Abstract
Chromosome instability and aneuploidy are hallmarks of cancer, but it is not clear how changes in the chromosomal content of a cell contribute to the malignant phenotype. Previously we have shown that we can readily isolate highly proliferative tumor cells and their revertants from highly invasive tumor cell populations, indicating how phenotypic shifting can contribute to malignant progression. Here we show that chromosome instability and changes in chromosome content occur with phenotypic switching. Further, we show that changes in the copy number of each chromosome quantitatively impose a proportional change in the chromosome transcriptome ratio. This correlation also applies to subchromosomal regions of derivative chromosomes. Importantly, we show that the changes in chromosome content and the transcriptome favor the expression of a large number of genes appropriate for the specific tumor phenotype. We conclude that chromosome instability generates the necessary chromosome diversity in the tumor cell populations and, therefore, the transcriptome diversity to allow for environment-facilitated clonal expansion and clonal evolution of tumor cell populations.
Keywords: aneuploidy, glioma, invasion, proliferation, HGF/SF
Nowell first proposed the clonal evolution of tumor cell populations to explain how malignant tumors arise over time (1). Tumor progression results from genetic variability within the tumor cell population that allows for clonal expansion of more aggressive tumor phenotypes (1, 2). Although chromosome instability and the resulting cytogenetic heterogeneity are the most readily recognized genetic events associated with tumor progression (3–5) and may be responsible for tumor evolution and progression, precisely how they contribute to the malignant phenotype is not clear. Invasion and proliferation are crucial requirements for tumor progression, and we have chosen to study these steps in glioblastoma tumor cells (6). Glioblastoma cells invade normal brain tissue (7); after surgical resection, residual invasive cells can quickly regain a proliferative phenotype, progressing to a more aggressive tumor (8). Because glioblastomas rarely metastasize from the CNS, the sequential selection of invasive and proliferative tumor cells constitutes the main theme for this tumor's progression (7). It is, therefore, critically important to understand the molecular mechanisms that permit high-frequency phenotypic switching and control tumor cell proliferation and invasion (6).
c-Met and its ligand, hepatocyte growth factor/scatter factor (HGF/SF), can regulate both proliferative and invasive phenotypes of glioblastoma tumor cells (9). We have previously shown that alternating between proliferative and invasive phenotypes was critically linked with switching between the Myc and Ras/MAPK pathways, respectively (6). Starting with a highly invasive population, DB-P, we were able to select two subclones, DB-A2 and DB-A6, that are highly proliferative or both invasive and proliferative, respectively. The parental DB-P cells and each subclone showed distinct in vitro and in vivo phenotypes and signaling pathways that correlated with their invasive or proliferative phenotypes [supporting information (SI) Table 4]. From the DB-A2 subclone, we further selected a highly invasive revertant, A2-BH7. Phenotypic switching is fundamental for malignant progression (6), and therefore it is important to understand the responsible mechanisms.
Glioblastomas characteristically show extensive regional cytogenetic heterogeneity (10, 11), and this diversity may be responsible for tumor evolution and progression (10, 12). Here we show that distinct changes in karyotype from chromosome instability accompany phenotypic switching. These changes, in turn, dictate changes in the chromosome transcriptome that provide the expression of individual genes that are necessary for the conversion between the invasive and proliferative phenotypes.
Results and Discussion
Karyotype Differences Accompany Switching of Glioblastoma Tumor Cell Phenotypes.
To determine whether chromosome instability is responsible for tumor cell phenotypic switching, we examined DB-P, DB-A2, DB-A6, and A2-BH7 cells (SI Table 4) by using spectral karyotyping (SKY) (SI Fig. 4). For each cell type, we determined the total number of copies of each chromosome [or derivative (der) chromosomes] on the basis of 10 metaphase cells (Tables 1 and 2, respectively). Each cell population had near-tetraploid karyotypes, but karyotypes were particularly different from the parental DB-P cells as well as distinct from one another (Tables 1 and 2).
Table 1.
Full chromosomes in DB-P and its subclones via SKY
| Chromosome | DB-P | DB-A2 | DB-A6 | A2-BH7 |
|---|---|---|---|---|
| 1 | 2(10) 20 | 2(10) 20 | 2(10) 20 | 2(10) 20 |
| 2 | 3(10) 30 | 4(5) 3(5) 35 | 3(9) 2(1) 29 | 4(7) 3(3) 37 |
| 3 | 3(10) 30 | 4(1) 3(9) 31 | 3(10) 30 | 2(10) 20 |
| 4* | 0 | 0 | 0 | 0 |
| 5 | 2(10) 20 | 2(9) 1(1) 19 | 2(7) 1(3) 17 | 2(10) 20 |
| 6 | 3(10) 30 | 3(10) 30 | 3(6) 2(4) 26 | 2(10) 20 |
| 7 | 3(10) 30 | 4(10) 40 | 5(1)3(4) 2(5) 27 | 3(10) 30 |
| 8 | 3(10) 30 | 4(10) 40 | 4(7) 3(3) 37 | 4(10) 40 |
| 9 | 3(9) 2(1) 29 | 2(10) 20 | 3(1)2(7) 1(2) 19 | 2(10) 20 |
| 10 | 3(10) 30 | 2(10) 20 | 2(10) 20 | 2(10) 20 |
| 11* | 4(2) 3(6) 2(2) 30 | 0 | 0 | 0 |
| 12 | 4(10) 40 | 4(9) 3(1) 39 | 4(8) 3(2) 38 | 4(10) 40 |
| 13 | 3(9) 2(1) 29 | 2(8) 1(2) 18 | 2(10) 20 | 2(10) 20 |
| 14 | 3(7) 2(3) 27 | 2(10) 20 | 2(10) 20 | 2(10) 20 |
| 15 | 5(5) 4(3) 3(2) 43 | 4(8) 3(2) 38 | 3(9) 2(1) 29 | 4(10) 40 |
| 16 | 1(10) 10 | 0 | 0 | 0 |
| 17 | 4(10) 40 | 4(9) 3(1) 39 | 4(7)3(2)2(1) 36 | 4(10) 40 |
| 18 | 3(9) 2(1) 29 | 3(10) 30 | 3(10) 30 | 3(10) 30 |
| 19 | 4(3) 3(7) 33 | 4(10) 40 | 4(10) 40 | 4(10) 40 |
| 20 | 5(8) 4(2) 48 | 4(9) 3(1) 39 | 4(10) 40 | 4(10) 40 |
| 21 | 8(1) 7(7) 6(2) 69 | 4(9) 3(1) 39 | 4(10) 40 | 4(4) 3(6) 34 |
| 22† | 4(6) 3(4) 36 | 4(10) 40 | 4(9) 3(1) 39 | 4(10) 40 |
| X | 4(9) 3(1) 39 | 4(9) 3(1) 39 | 4(6) 3(4) 36 | 4(10) 40 |
Bold, number of copies of chromosome (n) observed out of 10 metaphases. Italics, total number of copies of chromosome in 10 metaphases; this is the number used to determine the chromosome content ratio.
