider P58's lymphocyte apoptosis defect to fulfill one of the diagnostic criteria of ALPS. Because he also exhibits increased αβDNTs and expanded secondary lymphoid tissue, he would manifest all of the required features for a diagnosis of ALPS. Because his overall phenotype and genotype are distinctive, and clearly different from ALPS types I and II, we provisionally name this condition ALPS, type IV (type III represents undefined molecular pathogenesis).
The active NRAS phenotype we observed is different from homozygous deficiency of BIM in rodents, because the latter does not cause an increase in CD4−CD8− αβ T cells, and induces the expansion of other cell types, such as granulocytes (18). These differences may reflect either the residual expression of BIM or the stimulatory effects of NRAS on ERK and other downstream RAS effectors, which could have direct mitogenic effects that are not triggered by BIM modulation. Moreover, there are several regulatory levels between NRAS and BIM where other factors could account for the difference between the BIM knockout mice and P58.
The mechanism of BIM suppression under conditions of hyperactive NRAS is not clear. In other cellular models of hyperactive KRAS, BIM is down-regulated through phosphorylation, ubiquitination, and degradation by the proteasomal machinery (26). Here, we show that BIM protein levels are severely depressed in resting and activated T cells from P58 compared with normals, despite equivalent basal mRNA levels. Additionally, the usual up-regulation of BIM mRNA after IL-2 withdrawal was impaired in cells from P58. Also, BIM protein returned to almost normal levels on MEK/ERK blockage, but mRNA remained the same. In our preliminary experiments (data not shown), there was no change in BIM levels on proteasomal blockade in lymphocytes from P58. Taken together, these data suggest a double mechanism for BIM suppression via hyperactive NRAS: inhibition of up-regulation of mRNA and translational inhibition, but this remains to be formally shown. Our new understanding of NRAS suggests that RAS antagonists such as FTIs could have beneficial effects on disorders of lymphocyte homeostasis and autoimmunity in addition to cancer.
Materials and Methods
Cells and Treatments.
Patients were studied under an NIH Institutional Review Board-approved ALPS research protocol after obtaining informed consent. PBLs were isolated, activated, and cultivated as previously described (34). Activated T cells were cultured in 100 units/ml recombinant human IL-2 (Roche Applied Science, Indianapolis, IN) for at least 6 days, and for withdrawal studies, the cells were then washed three times with PBS and resuspended at 1 × 106 cells/ml in complete media without IL-2 and cultured for different periods of time. Other apoptosis assays used staurosporine (Calbiochem, EMD Biosciences, San Diego, CA), an agonistic anti-Fas antibody Apo1.3 (Alexis, San Diego, CA) or γ-irradiation. The level of apoptosis was determined by staining with 50 ng/ml propidium iodide, and 40 nM 3,3′-dihexyloxacarbocyanine iodide (DiOC6) (Calbiochem, EMD Biosciences) and flow cytometry using a constant time acquisition as described (34). Chemical inhibitors PD98059, U0126, LY294002, and FTI-277 were obtained from Calbiochem.
DNA and Protein Analyses.
Immunoblotting and confocal microscopic analysis used the following antibodies: anti-BIM, anti-BAK (Stressgen, Ann Arbor, MI); anti-BCL-2, anti-p27kip1, anti-MCL-1, anti-β-actin, anti-BAX, anti-cytochrome c (clone 7H8.2C12), anti-cytochrome c oxidase IV subunit II, anti-BCL-XL (BD Biosciences, San Jose, CA); anti-PUMA (Axxora, San Diego, CA); anti-AIF, anti-NRAS (clone F155) (Santa Cruz, Santa Cruz, CA), anti-cytochrome c (6H2.B4; BD Pharmingen, San Diego, CA), anti-HSP60 (E-1; Santa Cruz), and anti-BAX (NT; Upstate, Charlottesville, VA). Active GTP-bound NRAS was immunoprecipitated by using the EZ detection Ras activation kit (Pierce, Rockford, IL), according to the manufacturer's protocol. The immunoprecipitated proteins were separated by SDS/PAGE and probed with an anti-NRAS antibody (F-155; Santa Cruz). Quantitative PCR was done on a 7700 ABI PRISM instrument with all of the probes and primers purchased from the same company (Applied Biosystems, Foster City, CA) by using established procedures. The reactions were carried out and normalized to the 18S rRNA signal as described (35). The values were again normalized against a normal control value in each experiment, thus to calculate the fold changes in comparison with the normal. DNA sequencing was carried out on purified PCR products by using puReTaq Ready-To-Go PCR beads (Amersham Biosciences, Piscataway, NJ), and then directly sequenced by using ABI Prism BigDye (version 1.1) terminators and analyzed on an ABI 3100 Sequencer (Applied Biosystems).
