Abstract
Floodwaters in New Orleans from Hurricanes Katrina and Rita were observed to contain high levels of fecal indicator bacteria and microbial pathogens, generating concern about long-term impacts of these floodwaters on the sediment and water quality of the New Orleans area and Lake Pontchartrain. We show here that fecal indicator microbe concentrations in offshore waters from Lake Pontchartrain returned to prehurricane concentrations within 2 months of the flooding induced by these hurricanes. Vibrio and Legionella species within the lake were more abundant in samples collected shortly after the floodwaters had receded compared with samples taken within the subsequent 3 months; no evidence of a long-term hurricane-induced algal bloom was observed. Giardia and Cryptosporidium were detected in canal waters. Elevated levels of fecal indicator bacteria observed in sediment could not be solely attributed to impacts from floodwaters, as both flooded and nonflooded areas exhibited elevated levels of fecal indicator bacteria. Evidence from measurements of Bifidobacterium and bacterial diversity analysis suggest that the fecal indicator bacteria observed in the sediment were from human fecal sources. Epidemiologic studies are highly recommended to evaluate the human health effects of the sediments deposited by the floodwaters.
Keywords: Lake Pontchartrain, water quality
Hurricane Katrina made landfall along the U.S. Gulf Coast on August 29, 2005, resulting in extensive flooding in the City of New Orleans (NO), Louisiana, due to storm surge from adjacent Lake Pontchartrain (LP) and several levee failures. These floodwaters had been partially pumped back into LP when the city experienced additional flooding and levee failures from Hurricane Rita on September 24, 2005. Floodwaters completely receded by October 11, 2005. Much of the flooding occurred in urbanized and industrial areas, fueling concerns that a public health crisis could result from exposures to chemically and microbiologically contaminated floodwaters.
Preliminary investigations in mid-September 2005 documented high levels of microbial and toxicant contamination in the NO floodwaters (1–3). Reports of mean total coliform and total Escherichia coli levels as high as 8 × 108 CFU per 100 ml and 3 × 107 CFU per 100 ml, respectively (1) indicated the presence of sewage contamination and associated sewage-borne pathogens. Elevated concentrations of fecal coliforms have previously been observed in floodwaters of the NO region, but the 2005 event was characterized by an unusually large volume and long duration of human exposure (4). The most contaminated area tested near the Superdome also contained high levels of nonsewage pathogens, with an estimated Aeromonas spp. concentration of 1.7 × 108 per 100 ml (1). Concentrations of Vibrio spp. were not measured, but the temperature and salinity of the floodwaters would have been favorable for their growth, and the number of Vibrio infections reported in the month following Hurricane Katrina was higher than normal (5).
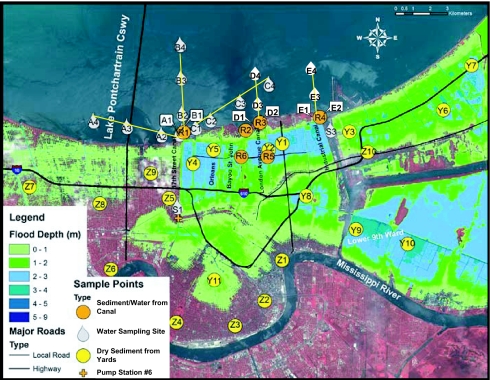
As shown in the satellite floodwater image, ≈34 billion liters of water covered NO as of August 30, 2005, with floodwater depths in several areas in excess of 3 m (Fig. 1). As these floodwaters were pumped back into LP, sediments were deposited throughout the flooded regions of the city (including the interior of homes). To a large extent, these sediments, now dried, remain even after 1 year following their deposition by the floodwaters. Therefore, the human population of the NO region can be exposed to microbial pollution through contact with the waters of LP and the dried sediments, which may be ingested or suspended in the air and inhaled. Long-term environmental impacts of this floodwater discharge on the microbial water quality of the lake, and the potential public health risk associated with exposures to the remaining sediment deposits, are unclear.
Fig. 1.
Floodwater depths and sampling sites. Color shadings indicate depth of floodwater (in meters) in NO as of August 30, 2005. Droplet symbols indicate the location of sites where only water samples were collected including LP water sample sites and canal sites. Circles indicate the location of sites where sediment samples were collected. For sites adjacent to canals (R1–R6), water samples were also collected. The cross shows the location of the 17th Street Canal Pump Station 6, the main discharge point for the inner city canals into LP.
The primary objective of this study was to document the microbial water quality of LP after the city was dewatered, and to evaluate canal waters and sediments within residential areas of NO as potential sources of microbial contaminants. This study differs from others in the timing of the measurements, which were conducted shortly after floodwaters receded from the city, and also in that sediments from the urban areas of the city were evaluated. This study is also unique in that a diverse group of microbes were measured, including traditional fecal indicators (enterococci and Escherichia coli) using three methods [membrane filtration (MF), chromogenic substrate (CS), and quantitative PCR (qPCR)]. A suite of nontraditional fecal indicators (FRNA coliphage, Clostridium perfringens, Bacteroidales, and Bifidobacterium adolescentis) was also evaluated. FRNA coliphage, C. perfringens, and Bacteroidales have been recommended as alternative indicators of sewage pollution, and more recently, Bifidobacterium adolescentis has been shown to grow exclusively in the intestines of humans (6, 7) and has been proposed for microbial source-tracking of human fecal contamination (8–10). Efforts were also made to measure pathogenic bacteria of the genus Vibrio and Legionella, with species detection as Vibrio vulnificus, Vibrio parahaemolyticus, Vibrio cholera, and Legionella pneumophila. Protozoa (Cryptosporidium and Giardia) were measured at the genus level. The study also included an innovative method of evaluating the possible origin of microbial contamination through microbial assemblage analysis as well as a preliminary screening for the presence of algal blooms.