*There are no full copies of chromosomes 4 and 11 in any of the cells. However, portions of chromosome 4 and 11 are represented in der chromosomes (see Table 2).
†In some metaphases, chromosome 22 is indistinguishable from marker chromosomes by SKY. Detailed examination with FISH showed a DB-A2/DB-P ratio of 1.02 (240/236 signals for chromosome 22 in 60 cells). Their value is used as chromosome content ratio.
Table 2.
Derivative chromosomes partitioning in DB-P cells and subclones
| Derivative chromosomes | DB-P | DB-A2 | DB-A6 | A2-BH7 |
|---|---|---|---|---|
| der(1)t(11;1;4)(?;p31-q43?;q?) | 1(10) 10 | 1(10) 10 | 1(9) 9 | 1(10) 10 |
| der(4)t(1;4)(?;q12) | 2(9) 1(1) 19 | 2(10) 20 | 2(8) 1(2) 18 | 2(10) 20 |
| der(4)t(4;5)(p14;q?) | 2(10) 20 | 2(10) 20 | 2(10) 20 | 2(10) 20 |
| der(5)t(16;5;4)(q12;p14-q23.3;p15) | 1(10) 10 | 2(10) 20 | 2(10) 20 | 2(10) 20 |
| der(7)t(6;7)(?;p22) | 2(10) 20 | 2(10) 20 | 2(9) 18 | 2(10) 20 |
| ins(9;13)(p12;?) | 2(9) 1(1) 19 | 2(10) 20 | 2(8) 1(2) 18 | 2(10) 20 |
| der(11)t(1;11)(q31;q13) | 1(9) 9 | 2(9) 1(1) 19 | 2(8) 1(2) 18 | 2(10) 20 |
| del(13)(q13) | 2(8) 1(2) 18 | 4(1) 2(9) 22 | 2(9) 1(1) 19 | 2(10) 20 |
| der(16)t(5;16)(p13;q12) | 2(10) 20 | 2(10) 20 | 2(8) 1(2) 18 | 1(10) 10 |
| der(3)t(3;13)(p21;q12) | 1(10) 10 | 0 | 0 | 0 |
| der(5)t(19;5;4)(p13?;p11-q23.3;p15) | 1(10) 10 | 0 | 0 | 0 |
| der(10)t(X;10)(p21;p11) | 1(10) 10 | 0 | 0 | 0 |
| der(13)t(4;13)(p10;q10) | 1(8) 8 | 0 | 0 | 0 |
| der(19)t(16;19)(q12;p13) | 1(8) 8 | 0 | 0 | 0 |
| del(15)(?) | 1(3) 3 | 1(1) 1 | 0 | 0 |
| der(11)t(4;5;11)(p15;q13-q23.3;p15) | 0 | 2(10) 20 | 2(10) 20 | 2(10) 20 |
| der(16)t(10;16)(q22;q22) | 0 | 1(10) 10 | 2(9) 1(1) 19 | 2(10) 20 |
| del(16)(p11.2)der(16)t(10;16)(q2;q22) | 0 | 1(10) 10 | 0 | 1(10) 10 |
| der(3)t(3;17)(p10;q11) | 0 | 0 | 1(10) 10 | 0 |
| der(3)t(3;18)(q21;p11.2) | 0 | 1(9) 9 | 0 | 1(10) 10 |
| der(5)t(5;15)(q21;q22) | 0 | 0 | 1(3) 3 | 0 |
| der(6)t(3;6)(q26?;q23) | 0 | 0 | 1(4) 4 | 0 |
| der(15)t(10;15)(?;q22) | 0 | 0 | 1(9) 9 | 0 |
Bold, number of copies of chromosome (n) observed out of 10 metaphases. Italics, total number of copies of chromosome in10 metaphases and the value used to determine the chromosome content ratio.
The Differences in DB-A2 and DB-P Cell Karyotypes Are Reflected in Their Transcriptome Ratios.