siRNA and Transient Transfections.
Activated PBLs were transfected with either a small interfering oligonucleotide RNA (siRNA) or a scrambled nonsilencing control oligo (nsRNA) by using the Nucleofection system (Amaxa, Koln, Germany). siRNAs were designed online by using the software BLOCK-iT RNAi designer and purchased from Invitrogen (San Diego, CA). Assessment of knockdown efficiency and experiments were performed 3 days later by immunoblotting. The sequences of the relevant oligoribonucleotides are described in SI Methods. Plasmids expressing human wild-type NRAS (kindly provided by Silvio Gutkind, National Institute of Dental and Craniofacial Research, NIH, Bethesda, MD) and G13D mutant and were transiently transfected into appropriate cells.
Microarray Analysis.
RNA expression in P58 was compared with that in two normal controls (NL1, NL2) at 0 and 24 h after IL-2 withdrawal from activated cycling T lymphocytes. Total RNA was hybridized to human Affymetrix (Santa Clara, CA) U133Plus2.0 microarrays following standard Affymetrix procedures (Affymetrix) and samples were stained and scanned in the Functional Genomics and Proteomics Facility by using the Affymetrix GeneChip Scanner. More detailed methods are available in SI Methods.
Supplementary Material
Acknowledgments
We thank C. Logun for technical assistance; T. Inaba (Hiroshima University, Hiroshima, Japan), J. Gavard (National Institutes of Health), and S. Gutkind for reagents; R. Germain, R. Siegel, and A. Snow for critical reading of the manuscript; and S. Patel for help with cytokine measurement. Cynthia P. Nix was supported by the Howard Hughes Medical Institute–NIH Research Scholar Program. This work was supported by the National Institute of Allergy and Infectious Diseases NIH Intramural Research Program.
Abbreviations
- ALPS
autoimmune lymphoproliferative syndrome
- DNT
double-negative T cell
- BCL-2
B cell lymphoma 2
- BIM
BCL-2-interacting mediator of cell death
- siRNA
small interfering RNA
- FTI
farnesyltransferase inhibitor
- PBL
peripheral blood lymphocyte.
Footnotes
The authors declare no conflict of interest.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE7345).
This article contains supporting information online at www.pnas.org/cgi/content/full/0702975104/DC1.