Sample Collection
Sample collection and analysis was completed through a collaborative response of six academic institutions including Louisiana State University (LSU) and three NSF/NIEHS National Centers for Oceans and Human Health at the University of Miami, University of Hawaii, and Woods Hole (Woods Hole Oceanographic Institution, Marine Biological Laboratory, Massachusetts Institute of Technology), with supporting collaborations from Florida International University and Georgia College and State University.
The period of intensive sample collection occurred during the 2 months immediately following the dewatering of NO. Given the results from this intensive monitoring period, sampling locations within the lake were modified and additional sites were identified within NO. Sampling within NO focused on collecting water samples from interior canals, and collecting sediments from canal banks and from floodwater sediments deposited within residential neighborhoods. After November 2005, the frequency of sample collection was decreased to monthly and ultimately to a quarterly basis.
Lake Transects.
During the months of October and November 2005, surface water samples were collected from southern LP along three transects (labeled A–C) radiating outward from the outlet of the 17th Street Canal. These samples were evaluated for traditional and nontraditional fecal indicators plus Vibrio and Legionella spp. Each transect was comprised of four stations along an ≈4-km track with stations identified by transect letter and a station number (1–4) assigned consecutively beginning with the station nearest shore. Transect A ran to the W–NW (stations A1–A4), transect B was located to the north, perpendicular to shore (stations B1–B4), and transect C ran NE (stations C1–C4) (Fig. 1). In January 2006, transects were redesigned to sample for gradients extending from the outlets of three canals along the south shore (the 17th Street, London Avenue, and Industrial Canals). Two near-shore stations were sampled for each new transect, one to the east and one directly in front of the outlet of the respective canal. Two additional stations were sampled along each transect at increasing distances from the outlets of the canals running perpendicular to the shoreline. The transect from the 17th Street Canal was comprised of previously sampled stations (A1, B1, B2, B3), whereas those from the London Avenue (stations D1–D4) and Industrial Canals (E1–E4), were new (Fig. 1).
Canal and Shoreline Water Sampling.
Water samples from interior canals and from the canal shorelines were collected during October and November 2005. Particular efforts were placed on collecting water samples from the 17th Street (site S1) and Industrial Canals (site S3). A distinguishing feature of the 17th Street Canal is the presence of a large pump station at its interior end, as this canal was designed to discharge waters from the interior portions of NO. These waters originate from rainfall runoff and from groundwater seepage as the city lies below sea level. The Industrial Canal is a navigational canal that receives flows from the Gulf of Mexico during incoming tide through the Mississippi River Gulf Outlet. Canal and shoreline water samples were analyzed for fecal indicators (traditional and nontraditional) and for Vibrio and Legionella spp. Microbial assemblage analysis was also evaluated for three samples (two from site S1 and one from site S3). Water samples (sites S1 and A1) were also collected during December 2006 for subsequent analysis of Cryptosporidium and Giardia.
Sediment Sampling.
Canal shoreline sediments from six locations and deposited floodwater sediments from three homes were collected during November 2005 and March 2006. The six shoreline sediment sites corresponded to the outfalls of major canals [17th Street Canal (R1), Bayou St. Johns (R2), London Avenue Canal (R3), and the Industrial Canal (R4)] and from the inner city canal sites (R5 and R6) corresponding to prehurricane sampling sites established by the Louisiana Department of Environmental Quality. The sites corresponding to deposited floodwater sediments were randomly chosen from within a targeted area. Targeted areas were based on depth of flooding and the specific canal breach that was the cause of flooding. The first residential site (Y1) corresponded to a home that received moderate flood depth (1.5–2 m) from multiple canal breaches. The second site (Y2) received deep flooding (≥3 m) directly from the London Avenue Canal breach; the third site (Y3) received deep flooding (≥3 m) from a breach on the Industrial Canal. In June 2006, additional sediment sampling sites were included to evaluate the impacts of hurricane-induced flooding on sediment sites. These additional sites were geographically distributed within residential neighborhoods from flooded (Y4–Y11) and nonflooded areas (Z1–Z10). Sediment samples were analyzed for enterococci, E. coli, and Bifidobacterium. Microbial assemblages were established for a sediment sample (Y3) showing elevated fecal indicator concentrations.
Regional and Local Surveys of Chlorophyll-a.