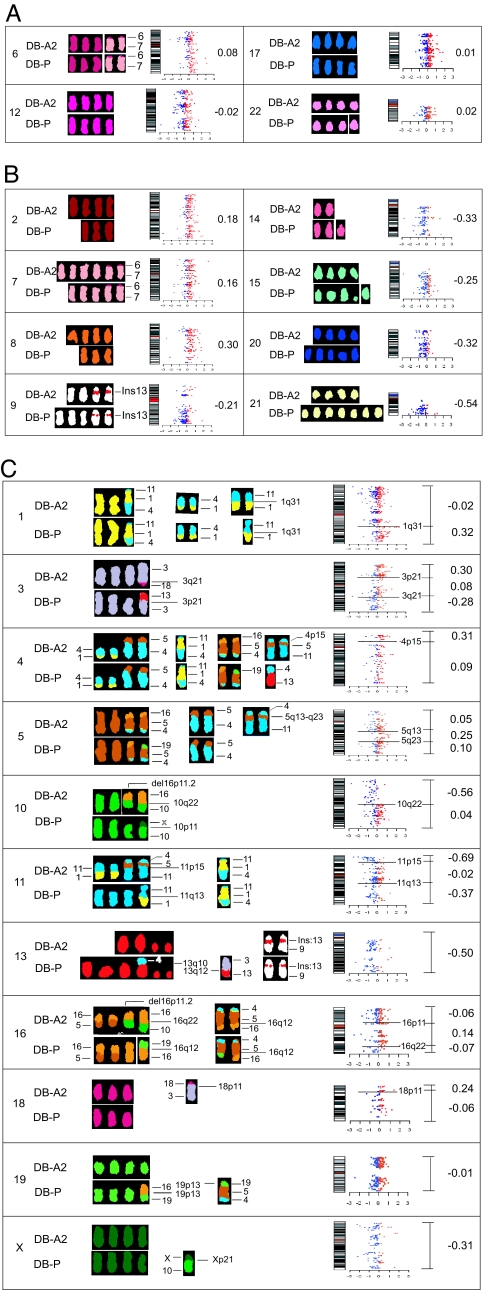
The significant differences in the karyotypes of each subclone led us to ask whether the changes in chromosome content mediated changes in chromosome transcriptome that could influence phenotype determination. Recent gene expression profiling studies have been used to assess the influence of chromosomal imbalance on overall gene expression (13–17). We used this approach to determine whether the ratio of the changes in chromosome content between parental DB-P cells and the subclones influenced the chromosome transcriptome ratios. Beginning with cDNA microarrays, individual gene expression differences between the DB-A2 subclone and the parental line, DB-P, were calculated as described (15). The transcriptome ratios were determined by averaging the relative expression ratios of each gene in a given chromosome or der subchromosomal region. These data are presented as log2-transformed to show the direction (+/−) and magnitude or are in linear scale to compare to the chromosome content ratio. The log2-transformed data were displayed as scatter plots on the respective chromosome (Fig. 1). In this way, the consequences of the chromosomal changes can be directly compared with changes in the transcriptome.
Fig. 1.
Comparisons of chromosome copy number and the chromosome transcriptome for DB-A2 and DB-P. Representative chromosomes (indicated by the numeral at the left) from DB-A2 and DB-P cells were aligned to compare the relative gain or loss of chromosome copies. Expression data for genes that map to each chromosome are displayed as scatter plots. Blue dots represent down-regulated genes and red dots represent up-regulated genes. The change in the transcriptome for each chromosome is in log2 units and is shown by the number to the right of each scatter plot. (A) Chromosomes without copy number changes. (B) Chromosomes with full copy gains or losses. (C) Chromosomes with derivative regions. Each scatter plot was subdivided according to gain or loss of the subchromosomal regions. The chromosome transcriptome ratio for each region is indicated by the mean values (in log2 units) to the right of the scatter plot. A correlation is observed between each derivative subchromosomal region and the chromosome transcriptome profile.
Comparison of Chromosome Content and Transcriptome for DB-A2 and DB-P.
For chromosomes 6, 12, 17, and 22, no changes in chromosome copy number were observed between the DB-A2 subclone and the parental DB-P (Table 1 and Fig. 1A). Concordantly, a comparison of the chromosome transcriptomes shows that the average of the number of up- and down-regulated genes on these chromosomes was largely unchanged (0.08, −0.02, 0.01, and 0.02 in log2 ratio units, respectively) (Fig. 1A). However, when the chromosome copy number changed between DB-A2 and DB-P, the chromosome transcriptome also changed in the same direction. Thus, for chromosomes 2, 7, and 8, which were higher in copy number in DB-A2 cells, the transcriptome ratios were 0.18, 0.16, and 0.30 in log2 ratio units, respectively (Fig. 1B). For chromosomes 9, 14, 15, 20, and 21, which were fewer in copy number in DB-A2, the corresponding ratios were −0.21, −0.33, −0.25, −0.32, and −0.54, respectively (Fig. 1B); chromosome 21 showed the greatest difference in copy number and the largest change in transcriptome ratio. These results show remarkable concordance between the chromosome copy number and the change in chromosome transcriptome, suggesting that the change in chromosome content can be responsible for the changes in expression of each gene as part of the transcriptome.
Subchromosomal Regions of Derivative Chromosomes Contribute to the Chromosome Transcriptome (DB-A2 vs. DB-P).
Many differences exist in derivative chromosomes between DB-P and the subclones (Table 2). Some derivatives were common to all cell lines, whereas others were present in a fraction of the subclones. Particularly interesting are the derivative chromosomes that are unique to the subclones, indicating that they were present in <1 in 10 metaphases in the parental DB-P cells (or perhaps new translocation events).
The surprising concordance between chromosome copy number and transcriptome changes led us to test whether subchromosomal regions in derivative chromosomes would also influence the chromosome transcriptome. Thus, the chromosome copy number (Table 1) is added to the subchromosomal region provided by the derivative copy (Table 2) to yield the chromosome content for that region. Such comparisons for DB-A2 and DB-P are shown (Fig. 1C). Thus, a net gain in chromosome 1q31-qter in DB-A2 from der (11)t (1:11) (Table 2) compared with DB-P increases the chromosome transcriptome ratio in this region to 0.32 in log2 units (Fig. 1C). Interestingly, the contributions of der (3)t (3, 13) and der (3)t (3, 18) to the DB-A2 and DB-P chromosome 3 transcriptomes have opposing effects in the regions of 3pter-p21 and 3q21-qter. Here the transcriptome ratio changes in opposite directions to 0.30 and −0.28 log2 units, respectively, but remains unchanged at 0.08 in the region between (Fig. 1C). Next, in chromosome 4, a gain in the 4p15 region from der (11)t (4, 5, 11) changes the chromosome 4 transcriptome ratio in this region for DB-A2 to 0.31. However, the same der chromosomes, der (11)t (4, 5, 11), also contribute to an increase in the chromosome 5 transcriptome in the 5q13-q23.3 region to 0.25.