References
- 1.Malumbres M, Barbacid M. Nat Rev Cancer. 2003;3:459–465. doi: 10.1038/nrc1097. [DOI] [PubMed] [Google Scholar]
- 2.Barbacid M. Annu Rev Biochem. 1987;56:779–827. doi: 10.1146/annurev.bi.56.070187.004023. [DOI] [PubMed] [Google Scholar]
- 3.Aoki Y, Niihori T, Kawame H, Kurosawa K, Ohashi H, Tanaka Y, Filocamo M, Kato K, Suzuki Y, Kure S, et al. Nat Genet. 2005;37:1038–1040. doi: 10.1038/ng1641. [DOI] [PubMed] [Google Scholar]
- 4.Rodriguez-Viciana P, Tetsu O, Tidyman WE, Step AL, Conger BA, Cruz MS, McCormick F, Paven KA. Science. 2006;3:1287–1290. doi: 10.1126/science.1124642. [DOI] [PubMed] [Google Scholar]
- 5.Schubbert S, Zenker M, Rowe SL, Boll S, Klein C, Bollag G, van der Burgt I, Musante L, Kalscheuer V, Wehner LE, et al. Nat Genet. 2006;38:331–336. doi: 10.1038/ng1748. [DOI] [PubMed] [Google Scholar]
- 6.Niihori T, Aoki Y, Narumi Y, Neri G, Cave H, Verloes A, Okamoto N, Hennekam RC, Gillessen-Kaesbach G, Wieczorek D, et al. Nat Genet. 2006;38:294–296. doi: 10.1038/ng1749. [DOI] [PubMed] [Google Scholar]
- 7.Tartaglia M, Pennacchio LA, Zhao C, Yadav KK, Fodale V, Sarkozy A, Pandit B, Oishi K, Martinelli S, Schackwitz W, et al. Nat Genet. 2007;39:75–79. doi: 10.1038/ng1939. [DOI] [PubMed] [Google Scholar]
- 8.Gelb BD, Tartaglia M. Hum Mol Genet. 2006;15:R220–R226. doi: 10.1093/hmg/ddl197. [DOI] [PubMed] [Google Scholar]
- 9.Oliveira JB, Fleisher T. Curr Opin Allergy Clin Immunol. 2004;4:497–503. doi: 10.1097/00130832-200412000-00005. [DOI] [PubMed] [Google Scholar]
- 10.Bidere N, Su HC, Lenardo MJ. Annu Rev Immunol. 2006;24:321–352. doi: 10.1146/annurev.immunol.24.021605.090513. [DOI] [PubMed] [Google Scholar]
- 11.Fisher GH, Rosenberg FJ, Straus SE, Dale JK, Middleton LA, Lin AY, Strober W, Lenardo MJ, Puck JM. Cell. 1995;81:935–946. doi: 10.1016/0092-8674(95)90013-6. [DOI] [PubMed] [Google Scholar]
- 12.Wang J, Zheng L, Lobito A, Chan FK, Dale J, Sneller M, Yao X, Puck JM, Straus SE, Lenardo MJ. Cell. 1999;98:47–58. doi: 10.1016/S0092-8674(00)80605-4. [DOI] [PubMed] [Google Scholar]
- 13.Wu J, Wilson J, He J, Xiang L, Schur PH, Mountz JD. J Clin Invest. 1996;98:1107–1113. doi: 10.1172/JCI118892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Holzelova E, Vonarbourg C, Stolzenberg MC, Arkwright PD, Selz F, Prieur AM, Blanche S, Bartunkova J, Vilmer E, Fischer A, et al. N Engl J Med. 2004;351:1409–1418. doi: 10.1056/NEJMoa040036. [DOI] [PubMed] [Google Scholar]
- 15.Strasser A. Nat Rev Immunol. 2005;5:189–200. doi: 10.1038/nri1568. [DOI] [PubMed] [Google Scholar]
- 16.Danial NN, Korsmeyer SJ. Cell. 2004;116:205–219. doi: 10.1016/s0092-8674(04)00046-7. [DOI] [PubMed] [Google Scholar]
- 17.Bleesing JJ, Brown MR, Straus SE, Dale JK, Siegel RM, Johnson M, Lenardo MJ, Puck JM, Fleisher TA. Blood. 2001;98:2466–2473. doi: 10.1182/blood.v98.8.2466. [DOI] [PubMed] [Google Scholar]