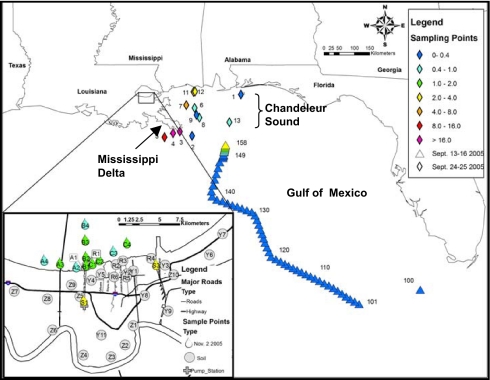
Water samples were collected during September 2005 along the Mississippi coast near the river delta and in Chandeleur Sound (which receives discharges from LP) to provide data concerning the relative abundance of algae (Fig. 2). Additional samples were collected during October and November 2005 along the initial, radial transects in LP.
Fig. 2.
Chlorophyll-a concentrations in LP, within the 17th Street and Industrial Canal of NO, in Chandeleur Sound–Mississippi River Delta area, and in the Gulf of Mexico after Hurricane Katrina. Samples were collected during two cruises (September 13–26 and 22–25, 2006) sponsored by National Oceanic and Atmospheric Administration. Data shown in Inset for Lake Pontchartrain were collected on November 2, 2005.
Results
For further details, see supporting information (SI) Text and SI Tables 1–5.
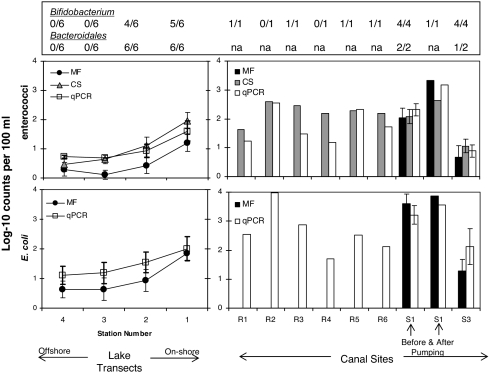
In the LP transects (characterized by salinities from 7 to 11 ppt), maximum recommended single sample exposure levels of enterococci (104 CFU per 100 ml for marine water) and E. coli (235 CFU per 100 ml for freshwater) (11) were only exceeded on occasion (as measured by qPCR and CS) at the stations closest to shore, and the levels dropped with distance away from shore (Fig. 3). None of the lake transect samples exceeded the single sample exposure levels as measured by the MF method. On occasion, the fecal indicator measurements at the lake sites located closest to shore exceeded the single sample exposure level. For the canal sites, single sample exposure levels were frequently exceeded regardless of method used. The counts observed within the canals were consistent with the concentrations observed in the near-shore sampling sites within LP suggesting that the interior portions of the city may be the source of the fecal indicator bacteria.
Fig. 3.
Fecal indicators, enterococci and E. coli, in LP transect B and canal sites including results from Bacteroidales analysis by PCR and Bifidobacterium. Results for the fecal indicators correspond to analysis using MF, CS, and qPCR. Error bars correspond to 95% confidence limits (Student's t test). Data points shown with error bars correspond to the average for samples collected between October 11 and November 12, 2005. For lake transects, the samples were also averaged over transects A, B, and C. Site S1 was sampled both before and after pumping at the 17th Street Canal Pump Station 6. For Bacteroidales and Bifidobacterium the number in the numerator corresponds to the number of positive samples and the number in the denominator corresponds to the total number of samples analyzed. “na” indicates not analyzed. See SI Tables 2 and 3.
During the sampling period FRNA coliphages and Bacteroidales by Taq nuclease assay (TNA) were not detected in near-shore and offshore water samples within the lake. Bacteroidales and Bifidobacterium were detected consistently by standard PCR in the near-shore samples, at sites 1 and 2 within transects A, B, and C, at the locations where fecal indicator bacteria counts were highest. Conversely, Bacteroidales and Bifidobacterium were consistently not detected in the offshore samples, at sites 3 and 4, in areas where fecal indicator counts were low. Bifidobacterium were also detected in shoreline waters adjacent to the canal outfalls (Fig. 3). During the sampling period, the highest C. perfringens levels were observed at near-shore sites 1 and 2 within transect A. Low to undetectable levels of C. perfringens were observed in offshore sites of transect A and in all sites within transects B and C (SI Table 2).
For the Industrial Canal (site S3), fecal indicator bacteria levels (enterococci: 9 CFU per 100 ml, E. coli: 140 CFU per 100 ml, means for all methods used collectively) were lower compared with the interior portions of the 17th Street Canal (site S1, enterococci: 380 CFU per 100 ml, E. coli: 4,600 CFU per 100 ml, means for all methods collectively). Concentrations were much lower, on average, at the outlet of the 17th Street Canal (sites A1, B1, and C1, mean enterococci: 70 CFU per 100 ml, E. coli: 120 CFU per 100 ml) relative to the interior site. During November 12, 2005, samples were taken at the effluent side of the 17th Street Canal pumping station (site S1), both before and after pumping. The relative abundance of enterococci in the canal water at this site increased by over an order of magnitude after pumping (mean of 93 CFU per 100 ml before pumping and 1,400 CFU per 100 ml after pumping), thereby suggesting that the source of fecal indicators to this canal is water from the interior portions of the city, which is then transported by the 17th Street Canal to LP. On the day that the pumps were active, lake shore-line water samples exceeded the U.S. Environmental Protection Agency single sample exposure levels at the mouths of Bayou St. John, the London Avenue Canal, the Industrial Canal, and at the interior canal sites within the London Avenue Canal and Bayou St. John (Fig. 3). Samples for Cryptosporidium and Giardia, also collected during a pumping event on the 17th Street Canal, showed detectable levels for both protozoans at site S1 (23 and 8 oocysts per 100 liters for Cryptosporidium and 15 and 16 cysts per 100 liter for Giardia). The levels of Cryptosporidium decreased to below detection limits (<0.5 oocyts per 100 liters) at the outlet of the 17th Street Canal, whereas Giardia remained elevated (10 cysts per 100 liter).