The chromosome transcriptome can also be influenced by multiple derivative chromosomes. Thus, in DB-A2, the absence of 10pter-q22 in der (16)t (10, 16) and der (16)(p11.2)der (16)t (10, 16) results in the chromosome 10 transcriptome ratio in this region of −0.56 compared with 0.04 in the remainder of the chromosome, where the content of DB-A2 and DB-P are the same (Fig. 1C). Likewise, gene expression on chromosome 11 is influenced by three chromosomes, der (1)t (11, 1, 4), der (11)t (1, 11), and der (11)t (4, 5, 11), resulting in a decrease of two different regions (11pter-11p15 and 11q13-qter) of −0.69 and −0.37, respectively. Chromosomes 13 and 16 changes are complex, consisting entirely of der chromosomes or a combination of whole chromosomes and derivatives (Fig. 1C). The chromosome 19 transcriptome ratio does not change with der (5)t (19;5;4;) and der (19)t (16, 19) (Table 2), but these two derivatives may compensate for the lower copy numbers of chromosome 19 in DB-P cells (Tables 1 and 2). The major exception we found in associating chromosome copy number changes with transcriptome changes is the significant decrease in the X transcriptome in DB-A2 cells, although the only region absent is pter-p21 (Fig. 1C), which can possibly be explained by X-inactivation.
We conclude that, whether comparing full chromosome content or segments of translocated regions of der chromosome(s), the chromosome transcriptomes' ratios reflect chromosome content.
Comparing Chromosome and Transcriptome Ratios of DB-A6 to DB-A2 (vs. DB-P).
We next compared the chromosome content changes and transcriptome ratios between DB-A6 and DB-P to see whether the results obtained with DB-A2 versus DB-P extended to other cell clonal isolates. The DB-A6 and DB-A2 karyotypes are more similar to each other than either is to the parental DB-P cells (Tables 1 and 2). The chromosome content ratios of DB-A6 to DB-P are the same as DB-A2 to DB-P with chromosomes 8, 9, 14, 20, and 21 (cf. Fig. 1B with SI Fig. 5B), and the chromosome transcriptome ratios are all virtually the same [(0.30/0.33), (−0.21/−0.22), (−0.33/−0.35), (−0.32/−0.28), and (−0.54/−0.55), respectively]. By contrast, DB-A6 and DB-A2 differ significantly in chromosomes 2, 7, and 15 content (Tables 1 and 2), and we observe very different transcriptome ratios [(0.18/0.11), (0.16/0.02), and (−0.25/-0.40), respectively; cf. Fig. 1B and SI Fig. 5 A and B]. For the six chromosomes with no obvious changes in chromosome copy number, three chromosomes (7, 12, and 22) show little change in chromosome transcriptome ratios (0.02, −0.05, and 0.07 respectively), two of them (chromosomes 2 and 6) show a marginal increase (0.11), and chromosome 18 shows a substantial increase (0.20) (SI Fig. 5A). These variations in the chromosome transcriptome ratio may result from undetectable cytogenetic changes or, for chromosome 18, be due to the low number of genes expressed from the chromosome (chromosome 18).
As with the DB-A2 comparison with DB-P cells, we also observe a strong influence of der chromosomes on the transcriptomes. The chromosome 15 transcriptome ratio in DB-A6 is influenced by der (15)t (10:15) (SI Fig. 5C). Likewise, the transcriptome ratios of chromosome 3 [(0.30, 0.08, −0.28) (Fig. 1C) and (0.15, 0.22, −0.24) (SI Fig. 5C)] and chromosome 16 [(−0.06, 0.14, −0.07) (Fig. 1C) and (0.30, 0.29, −0.13) (SI Fig. 5C)] are dramatically different among DB-A2, DB-A6, and DB-P due to derivative chromosomes (Table 2). These analyses further show that the content of chromosome governs the differences in the chromosome transcriptome ratio.
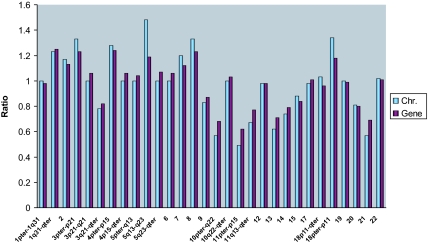
Global Comparison of Chromosome Content and Transcriptome Differences.
Up to this point, we have been comparing the numerical values of the chromosome content ratio to the transcriptome ratio in log2 units. We, therefore, converted the log2-transformed transcriptome ratios back to a linear scale and made the comparisons for all combinations of tumor cell populations for full chromosomes (SI Table 5) or for the DB-A2/DB-P comparison of subchromosomal regions of der chromosomes (SI Table 6). In Fig. 2, we present the ratios of the total number of chromosomes counted in 10 metaphase spreads (Tables 1 and 2) as histogram plots against the numerical ratios of respective chromosome transcriptomes converted from the log2 ratios. Further, the comparisons between chromosome content ratios and chromosome transcriptome ratios for DB-P parental and DB-A2, DB-A6, and A2-BH7 comparisons are presented in SI Fig. 6 and SI Table 5. These analyses show dramatically that the fold increase or decrease in the chromosome content ratio is virtually the same as the transcriptome ratio for all comparisons, and the chromosome content transcriptome ratios over all chromosomes average ≈1.0 (SI Table 5). Even the subchromosomal regions of derivative chromosomes, when considered as part of the copy number of a specific chromosome (Fig. 2 and SI Table 6), markedly influence the transcriptome ratio of that chromosome, and, dramatically, the numerical ratios of the chromosome content are virtually the same as the transcriptome of the specific chromosome region. We conclude that there is a direct quantitative correlation between chromosome content and a proportional change in the transcriptome.