- 18.Bouillet P, Metcalf D, Huang DC, Tarlinton DM, Kay TW, Kontgen F, Adams JM, Strasser A. Science. 1999;286:1735–1738. doi: 10.1126/science.286.5445.1735. [DOI] [PubMed] [Google Scholar]
- 19.Strasser A, Pellegrini M. Trends Immunol. 2004;25:610–615. doi: 10.1016/j.it.2004.08.012. [DOI] [PubMed] [Google Scholar]
- 20.Bloethner S, Chen B, Hemminki K, Muller-Berghaus J, Ugurel S, Schadendorf D, Kumar R. Carcinogenesis. 2005;26:1224–1232. doi: 10.1093/carcin/bgi066. [DOI] [PubMed] [Google Scholar]
- 21.Bos JL, Toksoz D, Marshall CJ, Verlaan-de Vries M, Veeneman GH, van der Eb AJ, van Boom JH, Janssen JW, Steenvoorden AC. Nature. 1985;315:726–730. doi: 10.1038/315726a0. [DOI] [PubMed] [Google Scholar]
- 22.Lubbert M, Mirro J, Jr, Miller CW, Kahan J, Isaac G, Kitchingman G, Mertelsmann R, Herrmann F, McCormick F, Koeffler HP. Blood. 1990;75:1163–1169. [PubMed] [Google Scholar]
- 23.Rodriguez-Viciana P, Tetsu O, Oda K, Okada J, Rauen K, McCormick F. Cold Spring Harb Symp Quant Biol. 2005;70:461–467. doi: 10.1101/sqb.2005.70.044. [DOI] [PubMed] [Google Scholar]
- 24.Vetter IR, Wittinghofer A. Science. 2001;294:1299–1304. doi: 10.1126/science.1062023. [DOI] [PubMed] [Google Scholar]
- 25.Ley R, Balmanno K, Hadfield K, Weston C, Cook SJ. J Biol Chem. 2003;278:18811–18816. doi: 10.1074/jbc.M301010200. [DOI] [PubMed] [Google Scholar]
- 26.Ley R, Ewings KE, Hadfield K, Cook SJ. Cell Death Differ. 2005;12:1008–1014. doi: 10.1038/sj.cdd.4401688. [DOI] [PubMed] [Google Scholar]
- 27.Ley R, Ewings KE, Hadfield K, Howes E, Balmanno K, Cook SJ. J Biol Chem. 2004;279:8837–8847. doi: 10.1074/jbc.M311578200. [DOI] [PubMed] [Google Scholar]
- 28.Tan TT, Degenhardt K, Nelson DA, Beaudoin B, Nieves-Neira W, Bouillet P, Villunger A, Adams JM, White E. Cancer Cell. 2005;7:227–238. doi: 10.1016/j.ccr.2005.02.008. [DOI] [PubMed] [Google Scholar]
- 29.Karp JE, Lancet JE. Curr Mol Med. 2005;5:643–652. doi: 10.2174/156652405774641052. [DOI] [PubMed] [Google Scholar]
- 30.Mor A, Philips MR. Annu Rev Immunol. 2006;24:771–800. doi: 10.1146/annurev.immunol.24.021605.090723. [DOI] [PubMed] [Google Scholar]
- 31.de Castro IP, Diaz R, Malumbres M, Hernandez MI, Jagirdar J, Jimenez M, Ahn D, Pellicer A. Cancer Res. 2003;63:1615–1622. [PubMed] [Google Scholar]
- 32.Johnson L, Greenbaum D, Cichowski K, Mercer K, Murphy E, Schmitt E, Bronson RT, Umanoff H, Edelmann W, Kucherlapati R, et al. Genes Dev. 1997;11:2468–2481. doi: 10.1101/gad.11.19.2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Esteban LM, Vicario-Abejon C, Fernandez-Salguero P, Fernandez-Medarde A, Swaminathan N, Yienger K, Lopez E, Malumbres M, McKay R, Ward JM, et al. Mol Cell Biol. 2001;21:1444–1452. doi: 10.1128/MCB.21.5.1444-1452.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bidere N, Snow AL, Sakai K, Zheng L, Lenardo MJ. Curr Biol. 2006;16:1666–1671. doi: 10.1016/j.cub.2006.06.062. [DOI] [PubMed] [Google Scholar]
- 35.Livak KJ, Schmittgen TD. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.