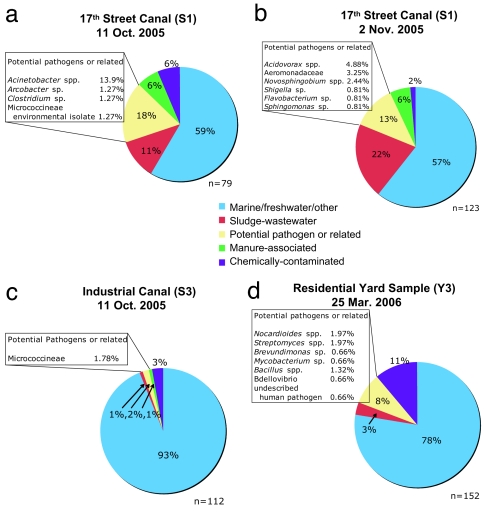
Bacterial assemblage data also showed fewer potential pathogens or related organisms in the Industrial Canal versus the 17th Street Canal and demonstrated a likely lower percentage of bacteria derived from contaminated (e.g., sludge, chemical, etc.) study sites (Fig. 4 a–c). Approximately 60% of the bacteria in samples collected from the 17th Street Canal showed top BLAST scores with microbes observed in uncontaminated aquatic environments; from 10 to 20% of the bacteria were typical of those found in sewage, with 13–18% associated with taxa that could be potentially pathogenic. The sample from the Industrial Canal, however, was characterized by a microbial community typical of uncontaminated aquatic environments (93%), with much smaller fractions associated with sewage sludge bacteria (1%) or potential pathogen groups (2%).
Fig. 4.
Comparative bacterial community analysis of samples collected from the 17th Street Canal on two separate occasions, from the Industrial Canal and a residential yard. Analyses are based on sequencing of random clones from libraries of PCR-amplified small-subunit rRNA gene fragments with sequences being binned into habitat types based on similarity to top scoring BLAST hits.
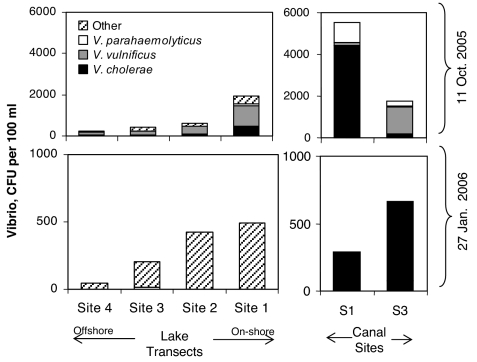
Concentrations of CFUs for putative Vibrio spp. averaged 1,200 CFU per 100 ml (range 130–5,600) in October 2005, with highest total counts found in the canals and at stations near the outlet of the 17th Street Canal, and the lowest counts found at the stations farthest from shore (Fig. 5). At that time, V. cholerae and V. vulnificus were present at nearly all sites and occasionally dominated the culturable Vibrio community, but V. parahaemolyticus was rare everywhere. V. cholerae was particularly dominant in the 17th Street Canal (79% of isolates), whereas V. vulnificus dominated the Industrial Canal (72% of isolates) and was most common on average, comprising 46% of isolates from all stations. The concentration of total putative Vibrio spp. was 4-fold lower on average in January 2006, with a mean of 300 CFU per 100 ml (range 32–680) and total CFUs on the Vibrio-selective media were again found to decline with distance from shore (Fig. 5). At that time, none of the three targeted species was common at any site, and V. vulnificus, in particular, was not detected in any sample.
Fig. 5.
Abundance of Vibrio species in LP transects and canal sites during the October 11, 2005, and January 27, 2006, sampling dates. Total Vibrio spp. abundance is based on total colony-forming units on TCBS medium. Abundances of V. cholerae, V. vulnificus, and V. parahaemolyticus are estimated from the proportion of total colony-forming units testing positive for each of these species by biochemical and genetic tests. “Other” refers to those colonies not classified as one of the three targeted pathogens. Data shown are representative of the data collected. For lake transects, the samples were also averaged over three available transects. See SI Table 4.
The majority (92%) of the transect water samples from October were positive for Legionella spp., but none of these was positive for L. pneumophila (SI Table 2). Lake samples collected in November, December, and January were characterized by a smaller proportion of positives (50%) for Legionella spp., but one of the 20 samples evaluated was positive for L. pneumophila (site B2, December 12, 2005). This positive sample was confirmed by sequence analysis of the recovered amplification product. By March 2006, the proportion of positive samples from the lake samples increased back to 100% with no positives for L. pneumophila, thereby suggesting that the presence of Legionella sp. may in part be affected by seasonal factors. The presence of Legionella was not correlated with levels of fecal indicator bacteria.