Fig. 2.
Comparing chromosomal copy number ratio to the chromosome transcriptome ratio. The copy numbers of each chromosome in DB-P and DB-A2 were determined by SKY and FISH and are presented in Tables 1 and 2. The ratios of chromosome numbers for DB-A2 versus DB-P were determined by comparing the total number of chromosome or subchromosomal region in 10 metaphases. The chromosome transcriptome ratios were obtained by averaging the expression of all available genes on each chromosome or subchromosome region, and values determined in log2 units were converted to numerical ratios.
Do Changes in Chromosome Content Provide the Transcriptome Changes Required for Phenotype Determination?
The major question of these analyses is whether changes in the chromosome transcriptome result in changes in the expression of specific genes that favor the invasive or proliferative phenotypes. Discriminant gene analysis was performed to identify specific genes/gene sets that had significant expression differences between DB-A2 and DB-P (SI Table 7). Expression of 89 genes that reside in autosomes are altered >2-fold in DB-A2 (all P values <10−4; SI Table 7). Twenty-seven of these genes have been implicated in the regulation of tumor growth or apoptosis (Table 3 and SI Table 7). Consistent with the rapid tumor growth and low invasion/migration phenotypes of DB-A2 cells (SI Table 4), all 22 genes down-regulated in DB-A2 are related to proinvasion, proapoptosis, or growth inhibition, whereas all five up-regulated genes are proproliferation or antiinvasion genes. Interestingly, 17 of the 22 down-regulated genes reside in chromosomes or subchromosomal regions that decrease in copy number in DB-A2, only three reside in chromosomes that increase, and two are located in chromosomes that do not change (Table 3). These data show that karyotypic changes are consistent with having a role in phenotypic conversion of DB-A2 cells.
Table 3.
Genes with significantly altered expression in the DB-A2/DB-P comparison
| Map | Gene | Function in tumorigenesis* | Chromosome ratio† | Gene ratio† | P value‡ |
|---|---|---|---|---|---|
| 4p16.3 | SOD3 | Proliferation | 1.28 | 5.24 | 1.52e−5 |
| 17q21.3 | HOB7 | Proliferation | 0.98 | 2.75 | 8.75e−9 |
| 2q35 | FN1 | Invasion | 1.17 | 0.12 | 4.51e−5 |
| 2q37 | COL6A3 | Invasion | 1.17 | 0.05 | 4.67e−5 |
| 11p15.5 | IFITM1 | Invasion | 0.49 | 0.17 | 3.34e−5 |
| 11p15.5 | IFITM2 | Invasion | 0.49 | 0.24 | 3.79e−5 |
| 13q33 | EFNB2 | Invasion | 0.49 | 0.38 | 2.33e−5 |
| 14q22 | BMP4 | Invasion | 0.74 | 0.15 | 4.70e−5 |
| 16q13 | MMP2 | Invasion | 1.43 | 0.17 | 3.56e−5 |
| 20q11.23 | MYL9 | Invasion | 0.81 | 0.08 | 7.38e−5 |
| 21q22.3 | COL6A1 | Invasion | 0.56 | 0.22 | 4.01e−5 |
| 1q21.3 | CKIP−1 | Apoptosis | 1.00 | 0.39 | 2.60e−5 |
| 10p13 | CUGBP2 | Apoptosis | 0.75 | 0.15 | 4.02e−5 |
| 14q22 | ZFP36L1 | Apoptosis | 0.74 | 0.38 | 2.04e−5 |
| 13q31.2 | STK24 | Apoptosis | 0.49 | 0.47 | 8.27e−6 |
| 20p13 | SMOX | Apoptosis | 0.81 | 0.42 | 1.49e−5 |
| 3q23 | RPB1 | Growth inhibition | 0.76 | 0.16 | 3.76e−5 |
| 6q24 | AKAP12 | Growth inhibition | 1.00 | 0.49 | 5.88e−6 |
| 9q34.1 | AK1 | Growth inhibition | 0.83 | 0.33 | 2.42e−5 |
| 10p13 | RSU1 | Growth inhibition | 0.75 | 0.45 | 9.85e−6 |
| 10p15 | AKR1X3 | Growth inhibition | 0.75 | 0.36 | 2.75e−5 |
| 11p15.2 | DKK3 | Growth inhibition | 0.49 | 0.45 | 9.02e−6 |
| 11q23.2 | TSLC1 | Growth inhibition | 0.67 | 0.44 | 1.38e−5 |
| 21q22.1 | DSCR1 | Anti-invasion | 0.56 | 0.47 | 6.58e−6 |
| 16q24.2 | CDH13 | Anti-invasion | 0.73 | 6.06 | 1.89e−17 |
| 18p11.3 | TGIF | Anti−invasion | 1.34 | 2.28 | 1.95e−7 |
| 7q32 | ARP3β | Anti−metastasis | 1.20 | 2.84 | 2.92e−9 |
*See SI Table 7 for references.
†Simple numerical chromosome copy number ratio between Dβ-A2 and DB-P cells.
‡P value for gene expression ratio.
Whereas the expression of a small number of genes (89 of 19,552) alters significantly (SI Table 7), the expression changes of the majority of genes track closely with chromosome content (SI Fig. 7 A–C). The significant changes in transcription of a small subset of genes may result from mutations or epigenetics that alter the transcriptome, but it is also possible that they result from changes in the chromosome content of other chromosomes that bear transcription regulator genes, and it would not be too surprising for gene expression changes to occur, surrounding the translocated regions of the der breakpoints. However, it is likely that these changes already exist and are delivered by the changes in chromosome content.
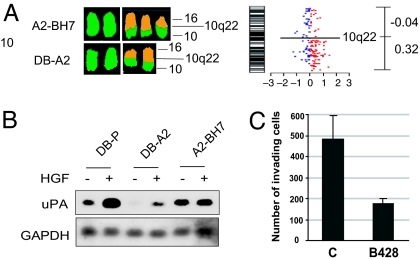
The gain of chromosome 7 and the loss of chromosome 10 are common cytogenetic alterations in glioblastoma (11) and may be directly related to malignant phenotype (18). In this study, we observed changes in chromosomes 7 and 10 in the subclones (Table 1). In the highly proliferative DB-A2 subclone, relative to DB-P, an increase in the copy number of chromosome 7 results in an increase in the transcriptome (0.16) (Fig. 1B). Further, chromosome 7 content in the invasive revertant of DB-A2, A2-BH7 (SI Fig. 8), was restored to the DB-P level, as was the transcriptome ratio of −0.17, offsetting the gain relative to DB-A2 (see Fig. 1B and SI Fig. 8). This result suggests that genes expressed on chromosome 7 may favor proliferation or antagonize invasion.
The parental clone DB-P has three copies of chromosome 10 plus one copy of der (10)t (X;10), whereas the proliferative DB-A2 subclone has two full copies of chromosome 10 plus two copies of der (16)t (10, 16), which contains the 10q22-qter region (Tables 1 and 2). Thus, two copies of 10p11-q22 and one copy of 10pter-p11 are absent from the DB-A2. However, in the invasive revertant A2-BH7, one copy of 10q22-qter is regained relative to DB-A2 (Table 2 and SI Fig. 8) and the chromosome 10 transcriptome ratio of this region increases to 0.32. In parallel, the expression of genes on 10pter-q22 (but not other parts of the same chromosome) was significantly down-regulated in DB-A2 relative to DB-P (Fig. 1C), whereas expression of genes on 10q22-qter were up-regulated in A2-BH7 relative to DB-A2 (SI Fig. 8). These results are consistent with the reports showing that the transfer of chromosome 10p suppresses oncogenic activity of glioma cells (19, 20). These data also suggest that the genes in these regions of chromosomes 7 and 10 are crucial for glioma tumor proliferation and invasion, as reported by others (11, 18). Thus, for these two chromosomes, our invasive and proliferative tumor cells (6) (SI Table 4) change in content (Tables 1 and 2). The analyses reveal how changes in karyotype and the transcriptome can mediate the gene expression changes necessary for phenotype determination.
We used literature-based searches and identified genes that are associated with glioma development. Gene expression comparisons between DB-P and DB-A2 cells indicated that three of the genes on chromosome 10p were significantly down-regulated in DB-A2 (Table 3 and SI Fig. 9) and none was significantly up-regulated. We show that, regardless of how similar the numerical ratios are for the respective chromosomes and chromosome transcriptomes (SI Tables 5 and 6), the individual gene ratios vary significantly in contributing to the transcriptome average (Table 3) (P < 10−4; SI Table 7). The chromosome content, therefore, in delivering the transcriptome in direct proportions, delivers a specific level of gene expression for each gene.
RSU-1 is associated with gliomas and is down-regulated in DB-A2 (Table 3) as part of chromosome 10p. This gene was isolated based on its ability to suppress v-RAS-induced transformation (21). More importantly, ectopic expression of RSU-1 in glioma cells inhibits cell proliferation, anchorage-independent growth, and tumorigenic activity in nude mice (21). Therefore, RSU-1 is a negative regulator of the proliferative phenotype in gliomas. Consistent with our analysis showing that the MAPK pathway is involved in the invasive phenotype (6) (SI Table 4), the decreased RSU-1 expression in DB-A2 cells may contribute to their highly proliferative and tumorigenic activity (22). Moreover, RSU-1 also enhances Erk-2 activity (21) and is consistent with elevated Erk-2 activity in response to HGF/SF in DB-P-invasive cells (6).
The der chromosomes that include chromosome 10 regions can also influence the phenotypes of DB-A2 and DB-P. The absence of the 10pter-10q22 region in DB-A2 cells as described above can enhance proliferation and tumorigenic activity, whereas the presence of 10q22-qter in A2-BH7 (Fig. 3, Table 2, and SI Fig. 8) can contribute to its invasive phenotype. cDNA microarray and Northern blotting indicated that the most significantly up-regulated gene in 10q22-qter is PLAU (Fig. 3 and data not shown). PLAU encodes urokinase-type plasminogen activator (uPA), a well known mediator of HGF/SF-induced cell invasion (23, 24). The role of uPA up-regulation in invasive phenotypic conversion in this study was confirmed with the uPA inhibitor, B428 (23), which significantly blocked HGF/SF-induced invasion in A2-BH7 cells (Fig. 3). In DB-P cells, uPA is HGF/SF-inducible, whereas, in A2-BH7 cells, it is constitutive, indicating that convergence to uPA expression has occurred through chromosome instability.
Fig. 3.
Chromosome change-associated up-regulation of uPA contributes to the invasive phenotype of A2-BH7. (A) Changes in A2-BH7 versus DB-A2 cells in chromosome number and chromosome transcriptome. (B) Northern blot analysis showing up-regulation of uPA in BH7 cells. (C) The uPA inhibitor, B428, blocked HGF-induced cell invasion through Matrigel. Cells (10,000 per insert) were loaded into Matrigel inserts and treated with 10 μM B428 or left untreated (controls, C) for 1 h before adding HGF/SF.
The loss of chromosomes 13, 14, 20, and 21 in DB-A2 relative to DB-P are also associated with the down-regulation of additional genes that have been implicated in glioma invasion (Table 3 and SI Fig. 9). These genes encode proteins that include extracellular matrix proteins COL6A1 (collagen type VI, α-1) (25), cytoskeleton-binding protein MYL9 (myosin-light polypeptide 9) (26), as well as EFNB2 (ephrin-B2) (27). Down-regulation of BMP4 (14q22), which promotes invasion and migration in malignant melanoma (28), may contribute to a low invasive phenotype of DB-A2. By contrast, the higher expression of these genes in DB-P cells is expected to enhance DB-P cell invasion.