Chlorophyll-a concentrations of the lake transect sites ranged from 0.79 μg/liter to 1.3 μg/liter during the November 8, 2005 sampling trip with no distinctive spatial pattern (Fig. 2). Samples collected from the canals were generally characterized by higher chlorophyll levels (3.7 μg/liter). These data showed no evidence of significant algal blooms occurring at the time of the November 2005 sampling, as concentrations were considerably lower than the 5–20 μg/liter of chlorophyll found in LP in 1997 (12). Samples along the Louisiana coast near the Mississippi delta and in Chandeleur Sound ranged from 4.0 to 42 μg/liter of chlorophyll-a 2–3 weeks after the hurricane (Fig. 2). These concentrations were similar to those found at earlier times (13–15), suggesting that algal blooms along the coast were not enhanced as a result of hurricane damage and flooding.
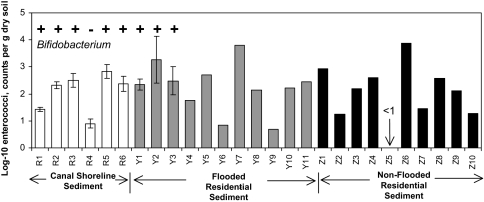
The shoreline sediments collected from the banks of the canals (sites R1–R6), demonstrated the presence of fecal indicators in the range of 0.7 to 970 most probable number (MPN)/g by both the qPCR and the CS assays (Fig. 6 and SI Table 5). Samples of dried floodwater sediments collected from residential neighborhoods on November 12, 2005, also demonstrated enterococci abundance at levels similar to the more contaminated canal shoreline sediments (270–980 MPN/g). Samples collected on March 25, 2006, ≈6 months after flooding, also showed viable enterococci present in the shoreline sediments and in dried floodwater sediments from all three residential home sites, with the highest levels measured at 1,040 CFU per g of sediment. The levels of indicator microbes appear to be related to the percent of organic matter of the sediments with all samples containing ≤3% organic matter characterized by indicator concentrations <150 MPN/g, whereas samples characterized by >11% organic content showed microbe levels >350 MPN/g. All sediment samples collected during March 2006 were also positive for Bifidobacterium, with the exception of the one sample (R4) that was characterized by a relatively low level (15 CFU/g) of enterococci and low percent of organic content (Fig. 6). Microbial assemblage data for a residential sediment sample (sample Y3 collected on March 25, 2006) showed that ≈80% of the bacteria were typical of those found in uncontaminated environments. Three percent were typical of those found in sewage sludge, with ≈8% characterized by genera that may be associated with pathogenic microbes (Fig. 4d). Additional samples collected during June 2006 from both flooded (11 sites) and nonflooded (10 sites) residential neighborhoods (see Fig. 1 for locations), showed levels of enterococci in excess of 1,000 MPN/g (Fig. 6). However, the distribution of enterococci abundance in NO sediments was heterogeneous and did not necessarily correlate with flooding history. Some of the highest enterococci counts were found in both floodwater-exposed sediments (i.e., site Y7 at 6,200 MPN/g, site Y2 at 1,900 MPN/g) and in nonflooded sediments (i.e., site Z1 at 840 CFU/g, site Z6 at 7,600 CFU/g). Conversely, some sediments that were flooded at depth for an extended period, including a site in the Lower Ninth Ward area, demonstrated relatively low enterococci abundance (i.e., site Y4 at 60 MPN/g, site Y6 at 7 MPN/g, and site Y9 at 5 MPN/g).
Fig. 6.
Abundance of enterococci in sediments of the NO area, including results from Bifidobacterium analysis for samples collected March 25, 2006. (+, positive; −, not detected). Enterococci results correspond to samples collected between November 12, 2005, and June 20, 2006, and analyzed using the CS method. Error bars correspond to 95% confidence limits (Student's t test). See SI Table 5.
Discussion
No evidence of a long-term algal bloom was observed in LP as a result of Hurricanes Katrina and Rita. Chlorophyll samples were collected from the lake starting in mid-October, after the dewatering of NO floodwaters was completed, so early blooms could have been missed. Even if an immediate bloom had occurred, evidence suggests that the lake would have recovered quickly, as no significant impact was noted 6 weeks after the first hurricane.
The decline in concentrations of Vibrio, or positive samples for Legionella, from October to January, is most likely attributable to normal seasonal cycles rather than recovery from a hurricane effect. Seasonal fluctuations in Vibrio spp. have been documented (16), with water temperature as a major driving variable (17). Average lake temperature in October was 25°C compared with 17°C in January, suggesting that induction into a viable but nonculturable state (18, 19) could account for part of the observed decline in Vibrio colony-forming units. The warm temperature (≈31°C) and low salinity (≈5–6 ppt) of floodwaters in downtown NO shortly after the storm (4) were conditions under which many Vibrio spp. would be expected to thrive, including the three targeted pathogens in this study, V. cholerae (20), V. parahaemolyticus (21), and V. vulnificus (22–24). Legionella and some Vibrio spp. have symbiotic relationships with other aquatic microbes. Their concentrations could therefore also vary in response to the general patterns of productivity among the plankton. Regardless of the cause, elevated concentrations of pathogenic Vibrio spp. in the lake waters flooding the city would have increased the risk of infections and thereby contributed to the spike in post-Katrina Vibrio-related infection and mortality (5).
The results of this study also indicate that fecal indicator bacteria in the offshore waters of LP, presumably introduced through the discharge of contaminated floodwaters from the city, had dissipated to low background levels within a few weeks after the dewatering process was complete. In contrast, levels of enterococci remained high in the near-shore waters of the lake and most canal waters, with pumping of inner city drainage resulting in an increase in indicator levels within the canals. Of note, the levels of fecal indicator bacteria (enterococci, E. coli, fecal coliform) were known to be high in NO interior-water discharges before the 2005 hurricane season, and the historic background levels of these fecal indicator bacteria in near-shore waters of LP have chronically exceeded recreational water quality standards for many decades (4, 25). The source of contamination appears to be the chronic discharge of contaminated water from the interior portions of the city. The detection of Cryptosporidium and Giardia in these waters is a strong indication of the poor quality. Efforts are needed in the region to improve the sanitary infrastructure. These improvements should focus on reducing sewage contamination of groundwater seepage and storm water drained from the NO region.
Fecal indicator bacteria in the sediments could not be attributed solely to the effects of Hurricanes Katrina and Rita, as counts of enterococci in both flooded and nonflooded areas were statistically the same. Elevated levels within sediments could have been due to sewage impacts or from environmental sources as there is a growing body of evidence that suggests persistence, and possibly regrowth, of fecal bacteria, particularly in subtropical and tropical environments in association with soils, marine sediments, and beach sand found near-shore (26–28). Because of this phenomenon, additional evidence is generally needed to determine the possible cause of the elevated indicator levels within sediments. For example, a nontraditional indicator, C. perfringens, was used to document fecal impacts to agricultural soils in North Carolina in response to a series of hurricane events (29). In the current study, the detection of B. adolescentis along with elevated concentrations of enterococci in the majority of sediment samples supports the hypothesis that sewage contamination is the source of the fecal indicator bacteria in the shoreline sediments. Additional samples should be evaluated to determine whether this cooccurrence is of statistical significance. The impacts from sewage (and possible presence of pathogens) is also supported by analysis of bacterial assemblage data which indicates that the sample sites characterized by higher levels of enterococci and E. coli (i.e., 17th Street Canal sites, flooded residential sediments, etc.) were also characterized by bacterial groups that have been commonly observed in sewage sludge, including the putative pathogens Bacillus spp. and Mycobacterium spp. (Fig. 4d). Again, only one sediment sample was evaluated in this case, and further work is needed to determine whether this cooccurrence is of significance.
Conclusion
The floodwater pumped from the city into the lake was a small fraction (≈0.5%) of the total lake volume, and the large dilution effect appears to have limited the environmental impact on the lake and substantially reduced health risks associated with exposure to lake water to levels typically observed before the hurricane. Water quality within the city and in nearshore waters of LP continue to be impacted by discharges from interior portions of the city that appear to be contaminated with fecal microbes, a chronic condition that persisted in the area both before and after Hurricanes Katrina and Rita. Sediments also remain a cause for concern because they may serve as a potential source of ongoing microbial exposure in near-shore waters and interior portions of NO. Given the history of this region with respect to extensive flooding of the city with sewage contaminated floodwaters induced by hurricanes, further investigation is needed to evaluate the microbial quality of floodwater sediments deposited in the NO area and to determine whether there is an elevated risk of exposure to human pathogens through contact, ingestion, and inhalation of these sediments.
Materials and Methods
Enterococci and E. coli were measured in water samples by using standard MF methods that included plating filters on mEI media (30) for enterococci and mTEC media (31) for E. coli. For enterococci, a commercially available CS method (Enterolert, IDEXX, Westbrook, ME) was also used, including the distribution of the sample into a series of wells for enumeration. Samples analyzed for enterococci and E. coli by qPCR included concentrating water samples with a polycarbonate filter, extracting the DNA, followed by use of qPCR testing kits containing lypholyzed reagents from Cepheid (Sunnyvale, CA). Sediment samples were analyzed by eluting the microbes and analyzing the eluate by CS and qPCR as described above. Volatile organics were measured for the sediment samples gravimetrically before and after ignition.