Our studies do not exclude any of the other mechanisms of tumor progression such as gain of function, loss of tumor suppressor function, or epigenetic changes, but aneuploidy can contribute to malignant progression by explaining how gene expression changes are delivered. Thus, genetic or epigenetic alterations, fueled by chromosome instability, can produce sufficient chromosome diversity in the tumor cell population to generate, in the proper environment, cells with the appropriate phenotype for malignant progression. Our data show that quantitative changes in the chromosome transcriptome are largely governed by changes in chromosome content, which we refer to as the one-to-one rule. The analysis of changes in specific genes and chromosomes is consistent with these changes being responsible for phenotype determination.
Materials and Methods
Cell Lines and SKY.
The origin of the DB-P (DBTRG parental cells), DB-A2, DB-A6, and A2-BH7 cells and the cell growth conditions have been described previously (6). For SKY, probe hybridization and detection were carried out according to the protocol provided with the SkyPaintTM kit (Applied Spectral Imaging, Midgal HáEmek, Israel). Metaphase images generated from cultured cells were captured by using a COOL-1300 SpectraCube imaging system (Applied Spectral Imaging) connected to an Olympus BX51 fluorescence microscope (Olympus, Tokyo, Japan). Analysis was performed by using SkyView software from Applied Spectral Imaging.
Preparation of RNA Samples for cDNA Array Analysis.
Cells at 80% of confluence were serum starved for 24 h and treated with HGF/SF (100 ng/ml) or without (control) for 3 h. The cells were solubilized in TRIzol (Gibco/BRL, Grand Island, NY), and total RNA was prepared according to the manufacturer's suggestion. Total RNA was recovered by precipitation in 2.5 M LiCl solution (Ambion, Austin, TX). RNA samples were reverse-transcribed and labeled for microarray analysis as described (www.microarray.vai.org/). Experiments were carried out at least in duplicate. Four arrays with DB-A2 or DB-A6 cells and two arrays with A2-BH7 HGF/SF-treated cells were processed by using the dye-swap-labeling technique (29). In all cases, RNA from DB-P cells cultured under normal conditions was used as a reference.
cDNA Microarray Data Generation.
Corning GAPS2 microarray slides (Corning, NY) spotted with 19,552 cDNA clones from the Research Genetics 40K Human Clone Set (Huntsville, AL) were prepared at the Van Andel Research Institute. Each hybridization was scanned by using a confocal fluorescent Scan Array Lite scanner (PerkinElmer Life and Analytical Services, Boston, MA) equipped with lasers operating at 532 nm (G, green) and 635 nm (R, red). Array features were identified and assigned background-corrected red and green fluorescence intensity values by using the GenePix Pro 5.0 image analysis software (Axon, Union City, CA) by using the default settings. Adjustment of gene expression values to compensate for experimental biases (normalization) was performed by using the within-print tip group-scaling technique as implemented in the limma BioConductor package (www.bioconductor.org) for the R environment (30, 31). Before normalization, spots were excluded if they had a signal lower than three times the SD of the global array background in either channel.
Gene Expression Data Analysis and the Chromosome Transcriptome.
Relative gene expression values between subclones were generated by subtraction of the mean log2-transformed expression value from the measured value for each subclone. Relative gene expression values were visualized by plotting the expression value based on the corresponding gene's chromosome mapping location by using the BioConductor idiogram package (32). The chromosome transcriptome value is determined by quantification of the overall relative gene expression in each chromosomal region. For this analysis, chromosomes were broken into segments based on visual inspection of the SKY data. The relative gene expression value for every gene located within each segment was calculated as described above. The average relative gene expression value was used to quantify the difference in the chromosome transcriptome for each segment. We also examined the data set for genes that had significant differential expression between the subclones. Discriminant genes were identified by using the empirical Bayes method as implemented in the limma package by using the default settings.
Supplementary Material
Acknowledgments
We thank Tony Hunter, George Klein, Stefan Imreh, and Beatrice Knudsen for critical reading of the manuscript and David Nadziejka and Michelle Reed-Bassett for assistance with preparation of the manuscript. This work was supported in part by the Michigan Life Sciences Corridor and the Jay and Betty Van Andel Foundation.
Abbreviations
- der
derivative
- HGF/SF
hepatocyte growth factor/scatter factor
- SKY
spectral karyotyping
- uPA
urokinase-type plasminogen activator.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0700631104/DC1.