Assays for alternative fecal indicators included measurements of FRNA coliphage by enrichment and culture on a host lawn of E. coli (32), C. perfringens by MF using mCP agar (33), fecal Bacteroidales using standard PCR and real-time PCR using the Taq nuclease assay (TNA) (34), and Bifidobacterium adolescentis by PCR (35). Some sediment samples were also assayed for Bifidobacterium. Bacterial diversity was evaluated through small subunit ribosomal RNA gene surveys using universal primer pairs that flank the V4–V8 region of the molecule and amplify SSU rRNA genes from all three domains of life (Bacteria, Archaea, and Eukarya). This manuscript reports only the results from bacterial sequence data. General bacterial diversity assessments help determine the presence of sequence types related to known bacterial pathogens or may be used to infer the source of bacteria by noting the environments in which similar organisms are found (such as within sewage). The three pathogenic bacterial species in the genus Vibrio (i.e., V. vulnificus, V. parahaemolyticus, and V. cholerae) were assayed in samples from the lake transects by filter plating on CHROMAgar Vibrio and TCBS agar. Colonies were enumerated, and a subset isolated for confirmatory identification based on their growth and color changes on selective media and by PCR testing with species-specific primers (36–39). Legionella species and L. pneumophila were assessed by PCR amplification (40, 41) with amplification of L. pneumophila targeting the mip gene (42). Cryptosporidium and Giardia were analyzed by filtration (Filtamax by IDEXX), elution, and purification using immuno-magnetic separation followed by epiflourescence microscopy (43). For further details, see SI Text.
Supplementary Material
Acknowledgments
We thank the Cepheid company, R. Noble and D. Blackwood from University of North Carolina (Chapel Hill, NC), for assistance with the Cepheid qPCR assays, and M. Wright, A. Abdelzaher, J. Rocca, M. Dennett, and D. Moran for their assistance with logistics and sample processing. This work was funded by National Science Foundation (NSF)–National Institute of Environmental Health Sciences (NIEHS) Oceans and Human Health Program (NSF Grants OCE0432368, OCE0432479, and OCE0430724 and NIEHS Grants P50 ES12736, ES012740, and ES012742), the NSF Small Grant for Exploratory Research Program (Grants OCE 0554402, OCE 0554674, OCE 0554850, and OCE0554768), the NSF Research Experiences for Undergraduates Program, and by the Georgia Sea Grant College Program (NA04OAR170033). Samples from the Gulf of Mexico were provided through National Oceanic and Atmospheric Administration. IDEXX provided Enterolert assay supplies free of charge.
Abbreviations
- NO
New Orleans
- LP
Lake Pontchartrain
- MF
membrane filtration
- CS
chromogenic substrate
- qPCR
quantitative PCR
- MPN
most probable number.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0610552104/DC1.
References
- 1.Presley SM, Rainwater TR, Austin GP, Platt SG, Zak JC, Cobb GP, Marsland EJ, Tian K, Zhang B, Anderson TA, et al. Environ Sci Technol. 2006;40:468–474. doi: 10.1021/es052219p. [DOI] [PubMed] [Google Scholar]
- 2.Stoeckel DM, Bushon RN, Demcheck DK, Skrobialowski SC, Kephart CM, Bertke EE, Mailot BE, Mize SV, Fendick RB., Jr . Bacteriological Water Quality in Lake Pontchartrain Basin LA, Following Hurricanes Katrina and Rita, Sep. 2005. Reston, VA: US Geol Surv; 2005. (Data Series 143). [Google Scholar]
- 3.Hou A, Laws EA, Gambrell RP, Bae H-S, Tan M., DeLaune RD, Li Y, Roberts H. Environ Sci Technol. 2006;40:5904–5910. doi: 10.1021/es060946u. [DOI] [PubMed] [Google Scholar]
- 4.Pardue JH, Moe WM, Mcinnis D, Thibodeaux LJ, Valsaraj KT, Maciasz E, Van Heerden I, Korevec N, Yuan QZ. Environ Sci Technol. 2005;39:8591–8599. doi: 10.1021/es0518631. [DOI] [PubMed] [Google Scholar]
- 5.Centers for Disease Control and Prevention. Morbid Mortal Wkly Rep. 2005;54:928–931. [Google Scholar]
- 6.Carson CA, Shear BL, Ellersieck MR, Schnell JD. Appl Environ Microbiol. 2003;69:1836–1839. doi: 10.1128/AEM.69.3.1836-1839.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mullie C, Odou MF, Singer E, Romond MB, Izard D. FEMS Microbiol Lett. 2003;222:129–136. doi: 10.1016/S0378-1097(03)00245-3. [DOI] [PubMed] [Google Scholar]
- 8.Kinzelman J, Ng C, Jackson E, Gradus S, Bagley R. Appl Environ Microbiol. 2003;69:92–96. doi: 10.1128/AEM.69.1.92-96.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Malathum K, Singh KV, Weinstock GM, Murray BE. J Clin Microbiol. 1998;36:211–215. doi: 10.1128/jcm.36.1.211-215.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nebra Y, Bonjoch X, Blanch AR. Appl Environ Microbiol. 2003;69:2651–2656. doi: 10.1128/AEM.69.5.2651-2656.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.US Environmental Protection Agency. Federal Register. 2004;69:67217–67243. [PubMed] [Google Scholar]
- 12.Turner RE, Dortch Q, Rabalais NN. Biogeochemistry. 2004;68:411–422. [Google Scholar]
- 13.Dortch Q, Whitledge TE. Cont Shelf Res. 1992;12:1293–1309. [Google Scholar]
- 14.Hitchcock GL, Wiseman WJ, Boicourt WC, Mariano AJ, Walker N, Nelsen TA, Ryan E. J Mar Syst. 1997;12:109–126. [Google Scholar]
- 15.Lohrenz SE, Dagg MJ, Whitledge TE. Cont Shelf Res. 1990;10:639–664. [Google Scholar]
- 16.Heidelberg JF, Heidelberg KB, Colwell RR. Appl Environ Microbiol. 2002;68:5488–5497. doi: 10.1128/AEM.68.11.5488-5497.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thompson JR, Randa MA, Marcelino LA, Tomita-Mitchell A, Lim E., Polz M. F. Appl Environ Micriobiol. 2004;70:4103–4110. doi: 10.1128/AEM.70.7.4103-4110.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Oliver JD. J Mircobiol. 2005;43:93–100. [Google Scholar]
- 19.Xu H-S, Roberts N, Singleton FL, Atwell RW, Grimes DJ, Colwell RR. Microbial Ecol. 1982;8:313–323. doi: 10.1007/BF02010671. [DOI] [PubMed] [Google Scholar]
- 20.Louis VR, Russek-Cohen E, Choopun N, Rivera ING, Gangle B, Jiang SC, Rubin A, Patz JA, Huq A, Colwell RR. Appl Environ Microbiol. 2003;69:2773–2785. doi: 10.1128/AEM.69.5.2773-2785.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.DePaola A, Nordstrom JL, Bowers JC, Wells JG, Cook DW. Appl Environ Microbiol. 2003;69:1521–1526. doi: 10.1128/AEM.69.3.1521-1526.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kelly MT. Appl Environ Microbiol. 1982;44:820–824. doi: 10.1128/aem.44.4.820-824.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lipp EK, Rodriguez-Palacios C, Rose JB. Hydrobiol. 2001;460:165–173. [Google Scholar]
- 24.Randa MA, Polz MF, Lim E. Appl Environ Microbiol. 2004;70:5469–5476. doi: 10.1128/AEM.70.9.5469-5476.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jin G, Englande AJ, Bradford H, Jeng H. Water Environ Res. 2004;76:245–255. doi: 10.2175/106143004x141807. [DOI] [PubMed] [Google Scholar]
- 26.Hardina CM, Fujioka RS. Environ Toxicol Water Qual. 1991;6:185–195. [Google Scholar]
- 27.Solo-Gabriele HM, Wolfert MA, Desmarais TR, Palmer CJ. Appl Environ Microbiol. 2000;66:230–237. doi: 10.1128/aem.66.1.230-237.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Anderson KL, Whitlock JE, Harwood VJ. Appl Environ Microbiol. 2005;71:3041–3048. doi: 10.1128/AEM.71.6.3041-3048.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Casteel MJ, Sobsey MD, Mueller JP. J Environ Sci Health A. 2006;41:173–184. doi: 10.1080/10934520500351884. [DOI] [PubMed] [Google Scholar]
- 30.US Environmental Protection Agency. Method 1600. 2002 (EPA Pub 821-R- 02-022) [Google Scholar]
- 31.US Environmental Protection Agency. Method 1103.1. 2002 (EPA Pub 821-R-02-020) [Google Scholar]
- 32.Sobsey MD, Yates MV, Hsu FC, Lovelace G, Battigelli D, Margolin A, Pillai SD, Nwachuku N. Wat Sci Technol. 2004;50:211–217. [PubMed] [Google Scholar]
- 33.US Environmental Protection Agency. ICR Microbial Laboratory Manual. 1996 (EPA Pub 600-R-95-178) [Google Scholar]
- 34.Dick LK, Field KG. Appl Environ Microbiol. 2004;70:5695–5697. doi: 10.1128/AEM.70.9.5695-5697.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.King EL, Bachoon DS, Gates KW. J Microbiol Methods. 2007;68:76–81. doi: 10.1016/j.mimet.2006.06.008. [DOI] [PubMed] [Google Scholar]
- 36.Lane DJ. In: Nucleic Acid Techniques in Bacterial Systematics. Stackebrandt E, Goodfellow M, editors. New York: Wiley; 1991. pp. 115–175. [Google Scholar]
- 37.Brasher CW, De Paola A, Jones DD, Bej AK. Curr Microbiol. 1998;37:101–107. doi: 10.1007/s002849900346. [DOI] [PubMed] [Google Scholar]
- 38.Bej AK, Patterson DP, Brasher CW, Vickery MC, Jones DD, Kaysner CA. J Microbiol Methods. 1999;36:215–225. doi: 10.1016/s0167-7012(99)00037-8. [DOI] [PubMed] [Google Scholar]
- 39.Chun J, Huq A, Colwell RR. Appl Environ Microbiol. 1999;65:2202–2208. doi: 10.1128/aem.65.5.2202-2208.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jonas D, Rosenbaum A, Weyrich S, Bhakdi S. J Clin Microbiol. 1995;33:1247–1252. doi: 10.1128/jcm.33.5.1247-1252.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Miyamoto H, Yamamoto H, Arima K, Fujii J, Maruta K, Izu K. Appl Environ Microbiol. 1997;63:2489–2494. doi: 10.1128/aem.63.7.2489-2494.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wilson DA, Yen-Lieberman B, Reischl U, Gordon SM, Procop GW. J Clin Microbiol. 2003;41:3327–3330. doi: 10.1128/JCM.41.7.3327-3330.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.US Environmental Protection Agency. Method 1623. 2001 (EPA Pub 821-R-01-025) [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.