References
- 1.Nowell PC. Science. 1976;194:23–28. doi: 10.1126/science.959840. [DOI] [PubMed] [Google Scholar]
- 2.Chambers AF, Groom AC, MacDonald IC. Nat Rev Cancer. 2002;2:563–572. doi: 10.1038/nrc865. [DOI] [PubMed] [Google Scholar]
- 3.Rajagopalan H, Lengauer C. Nature. 2004;432:338–341. doi: 10.1038/nature03099. [DOI] [PubMed] [Google Scholar]
- 4.Barrett MT, Sanchez CA, Prevo LJ, Wong DJ, Galipeau PC, Paulson TG, Rabinovitch PS, Reid BJ. Nat Genet. 1999;22:106–109. doi: 10.1038/8816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Duesberg P. Science. 2005;307:41. doi: 10.1126/science.307.5706.41d. [DOI] [PubMed] [Google Scholar]
- 6.Gao CF, Xie Q, Su YL, Koeman J, Khoo SK, Gustafson M, Knudsen BS, Hay R, Shinomiya N, Vande Woude GF. Proc Natl Acad Sci USA. 2005;102:10528–10533. doi: 10.1073/pnas.0504367102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Maher EA, Furnari FB, Bachoo RM, Rowitch DH, Louis DN, Cavenee WK, DePinho RA. Genes Dev. 2001;15:1311–1333. doi: 10.1101/gad.891601. [DOI] [PubMed] [Google Scholar]
- 8.Giese A, Bjerkvig R, Berens ME, Westphal M. J Clin Oncol. 2003;21:1624–1636. doi: 10.1200/JCO.2003.05.063. [DOI] [PubMed] [Google Scholar]
- 9.Koochekpour S, Jeffers M, Rulong S, Taylor G, Klineberg E, Hudson EA, Resau JH, Vande Woude GF. Cancer Res. 1997;57:5391–5398. [PubMed] [Google Scholar]
- 10.Coons SW, Johnson PC, Shapiro JR. Cancer Res. 1995;55:1569–1577. [PubMed] [Google Scholar]
- 11.Loeper S, Romeike BF, Heckmann N, Jung V, Henn W, Feiden W, Zang KD, Urbschat S. Cytogenet Cell Genet. 2001;94:1–8. doi: 10.1159/000048773. [DOI] [PubMed] [Google Scholar]
- 12.Wolman SR. Cancer Genet Cytogenet. 1986;19:129–140. doi: 10.1016/0165-4608(86)90380-8. [DOI] [PubMed] [Google Scholar]
- 13.Phillips JL, Hayward SW, Wang Y, Vasselli J, Pavlovich C, Padilla-Nash H, Pezullo JR, Ghadimi BM, Grossfeld GD, Rivera A, et al. Cancer Res. 2001;61:8143–8149. [PubMed] [Google Scholar]
- 14.Pollack JR, Sorlie T, Perou CM, Rees CA, Jeffrey SS, Lonning PE, Tibshirani R, Botstein D, Borresen-Dale AL, Brown PO. Proc Natl Acad Sci USA. 2002;99:12963–12968. doi: 10.1073/pnas.162471999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Furge KA, Lucas KA, Takahashi M, Sugimura J, Kort EJ, Kanayama HO, Kagawa S, Hoekstra P, Curry J, Yang XJ, Teh BT. Cancer Res. 2004;64:4117–4121. doi: 10.1158/0008-5472.CAN-04-0534. [DOI] [PubMed] [Google Scholar]
- 16.Harding MA, Arden KC, Gildea JW, Gildea JJ, Perlman EJ, Viars C, Theodorescu D. Cancer Res. 2002;62:6981–6989. [PubMed] [Google Scholar]
- 17.Upender MB, Habermann JK, McShane LM, Korn EL, Barrett JC, Difilippantonio MJ, Ried T. Cancer Res. 2004;64:6941–6949. doi: 10.1158/0008-5472.CAN-04-0474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Steilen-Gimbel H, Henn W, Kolles H, Moringlane JR, Feiden W, Steudel WI, Zang KD. Genes Chromosomes Cancer. 1996;16:180–184. doi: 10.1002/(SICI)1098-2264(199607)16:3<180::AID-GCC4>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 19.Steck PA, Ligon AH, Cheong P, Yung WK, Pershouse MA. Genes Chromosomes Cancer. 1995;12:255–261. doi: 10.1002/gcc.2870120404. [DOI] [PubMed] [Google Scholar]
- 20.Kon H, Sonoda Y, Kumabe T, Yoshimoto T, Sekiya T, Murakami Y. Oncogene. 1998;16:257–263. doi: 10.1038/sj.onc.1201488. [DOI] [PubMed] [Google Scholar]
- 21.Masuelli L, Cutler ML. Mol Cell Biol. 1996;16:5466–5476. doi: 10.1128/mcb.16.10.5466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tsuda T, Marinetti MR, Masuelli L, Cutler ML. Oncogene. 1995;11:397–403. [PubMed] [Google Scholar]
- 23.Jeffers M, Rong S, Vande Woude GF. Mol Cell Biol. 1996;16:1115–1125. doi: 10.1128/mcb.16.3.1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xie Q, Gao CF, Shinomiya N, Sausville E, Hay R, Gustafson M, Shen Y, Wenkert D, Vande Woude GF. Oncogene. 2005;24:3697–3707. doi: 10.1038/sj.onc.1208499. [DOI] [PubMed] [Google Scholar]
- 25.Chintala SK, Gokaslan ZL, Go Y, Sawaya R, Nicolson GL, Rao JS. Clin Exp Metastasis. 1996;14:358–366. doi: 10.1007/BF00123395. [DOI] [PubMed] [Google Scholar]
- 26.Murata K, Hirano K, Villa-Moruzzi E, Hartshorne DJ, Brautigan DL. Mol Biol Cell. 1997;8:663–673. doi: 10.1091/mbc.8.4.663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nakada M, Niska JA, Miyamori H, McDonough WS, Wu J, Sato H, Berens ME. Cancer Res. 2004;64:3179–3185. doi: 10.1158/0008-5472.can-03-3667. [DOI] [PubMed] [Google Scholar]
- 28.Rothhammer T, Poser I, Soncin F, Bataille F, Moser M, Bosserhoff AK. Cancer Res. 2005;65:448–456. [PubMed] [Google Scholar]
- 29.Churchill GA. Nat Genet. 2002;(32) Suppl:490–495. doi: 10.1038/ng1031. [DOI] [PubMed] [Google Scholar]
- 30.Yang YH, Dudoit S, Luu P, Lin DM, Peng V, Ngai J, Speed TP. Nucleic Acids Res. 2002;30:e15. doi: 10.1093/nar/30.4.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Smyth GK, Michaud J, Scott HS. Bioinformatics. 2005;21:2067–2075. doi: 10.1093/bioinformatics/bti270. [DOI] [PubMed] [Google Scholar]
- 32.Furge KA, Dykema KJ, Ho C, Chen X. BMC Genomics. 2005;6:67. doi: 10.1186/1471-2164-6-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.